Biofuel Production Based on Carbohydrates from Both Brown and Red Macroalgae: Recent Developments in Key Biotechnologies
Abstract
:1. Introduction
| Country | % |
|---|---|
| China | 53.97 |
| Indonesia | 27.40 |
| Philippines | 7.36 |
| Korea | 4.30 |
| Japan | 1.85 |
| Malaysia | 1.39 |
| Total | 96.27 |
| The Farmed Algae | Million Wet Tonnes |
|---|---|
| Eucheuma red algae | 8.30 |
| Japanese kelp (brown algae, Laminaria japonica) | 5.65 |
| Seaweed species not identified | 2.75 |
| Gracilaria spp. (red algae) | 2.75 |
| Wakame (brown algae, Undaria pinnatifida) | 2.10 |
| Porphyra spp. (red algae) | 1.75 |
| Other seaweeds and microalgae | 1.75 |
| Total | 25.50 |
2. Ethanol Production from Carbohydrates in Brown Macroalgae
2.1. Carbohydrates in Brown Macroalgae
| Brown Macroalgae a | Carbohydrates | Parts | Concentration (%) |
|---|---|---|---|
| L. cloustoni | Laminarin | Fronds | 1–32 |
| Whole plant | 0–18 | ||
| Alginate | Fronds | 8–18 | |
| Stipes | 19–23.5 | ||
| Mannitol | Fronds | 8–26 | |
| Stipes | 5–10 | ||
| L. digitata | Laminarin | Fronds | 1–15 |
| Whole plant | 1–12 | ||
