1. Introduction
The Golgi Apparatus (GA), an important eukaryotic organelle involved in the metabolism of numerous proteins [
1], is a major collection and dispatch station for numerous proteins destined for secretion, plasma membranes and lysosomes [
2,
3]. The main function of the GA is to store, package and distribute proteins [
4]. In plant cells, the GA further serves as the site at which the complex polysaccharides of the cell wall are synthesized [
5]. The GA is comprised of three distinct membrane-bounded cisternae located between the endoplasmic reticulum and the cell surface, including
cis-Golgi,
media-Golgi, and
trans-Golgi [
6]. The multiple classes of cisternae differ in structure, composition, and function. The
cis-Golgi and
trans-Golgi are thought to be specialised cisternae leading proteins in and out of the GA [
7]. The
cis-Golgi functions as the receiving end for the biosynthetic output from the endoplasmic reticulum [
4]. The function of the
trans-Golgi is to sort and ship proteins to their intended destinations [
8]. Many different glycosyltransferases and other proteins are retained preferentially in a sub-Golgi apparatus to perform their various synthetic activities. Although the basic mechanism of the GA processing is known, how Golgi cisternae transports biosynthetic secretory cargo, and how resident Golgi proteins are localized to particular sets of cisternae, remain important and fascinating questions that await resolution [
9]. Hence, to elucidate functions of the GA involved in various cellular processes, an initial but crucial step is to identify the protein composition of the subcellular compartments of the GA.
As indicated in [
6], defects in Golgi apparatus can result in neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) [
10], Parkinson’s disease [
2], and Alzheimer’s disease (AD) [
11]. The accumulation and aggregation of
β-amyloid (A
β) protein is one of the characteristic hallmarks of AD [
12,
13]. The Group 9 complexes presented in [
14] have great potential as inhibitors of A
β1-40 peptide aggregation that is linked to neurodegeneration in AD patients. Protein S-nitrosylation might represent a potentially viable therapeutic target for a wide range of neurodegenerative diseases [
15]. As neuroprotective and anti-inflammatory therapies have largely proved unsatisfactory, considerable effort will be needed to make progress towards effective therapies for neurodegenerative diseases [
16]. As demonstrated in [
17], dysfunction of Golgi apparatus and its cisternae can give rise to muscular dystrophy, diabetes, cancers and other inheritable diseases. In addition, the GA is considered as an early target of the neurodegenerative diseases [
18]. The GA is a major cargo sorting and glycosylation station [
19]. Glycans have also been proved to be associated with a number of epidemic diseases such as some inherited diseases, cancers and diabetes. However, the corresponding molecular clues are only just being elucidated [
17]. Accurate identification of protein subGolgi localizations could provide useful clues to clarify the contribution of GA dysfunction to diseases, which will significantly impact our ability to develop more effective therapies for diseases and spur further research into the links between glycosylation and disease pathology.
Recently, a substantial amount of machine learning methods for predicting protein subcellular locations have been developed [
20,
21,
22]. However, few methods have been reported for predicting protein subGolgi localizations (
cis-Golgi
vs. trans-Golgi). In 2011, Ding
et al. [
6] employed a special mode of pseudo amino acid composition (increment of diversity) with the modified Mahalanobis discriminant to predict the types of Golgi-resident proteins. The accuracy obtained by the jackknife test was 74.7% in discriminating
cis-Golgi proteins from
trans-Golgi proteins. In 2013, Ding
et al. [
4] further extended their work, and presented a discriminative computational framework using
g-gap dipeptide based protein features followed by support vector machine. The analysis of variance (ANOVA) was employed to obtain the optimal features. By the jackknife cross-validation, this method achieved an accuracy of 0.854 and an area under the receiver operating characteristic curve of 0.878. In this paper, we follow the pioneer studies aiming to further improve the prediction performance of protein subGolgi localizations (
cis-Golgi
vs. trans-Golgi).