| Alginate | Fronds | 17–25.5 | |
| Stipes | 28–33.5 | ||
| Mannitol | Fronds | 5–23 | |
| Stipes | 5–12 | ||
| L. saccharina | Laminarin | Fronds | 1–21 |
| Whole plant | 1–18 | ||
| Alginate | Fronds | 12.5–20 | |
| Stipes | 19.5–25 | ||
| Mannitol | Fronds | 9–23 | |
| Stipes | 6–12 |
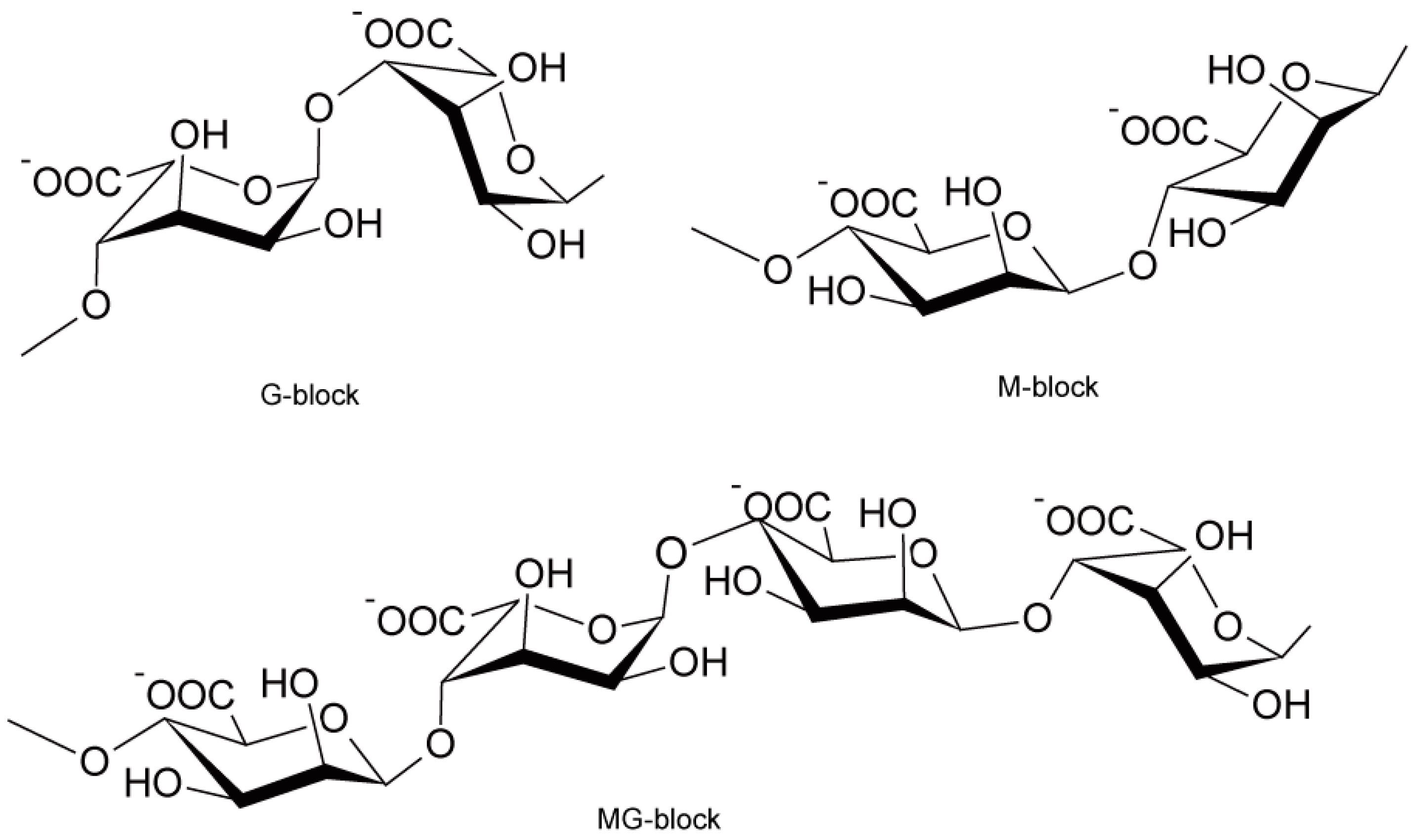
2.2. Key Biotechnologies for Production of Ethanol from Mannitol
2.2.1. Ethanol Production from Mannitol Using Microorganisms Other than S. cerevisiae
2.2.2. Ethanol Production from Mannitol Using S. cerevisiae
2.3. Key Biotechnology to Produce Ethanol from Alginate
2.3.1. Alginate Metabolism
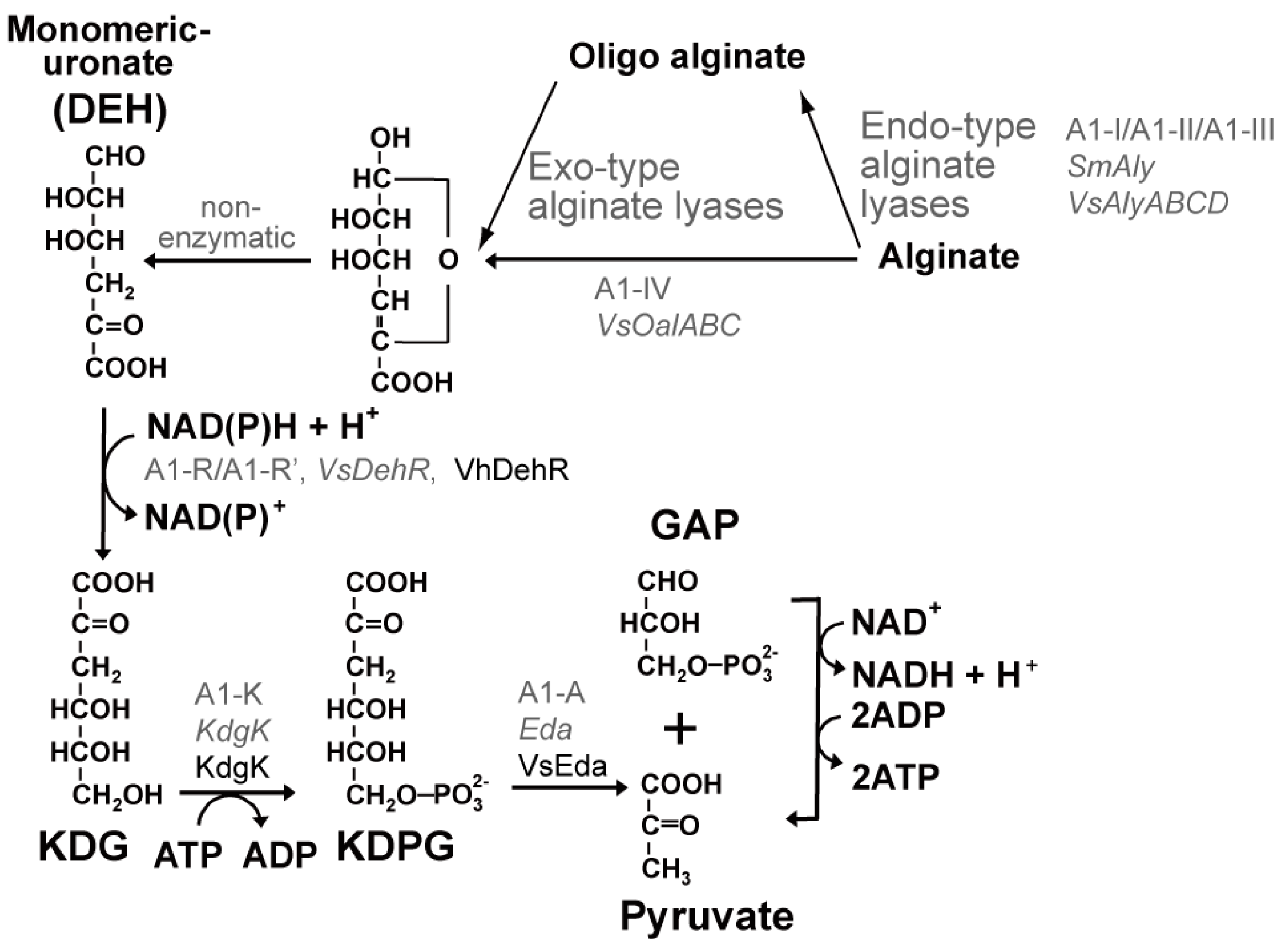
| Protein | Source Organism | NADPH | NADH | References | ||
|---|---|---|---|---|---|---|
| Km (mM) | Kcat a or Vmax b | Km (mM) | Kcat a or Vmax b | |||
| A1-R a | Sphingomonas sp. A1 | 0.009 | 220 | 0.192 | 25 | [36] |
| A1-R’ a | Sphingomonas sp. A1 | 0.272 | 233 | 0.024 | 274 | [36] |
| VsDehR b,c | V. splendidus 12B01 | 2.83 | 0.08 | 0.22 | 0.12 | [20] |
| AtDehR b,d | A. tumefaciens C58 | 0.08 | 0.05 | 8.60 | 0.12 | [20] |
| VhDehR b,c | Vibrio harveyi | 0.75 | 0.13 | 0.37 | 0.13 | [20] |
2.3.2. Ethanol Production from Alginate Utilizing Bioengineered Sphingomonas sp. A1
2.3.3. Ethanol Production from Both Alginate and Mannitol Utilizing Bioengineered E. coli
2.3.4. Ethanol Production from Both Alginate and Mannitol Utilizing Bioengineered S. cerevisiae
2.3.5. The Key Enzyme for Saccharification of Alginate, an Exo-Type Alginate Lyase
3. Ethanol Production from Red Macroalgae
3.1. Carbohydrates in Red Macroalgae
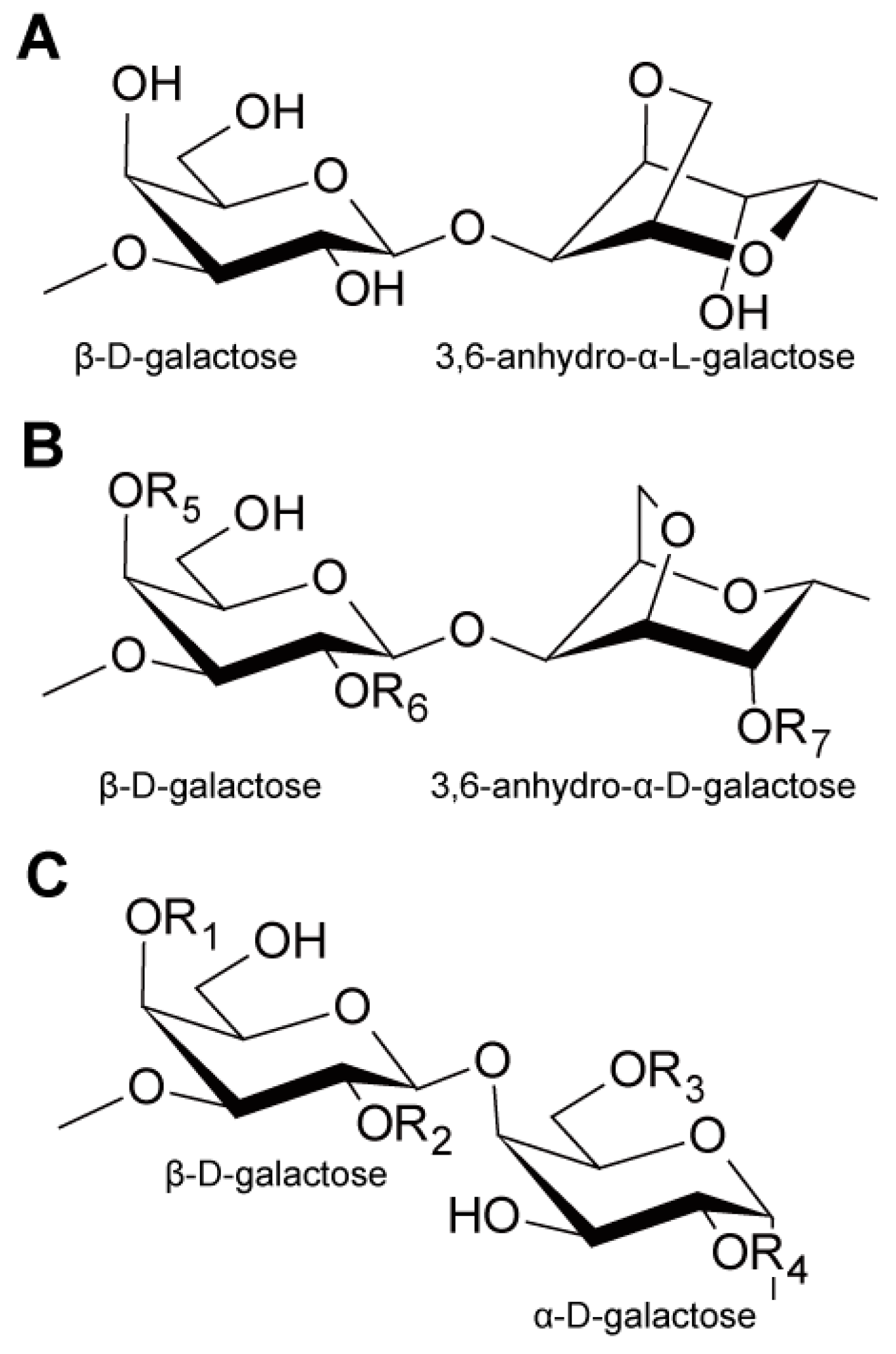
3.2. Key Biotechnology to Utilize Agar and Carrageenan for Ethanol Production: Utilization of 3,6-Anhydro-α-l-Galactose (AHG)
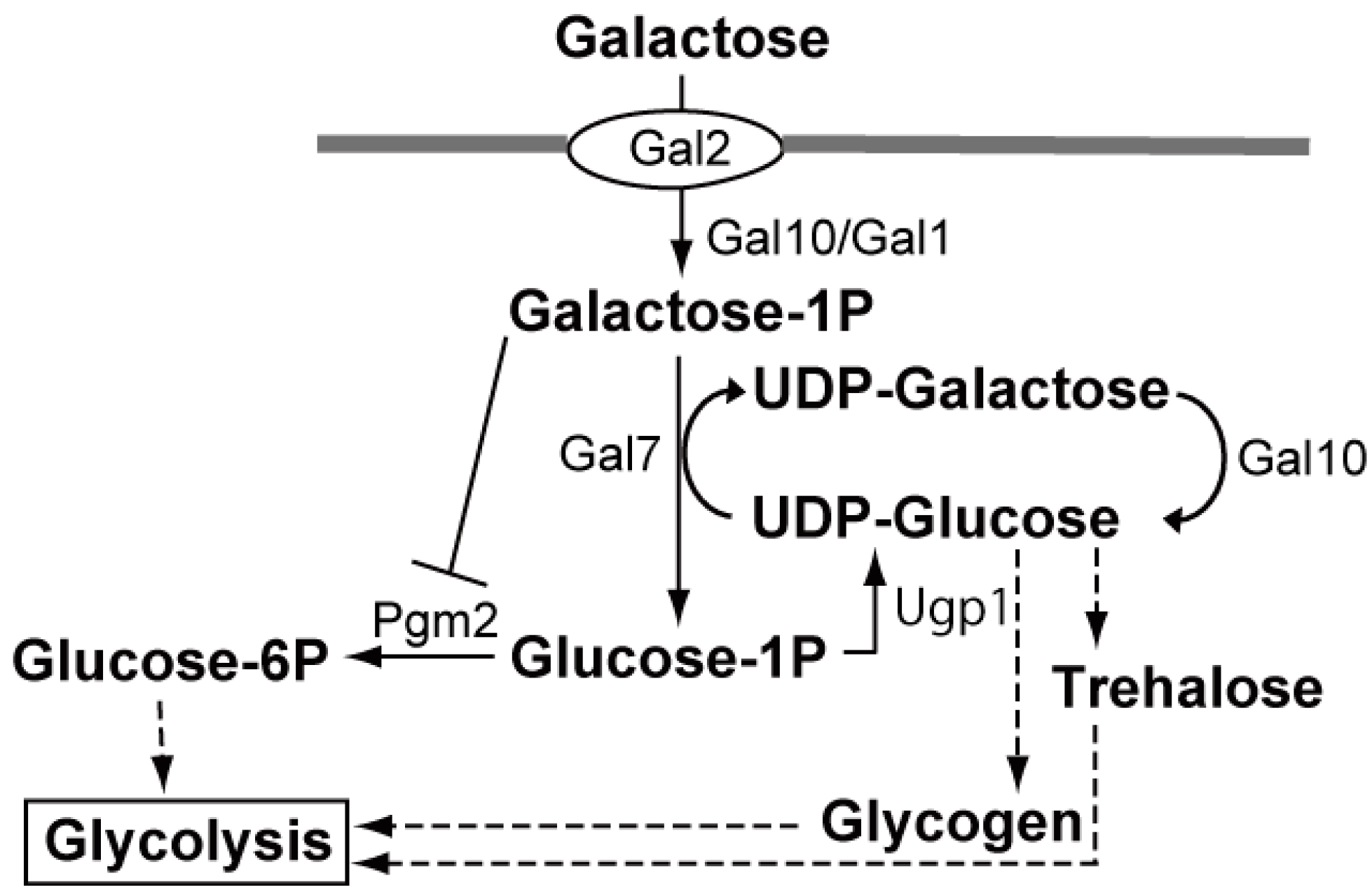
3.3. Key Biotechnology to Utilize Agar and Carrageenan for Ethanol Production: Improvement in the Utilization of Galactose
3.4. Key Biotechnology to Utilize Agar and Carrageenan for Ethanol Production: Methods for Saccharification of Agar and Carrageenan
4. Conclusions and Perspectives
| Strains | Concentration of Sugars in the Medium | Concentration of Ethanol Produced | Reference |
|---|---|---|---|
| Z. palmae | 38 g·L−1 of mannitol | 12 g·L−1 | [15] |