The aforementioned methods were trained on relatively small datasets with no more than 150 GA proteins. Predictors trained on a dataset of limited size and coverage often fail to identify protein attributes. Recent breakthrough of proteomic techniques has resulted in a rapid growth of newly discovered protein sequences. Therefore, the benchmark datasets used in the previous methods definitely need to be updated. In addition, the dataset is highly imbalanced in [
4],
i.e., the fraction of
trans-Golgi proteins is relatively small compared with that of
cis-Golgi proteins. For an imbalanced dataset, a classifier would tend to predict most of the incoming data belonging to the majority class [
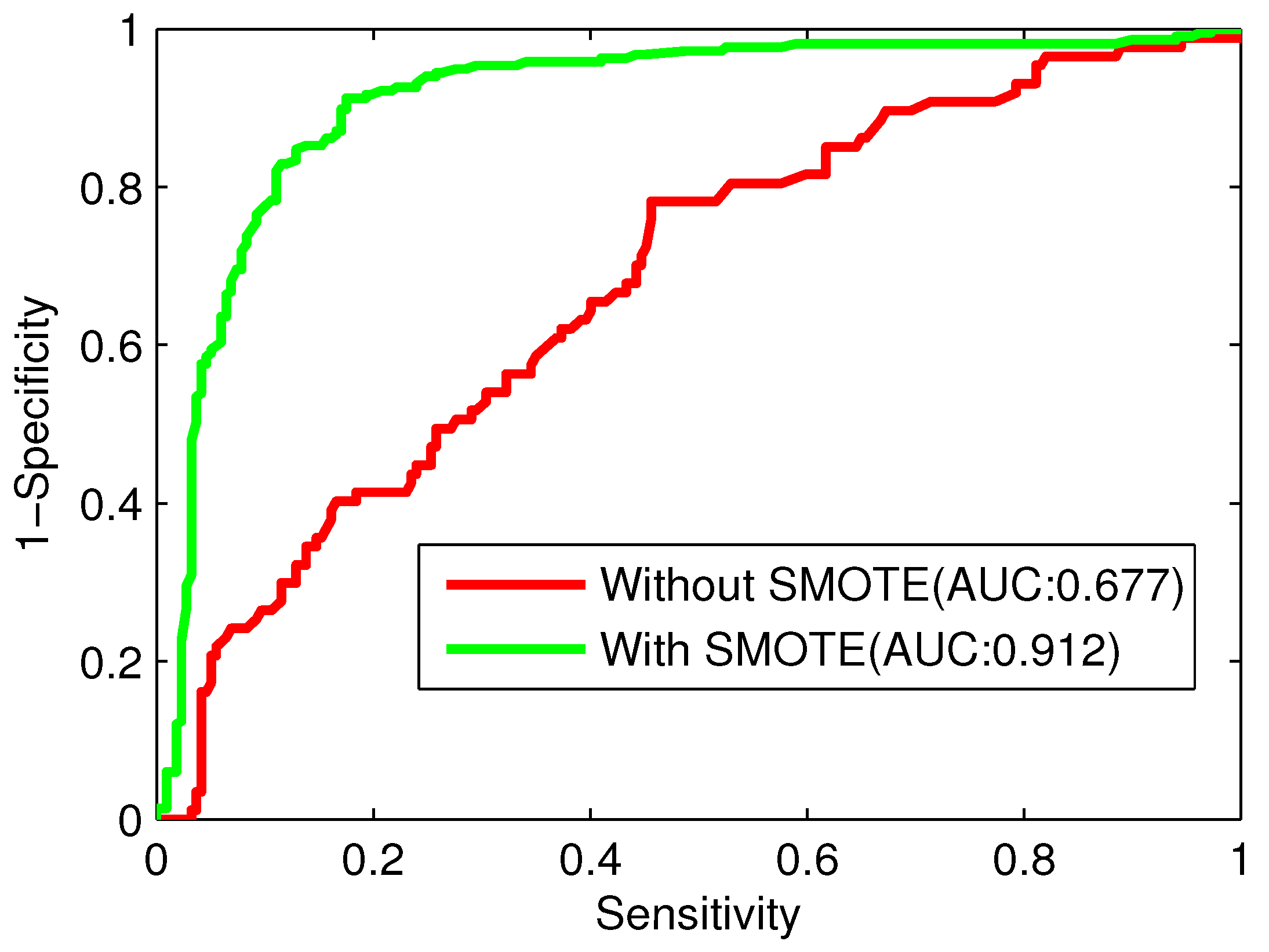
23]. In this study, we attempt to rebuild training sets through the SMOTE (Synthetic Minority Over-sampling Technique) to solve this imbalanced data problem.
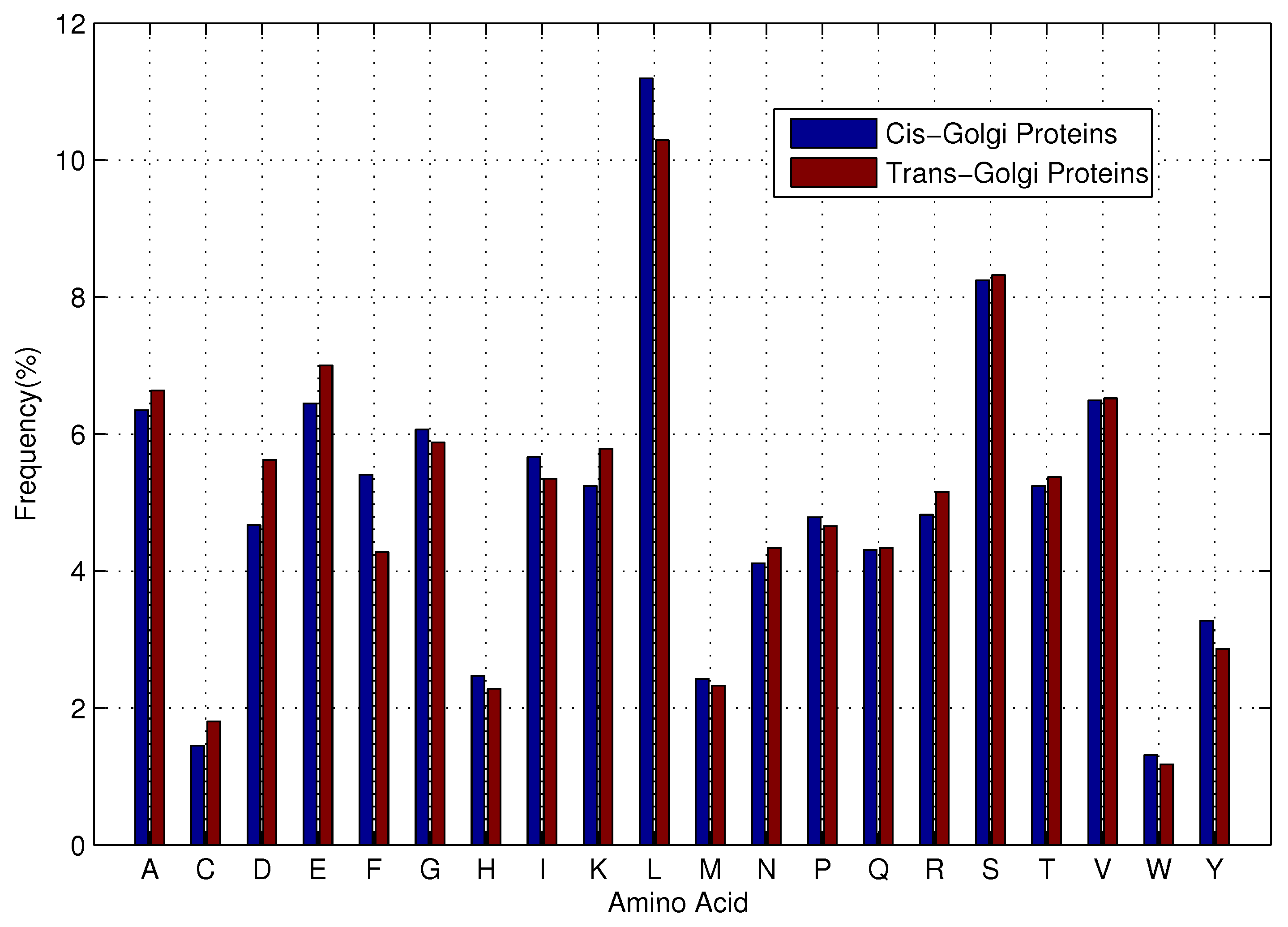
The previous predictors to discriminate
cis-Golgi proteins from
trans-Golgi proteins applied only information concerning the composition of the protein chain. Evolutionary-based features have not been adequately explored, which have been successfully applied in protein attribute predictions [
24,
25,
26]. These evolutionary-based features are extracted from the Position Specific Scoring Matrix (PSSM). Based on the concept of Common Spatial Patterns (CSP), a novel feature extraction technique is proposed in this study to extract three sets of features from PSSM-Dipeptide Composition (PSSM-DC), Bi-gram PSSM, and Evolutionary Difference-PSSM (ED-PSSM).