| P. angophorae | 40 g·L−1 of mannitol | 14.4 g·L−1 | [22] |
| E. coli KO11 | 75 g·L−1 of mannitol | 25.8 g·L−1 | [24] |
| S. paradoxus NBRC 0259-3 | 100 g·L−1 of mannitol | 45 g·L−1 | [25] |
| S. cerevisiae MK4416 | 100 g·L−1 of mannitol | 40 g·L−1 | [21] |
| Bioengineered Sphingomonas sp. A1 | (50 g + 10 g)·L−1 of sodium alginate | 13 g·L−1 | [37] |
| Bioengineered E. coli BAL1611 | 50 g·L−1 of a sugar mixture (alginate, mannitol, and glucose at a ratio of 5:8:1) | 20 g·L−1 | [18,48] |
| Bioengineered E. coli BAL1611 | 130 g·L−1 of dry milled brown macroalgae (L. japonica, kombu) | 35–41 g·L−1 | [18,48] |
| Bioengineered S. cerevisiae BAL3215 | 98 g·L−1 of sugar (1:2 molar ratio of DEH:mannitol) | 36.2 g·L−1 | [20] |
| Bioengineered E. coli KO11 | 3.2 g·L−1 of AHG and 4.1 g·L−1 of galactose | 1.4 g·L−1 | [64] |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Population Division of the Department of Economic and Social Affairs of the United Nations Secretariat. In World Population Prospects: The 2012 Revision, Highlights and Advance Tables; United Nations: New York, NY, USA, 2013.
- Adams, J.M.; Gallagher, J.A.; Donnison, I.S. Fermentation study on Saccharina latissima for bioethanol production considering variable pre-treatments. J. Appl. Phycol. 2009, 21, 569–574. [Google Scholar] [CrossRef]
- Yanagisawa, M.; Kawai, S.; Murata, K. Strategies for the production of high concentrations of bioethanol from seaweeds: Production of high concentrations of bioethanol from seaweeds. Bioengineered 2013, 4, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Borines, M.G.; de Leon, R.L.; Cuello, J.L. Bioethanol production from the macroalgae Sargassum spp. Bioresour. Technol. 2013, 138, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, M.; Nakamura, K.; Ariga, O.; Nakasaki, K. Production of high concentrations of bioethanol from seaweeds that contain easily hydrolyzable polysaccharides. Process Biochem. 2011, 46, 2111–2116. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Gleisner, R.; Scott, C.; Luo, X.L.; Tian, S. High titer ethanol production from simultaneous enzymatic saccharification and fermentation of aspen at high solids: A comparison between SPORL and dilute acid pretreatments. Bioresour. Technol. 2011, 102, 8921–8929. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Z.; Han, Y.J.; Xu, J. Simultaneous saccharification and fermentation of steam exploded wheat straw pretreated with alkaline peroxide. Process Biochem. 2008, 43, 1462–1466. [Google Scholar] [CrossRef]
- Varga, E.; Klinke, H.B.; Reczey, K.; Thomsen, A.B. High solid simultaneous saccharification and fermentation of wet oxidized corn stover to ethanol. Biotechnol. Bioeng. 2004, 88, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization. The State of World Fisheries and Aquaculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014. [Google Scholar]
- Wei, N.; Quarterman, J.; Jin, Y.S. Marine macroalgae: An untapped resource for producing fuels and chemicals. Trends Biotechnol. 2013, 31, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Myklestad, S. β-1,3-Glucans in Diatoms and Brown Seaweeds; Cambridge University Press: Cambridge, UK, 1978. [Google Scholar]
- Read, S.M.; Currie, G.; Bacic, A. Analysis of the structural heterogeneity of laminarin by electrospray-ionisation-mass spectrometry. Carbohydr. Res. 1996, 281, 187–201. [Google Scholar] [CrossRef]
- Rinaudo, M. Seaweed polysaccharides. In Comprehensive Glycoscience: From Chemistry to System Biology; Kamerling, J.P., Boons, G.-J., Lee, Y.C., Suzuki, A., Taniguchi, N., Voragen, A.G.J., Eds.; Elsevier: Oxford, UK, 2007; Volume 2, pp. 691–735. [Google Scholar]
- Black, W.A.P. The seasonal variation in weight and chemical composition of the common British Laminariaceae. J. Mar. Biol. Assoc. UK 1950, 29, 45–72. [Google Scholar] [CrossRef]
- Horn, S.J.; Aasen, I.M.; Østgaard, K. Production of ethanol from mannitol by Zymobacter palmae. J. Ind. Microbiol. Biotechnol. 2000, 24, 51–57. [Google Scholar] [CrossRef]
- Larsen, B.; Salem, D.M.S.A.; Sallam, M.A.E.; Mishrikey, M.M.; Beltagy, A.I. Characterization of the alginates from algae harvested at the Egyptian Red Sea coast. Carbohydr. Res. 2003, 338, 2325–2336. [Google Scholar] [CrossRef]
- Kimura, T.; Ueda, K.; Kuroda, R.; Akao, T.; Shinohara, N.; Ushirokawa, T.; Fukagawa, A.; Akimoto, T. The seasonal variation in polysaccharide content of brown alga akamoku Sargassum horneri collected off Oshima Island (Fukuoka Prefecture). Nippon Suisan Gakkaishi 2007, 73, 739–744. [Google Scholar] [CrossRef]
- Wargacki, A.J.E.; Leonard, M.N.; Win, D.D.; Regitsky, C.N.S.; Santos, P.B.; Kim, S.R.; Cooper, R.M.; Raisner, A.; Herman, A.B.; Sivitz, A.; et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 2012, 335, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Quain, D.E.; Boulton, C.A. Growth and metabolism of mannitol by strains of Saccharomyces cerevisiae. J. Gen. Microbiol. 1987, 133, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Enquist-Newman, M.; Faust, A.M.; Bravo, D.D.; Santos, C.N.; Raisner, R.M.; Hanel, A.; Sarvabhowman, P.; Le, C.; Regitsky, D.D.; Cooper, S.R.; et al. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature 2014, 505, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Chujo, M.; Yoshida, S.; Ota, A.; Murata, K.; Kawai, S. Acquisition of the ability to assimilate mannitol by Saccharomyces cerevisiae through dysfunction of the general corepressor Tup1-Cyc8. Appl. Environ. Microbiol. 2015, 81, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Schneider, H. Ethanol production from xylitol and some other polyols by Pichia angophorae. Biotechnol. Lett. 1987, 9, 581–584. [Google Scholar] [CrossRef]
- Horn, S.J.; Aasen, I.M.; Ostgaard, K. Ethanol production from seaweed extract. J. Ind. Microbiol. Biotechnol. 2000, 25, 249–254. [Google Scholar] [CrossRef]
- Kim, N.J.; Li, H.; Jung, K.; Chang, H.N.; Lee, P.C. Ethanol production from marine algal hydrolysates using Escherichia coli KO11. Bioresour. Technol. 2011, 102, 7466–7469. [Google Scholar] [CrossRef] [PubMed]