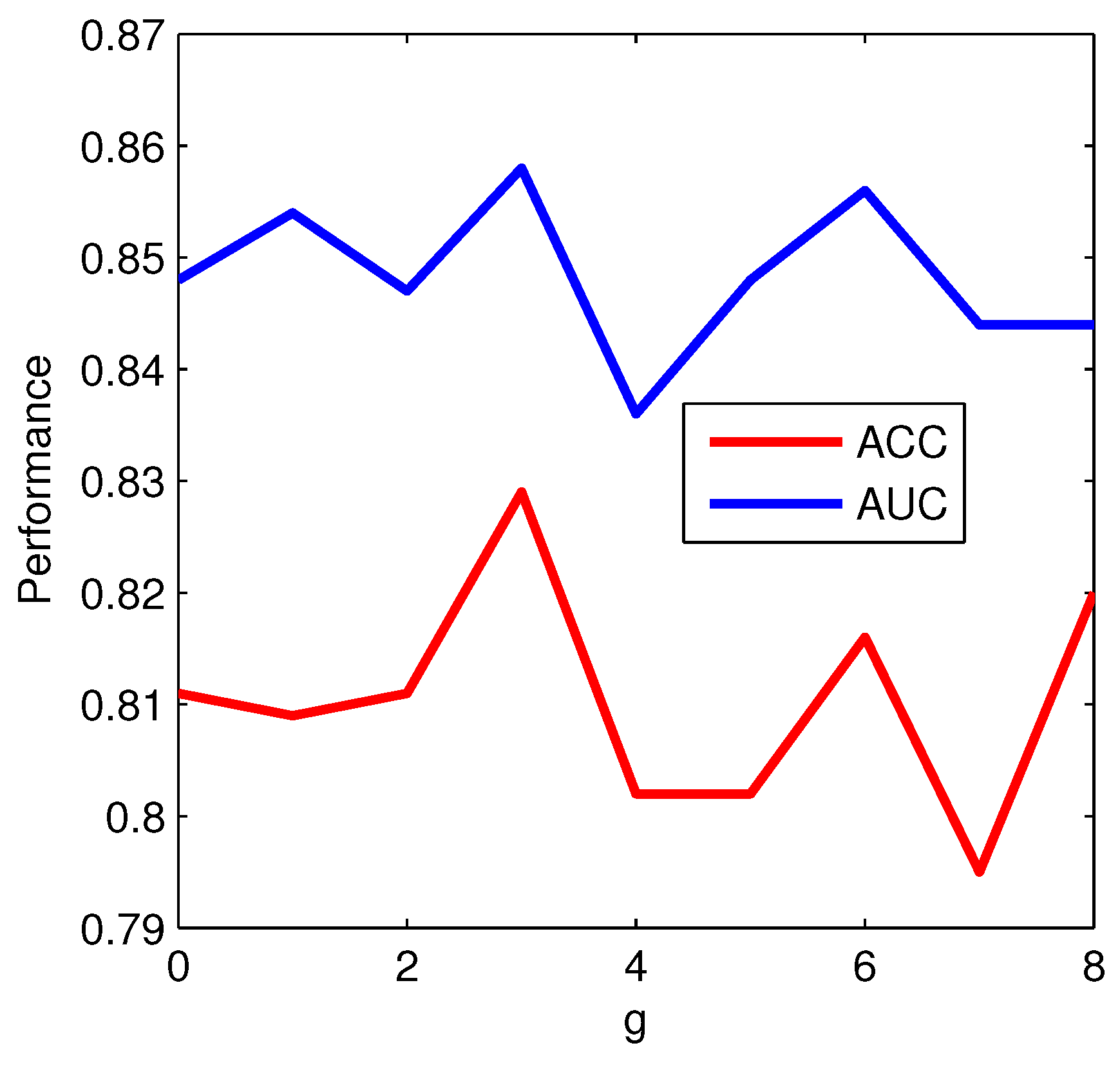
g-gap dipeptide based features have attained good results in previous studies [
4,
6] for this task. We improve the prediction accuracy by further incorporating the three informative evolutionary patterns and
g-gap dipeptide based features. The hybrid feature representation, containing evolutionary and sequence order information, can effectively analyze protein sequences. However, it leads to the feature vector with a high dimension. In order to reduce computation complexity and feature redundancy, the method of Random Forest-Recursive Feature Elimination (RF-RFE) is employed to find the optimal feature subset.