- Ota, A.; Kawai, S.; Oda, H.; Iohara, K.; Murata, K. Production of ethanol from mannitol by the yeast strain Saccharomyces paradoxus NBRC 0259. J. Biosci. Bioeng. 2013, 116, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Q.; Bai, F.W. Yeast flocculation: New story in fuel ethanol production. Biotechnol. Adv. 2009, 27, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.A. The catabolism of acyclic polyols by yeasts. J. Gen. Microbiol. 1968, 52, 131–159. [Google Scholar] [CrossRef] [Green Version]
- Perfect, J.R.; Rude, T.H.; Wong, B.; Flynn, T.; Chaturvedi, V.; Niehaus, W. Identification of a Cryptococcus neoformans gene that directs expression of the cryptic Saccharomyces cerevisiae mannitol dehydrogenase gene. J. Bacteriol. 1996, 178, 5257–5262. [Google Scholar] [PubMed]
- Treitel, M.A.; Carlson, M. Repression by SSN6-TUP1 Is directed by MIG1, a repressor activator protein. Proc. Natl. Acad. Sci. USA 1995, 92, 3132–3136. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Kawai, S.; Mikami, B.; Hashimoto, W. Superchannel of bacteria: Biological significance and new horizons. Biosci. Biotechnol. Biochem. 2008, 72, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Takase, R.; Ochiai, A.; Mikami, B.; Hashimoto, W.; Murata, K. Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1. Biochim. Biophys. Acta 2010, 1804, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Preiss, J.; Ashwell, G. Alginic acid metabolism in bacteria I. Enzymatic formation of unsaturated oligosac-charides and 4-deoxy-l-erythro-5-hexoseulose uronic acid. J. Biol. Chem. 1962, 237, 309–316. [Google Scholar] [PubMed]
- Preiss, J.; Ashwell, G. Alginic acid metabolism in bacteria II. The enzymatic reduction of 4-deoxy-l-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-d-gluconic acid. J. Biol. Chem. 1962, 237, 317–321. [Google Scholar] [PubMed]
- Pouyssegur, J.; Stoeber, F. Study of the common degradative pathway of hexuronates in Escherichia coli K 12. Purification, properties and individuality of 2-keto-3-deoxy-d-gluconnokinase. Biochimie 1971, 53, 771–781. [Google Scholar] [CrossRef]
- Egan, S.E.; Fliege, R.; Tong, S.; Shibata, A.; Wolf, R.E., Jr.; Conway, T. Molecular characterization of the Entner–Doudoroff pathway in Escherichia coli: Sequence analysis and localization of promoters for the edd-eda operon. J. Bacteriol. 1992, 174, 4638–4646. [Google Scholar] [PubMed]
- Takase, R.; Mikami, B.; Kawai, S.; Murata, K.; Hashimoto, W. Structure-based conversion of the coenzyme requirement of a short-chain dehydrogenase/reductase involved in bacterial alginate metabolism. J. Biol. Chem. 2014, 289, 33198–33214. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Yoneyama, F.; Kawai, S.; Hashimoto, W.; Murata, K. Bioethanol production from marine biomass alginate by genetically engineered bacteria. Energy Environ. Sci. 2011, 4, 2575–2581. [Google Scholar] [CrossRef]
- Hashimoto, W.; Miyake, O.; Momma, K.; Kawai, S.; Murata, K. Molecular identification of oligoalginate lyase of Sphingomonas sp. strain A1 as one of the enzymes required for complete depolymerization of alginate. J. Bacteriol. 2000, 182, 4572–4577. [Google Scholar] [CrossRef] [PubMed]
- Miyake, O.; Hashimoto, W.; Murata, K. An exotype alginate lyase in Sphingomonas sp. A1: Overexpression in Escherichia coli, purification, and characterization of alginate lyase IV (A1-IV). Protein Expr. Purif. 2003, 29, 33–41. [Google Scholar] [CrossRef]
- Yoon, H.J.; Hashimoto, W.; Miyake, O.; Okamoto, M.; Mikami, B.; Murata, K. Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp A1 alginate lyases. Protein Expr. Purif. 2000, 19, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Inose, T.; Hisano, T.; Abe, S.; Yonemoto, Y.; Yamashita, T.; Takagi, M.; Sakaguchi, K.; Kimura, A.; Imanaka, T. Bacterial alginate lyase: Enzymology, genetics and application. J. Ferment. Bioeng. 1993, 76, 427–437. [Google Scholar] [CrossRef]
- Yonemoto, Y.; Murata, K.; Kimura, A.; Yamaguchi, H.; Okayama, K. Bacterial alginate lyase: Characterization of alginate lyase-producing bacteria and purification of the enzyme. J. Ferment. Bioeng. 1991, 72, 152–157. [Google Scholar] [CrossRef]
- Momma, K.; Mishima, Y.; Hashimoto, W.; Mikami, B.; Murata, K. Direct evidence for Sphingomonas sp A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: Molecular insights into alginate transport in the periplasm. Biochemistry 2005, 44, 5053–5064. [Google Scholar] [CrossRef] [PubMed]
- Momma, K.; Okamoto, M.; Mishima, Y.; Mori, S.; Hashimoto, W.; Murata, K. A novel bacterial ATP-binding cassette transporter system that allows uptake of macromolecules. J. Bacteriol. 2000, 182, 3998–4004. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, W.; Kawai, S.; Murata, K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng. Bugs 2010, 1, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, Y.; Itoh, T.; Kaneko, A.; Nishitani, Y.; Mikami, B.; Hashimoto, W.; Murata, K. Structure of a bacterial ABC transporter involved in the import of an acidic polysaccharide alginate. Structure 2015, 23, 1643–1654. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Yoshida, S.; Murata, K.; Kawai, S. Regulation of pH attenuates toxicity of a byproduct produced by an ethanologenic strain of Sphingomonas sp. A1 during ethanol fermentation from alginate. Bioengineered 2014, 5, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.N.; Regitsky, D.D.; Yoshikuni, Y. Implementation of stable and complex biological systems through recombinase-assisted genome engineering. Nat. Commun. 2013, 4, 2503. [Google Scholar] [CrossRef] [PubMed]
- Klemm, P.; Hjerrild, L.; Gjermansen, M.; Schembri, M.A. Structure-function analysis of the self-recognizing Antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 2004, 51, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Larsson, C.; van Maris, A.; Pronk, J. Metabolic engineering of yeast for production of fuels and chemicals. Curr. Opin. Biotechnol. 2013, 24, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, A.; Hashimoto, W.; Murata, K. A biosystem for alginate metabolism in Agrobacterium tumefaciens strain C58: Molecular identification of Atu3025 as an exotype family PL-15 alginate lyase. Res. Microbiol. 2006, 157, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Kado, C.I. Historical account on gaining insights on the mechanism of crown gall tumorigenesis induced by Agrobacterium tumefaciens. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Chung, J.H.; Wang, D.; Lee, J.; Woo, H.C.; Choi, I.G.; Kim, K.H. Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2–40. Appl. Microbiol. Biotechnol. 2012, 93, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Jagtap, S.; Nair, S.K. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J. Biol. Chem. 2014, 289, 8645–8655. [Google Scholar] [CrossRef] [PubMed]
- Wang da, M.; Kim, H.T.; Yun, E.J.; Kim do, H.; Park, Y.C.; Woo, H.C.; Kim, K.H. Optimal production of 4-deoxy-l-erythro-5-hexoseulose uronic acid from alginate for brown macro algae saccharification by combining endo- and exo-type alginate lyases. Bioprocess Biosyst. Eng. 2014, 37, 2105–2111. [Google Scholar] [CrossRef] [PubMed]
- Ekborg, N.A.; Gonzalez, J.M.; Howard, M.B.; Taylor, L.E.; Hutcheson, S.W.; Weiner, R.M. Saccharophagus degradans gen. nov., sp. nov., a versatile marine degrader of complex polysaccharides. Int. J. Syst. Evol. Microbiol. 2005, 55, 1545–1549. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, S.S.; Hehemann, J.H.; Polz, M.F.; Lee, J.K.; Zhao, H. Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl. Environ. Microbiol. 2014, 80, 4207–4214. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, A.; Yamasaki, M.; Mikami, B.; Hashimoto, W.; Murata, K. Crystal structure of exotype alginate lyase Atu3025 from Agrobacterium tumefaciens. J. Biol. Chem. 2010, 285, 24519–24528. [Google Scholar] [CrossRef] [PubMed]
- Hutcheson, S.W.; Zhang, H.; Suvorov, M. Carbohydrase systems of Saccharophagus degradans degrading marine complex polysaccharides. Mar. Drugs 2011, 9, 645–665. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Yokoi, T.; Shibata, T.; Morisaka, H.; Kuroda, K.; Ueda, M. Engineered yeast whole-cell biocatalyst for direct degradation of alginate from macroalgae and production of non-commercialized useful monosaccharide from alginate. Appl. Microbiol. Biotechnol. 2015, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.P.; Chang, L.L.; Chang, S.N.; Wang, E.C.; Hwang, L.C.; Chen, Y.H.; Wang, Y.M. Successful preparation and characterization of biotechnological grade agarose from indigenous Gelidium amansii of Taiwan. Process Biochem. 2012, 47, 550–554. [Google Scholar] [CrossRef]
- Duckworth, M.; Yaphe, W. The structure of agar. Part I. Fractionation of a complex mixture of polysaccharides. Carbohydr. Res. 1971, 16, 189–197. [Google Scholar] [CrossRef]
- Campo, V.L.; Kawano, D.F.; da Silva, D.B.; Carvalho, I. Carrageenans: Biological properties, chemical modifications and structural analysis—A review. Carbohydr. Polym. 2009, 77, 167–180. [Google Scholar] [CrossRef]
- Yun, E.J.; Lee, S.; Kim, H.T.; Pelton, J.G.; Kim, S.; Ko, H.J.; Choi, I.G.; Kim, K.H. The novel catabolic pathway of 3,6-anhydro-l-galactose, the main component of red macroalgae, in a marine bacterium. Environ. Microbiol. 2015, 17, 1677–1688. [Google Scholar] [CrossRef] [PubMed]
- De Ley, J.; Doudoroff, M. The metabolism of d-galactose in Pseudomonas saccharophila. J. Biol. Chem. 1957, 227, 745–757. [Google Scholar] [PubMed]
- Wong, T.Y.; Yao, X.T. The DeLey-Doudoroff pathway of galactose metabolism in Azotobacter vinelandii. Appl. Environ. Microbiol. 1994, 60, 2065–2068. [Google Scholar] [PubMed]
- Hong, K.K.; Vongsangnak, W.; Vemuri, G.N.; Nielsen, J. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc. Natl. Acad. Sci. USA 2011, 108, 12179–12184. [Google Scholar] [CrossRef] [PubMed]
- Ostergaard, S.; Olsson, L.; Johnston, M.; Nielsen, J. Increasing galactose consumption by Saccharomyces cerevisiae through metabolic engineering of the GAL gene regulatory network. Nat. Biotechnol. 2000, 18, 1283–1286. [Google Scholar] [PubMed]
- Bro, C.; Knudsen, S.; Regenberg, B.; Olsson, L.; Nielsen, J. Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: Example of transcript analysis as a tool in inverse metabolic engineering. Appl. Environ. Microbiol. 2005, 71, 6465–6472. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Hong, M.E.; Jung, S.C.; Ha, S.J.; Yu, B.J.; Koo, H.M.; Park, S.M.; Seo, J.H.; Kweon, D.H.; Park, J.C.; et al. Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol. Bioeng. 2011, 108, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Tschopp, J.F.; Emr, S.D.; Field, C.; Schekman, R. GAL2 codes for a membrane-bound subunit of the galactose permease in Saccharomyces cerevisiae. J. Bacteriol. 1986, 166, 313–318. [Google Scholar] [PubMed]
- Holden, H.M.; Rayment, I.; Thoden, J.B. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 2003, 278, 43885–43888. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.; Timson, D.J. Characterization of the Saccharomyces cerevisiae galactose mutarotase/UDP-galactose 4-epimerase protein, Gal10p. FEMS Yeast Res. 2007, 7, 366–371. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thoden, J.B.; Sellick, C.A.; Timson, D.J.; Reece, R.J.; Holden, H.M. Molecular structure of Saccharomyces cerevisiae Gal1p, a bifunctional galactokinase and transcriptional inducer. J. Biol. Chem. 2005, 280, 36905–36911. [Google Scholar] [CrossRef] [PubMed]
- Christacos, N.C.; Marson, M.J.; Wells, L.; Riehman, K.; Fridovich-Keil, J.L. Subcellular localization of galactose-1-phosphate uridylyltransferase in the yeast Saccharomyces cerevisiae. Mol. Genet. Metab. 2000, 70, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, S.; Ghatak, J.; Mukherji, S.; Bhattacharjee, H.; Bhaduri, A. UDPgalactose 4-epimerase from Saccharomyces cerevisiae. A bifunctional enzyme with aldose 1-epimerase activity. Eur. J. Biochem. 2004, 271, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, J.P.; Kraemer, W.F.; Joshi, J.G. Purification and properties of phosphoglucomutase from Fleischmann’s yeast. Eur. J. Biochem. 1975, 57, 115–126. [Google Scholar] [CrossRef] [PubMed]
- De Jongh, W.A.; Bro, C.; Ostergaard, S.; Regenberg, B.; Olsson, L.; Nielsen, J. The roles of galactitol, galactose-1-phosphate, and phosphoglucomutase in galactose-induced toxicity in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2008, 101, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Ostergaard, S.; Walloe, K.O.; Gomes, S.G.; Olsson, L.; Nielsen, J. The impact of GAL6, GAL80, and MIG1 on glucose control of the GAL system in Saccharomyces cerevisiae. FEMS Yeast Res. 2001, 1, 47–55. [Google Scholar] [PubMed]
- Nehlin, J.O.; Carlberg, M.; Ronne, H. Control of yeast GAL genes by MIG1 repressor—A transcriptional cascade in the glucose response. EMBO J. 1991, 10, 3373–3377. [Google Scholar] [PubMed]
- Yun, E.J.; Choi, I.G.; Kim, K.H. Red macroalgae as a sustainable resource for bio-based products. Trends Biotechnol. 2015, 33, 247–249. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Hong, J.Y.; Jang, H.C.; Oh, S.G.; Kim, S.H.; Yoon, J.J.; Kim, Y.J. Use of Gelidium amansii as a promising resource for bioethanol: A practical approach for continuous dilute-acid hydrolysis and fermentation. Bioresour. Technol. 2012, 108, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Lee, S.; Kim, K.H.; Choi, I.G. The complete enzymatic saccharification of agarose and its application to simultaneous saccharification and fermentation of agarose for ethanol production. Bioresour. Technol. 2012, 107, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, H.T.; Yun, E.J.; Lee, A.R.; Kim, S.R.; Kim, J.H.; Choi, I.G.; Kim, K.H. A novel agarolytic β-galactosidase acts on agarooligosaccharides for complete hydrolysis of agarose into monomers. Appl. Environ. Microbiol. 2014, 80, 5965–5973. [Google Scholar] [CrossRef] [PubMed]
- Barbeyron, T.; Michel, G.; Potin, P.; Henrissat, B.; Kloareg, B. l-Carrageenases constitute a novel family of glycoside hydrolases, unrelated to that of κ-carrageenases. J. Biol. Chem. 2000, 275, 35499–35505. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Ishii, J.; Kondo, A. Rational design and evolutional fine tuning of Saccharomyces cerevisiae for biomass breakdown. Curr. Opin. Chem. Biol. 2015, 29, 1–9. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawai, S.; Murata, K. Biofuel Production Based on Carbohydrates from Both Brown and Red Macroalgae: Recent Developments in Key Biotechnologies. Int. J. Mol. Sci. 2016, 17, 145. https://doi.org/10.3390/ijms17020145
Kawai S, Murata K. Biofuel Production Based on Carbohydrates from Both Brown and Red Macroalgae: Recent Developments in Key Biotechnologies. International Journal of Molecular Sciences. 2016; 17(2):145. https://doi.org/10.3390/ijms17020145
Chicago/Turabian StyleKawai, Shigeyuki, and Kousaku Murata. 2016. "Biofuel Production Based on Carbohydrates from Both Brown and Red Macroalgae: Recent Developments in Key Biotechnologies" International Journal of Molecular Sciences 17, no. 2: 145. https://doi.org/10.3390/ijms17020145
APA StyleKawai, S., & Murata, K. (2016). Biofuel Production Based on Carbohydrates from Both Brown and Red Macroalgae: Recent Developments in Key Biotechnologies. International Journal of Molecular Sciences, 17(2), 145. https://doi.org/10.3390/ijms17020145