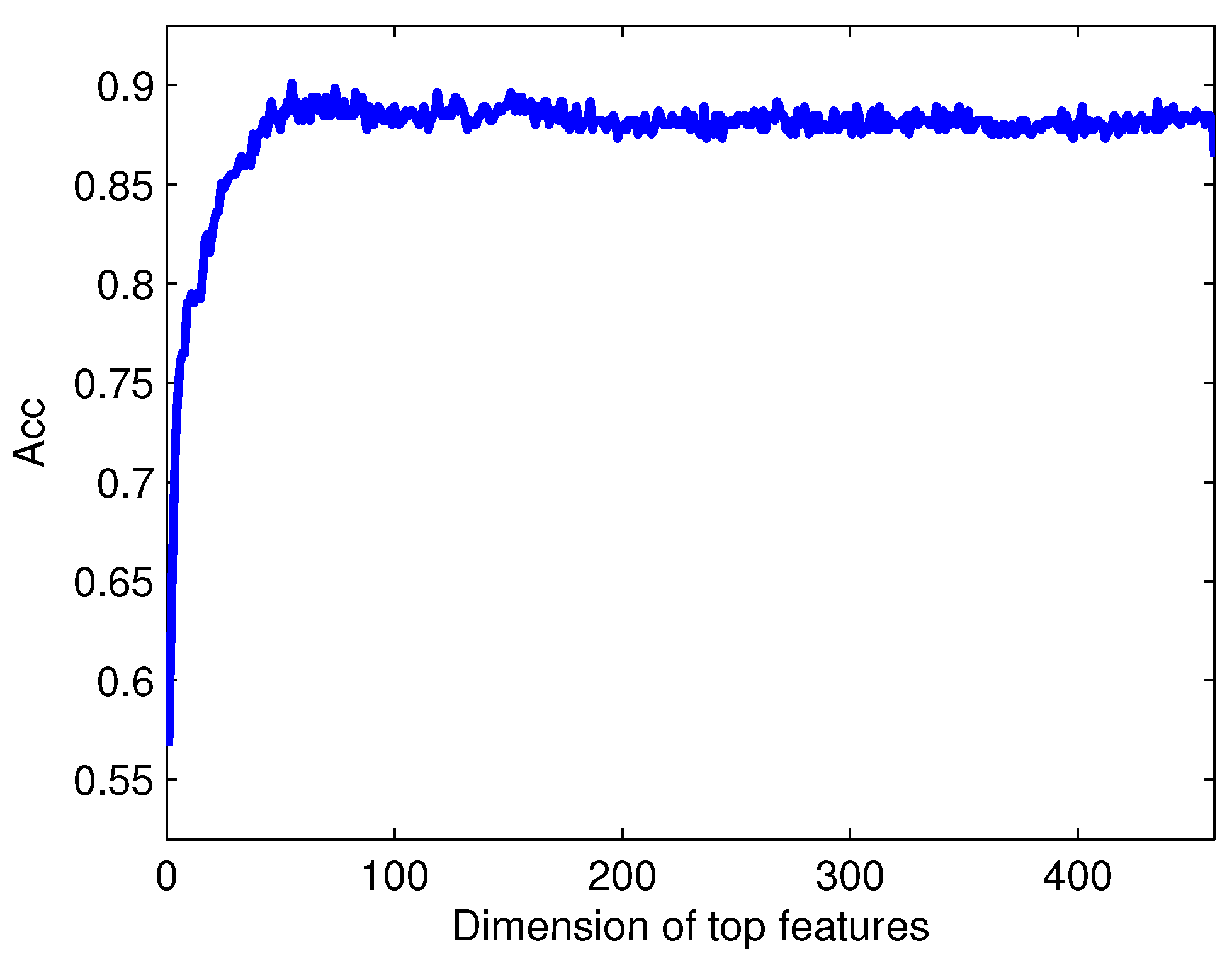
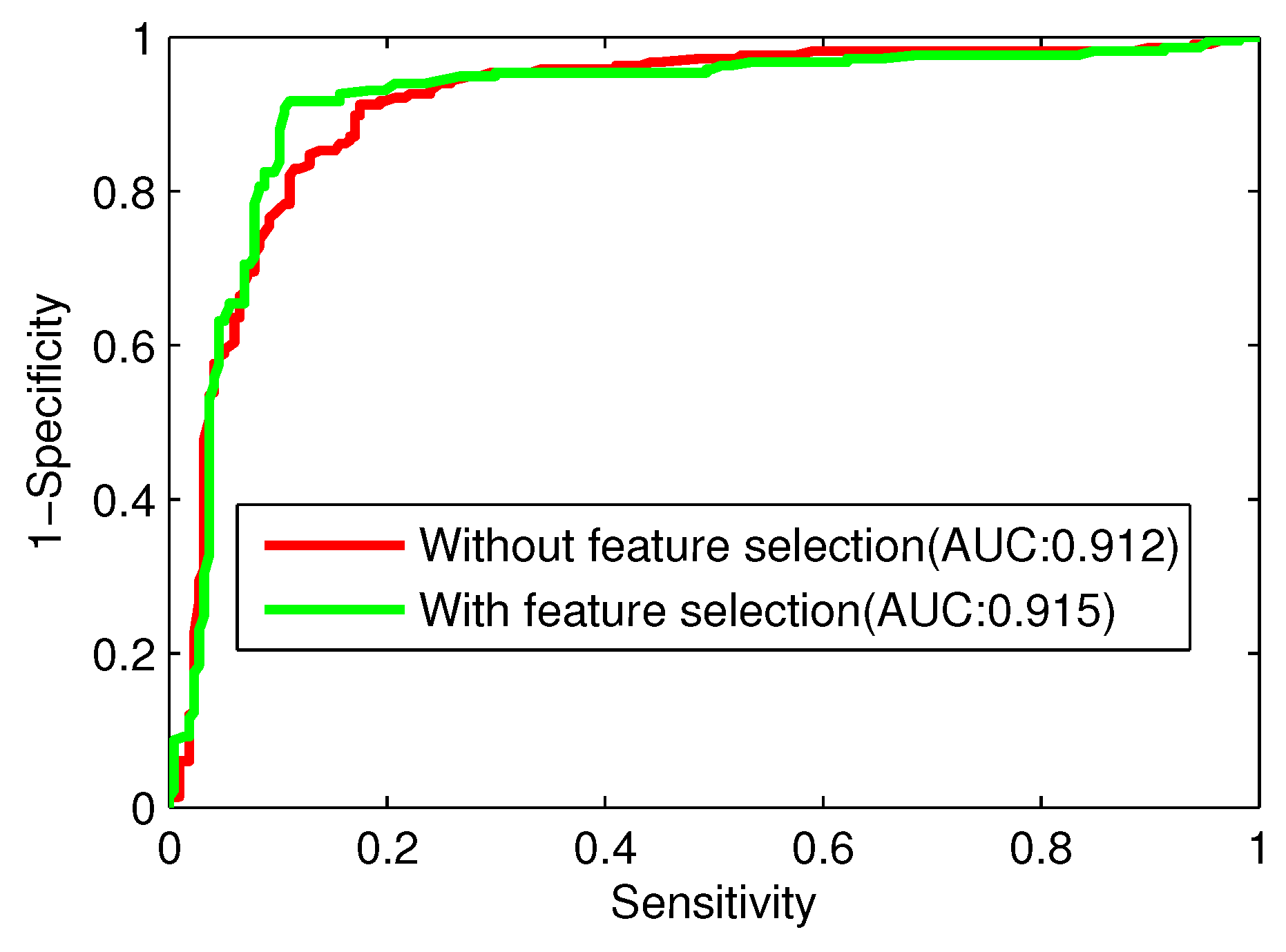
There are three major problems in the task of computational protein function prediction, including the construction of datasets, the extraction of protein representations, and the choice of classification algorithms [
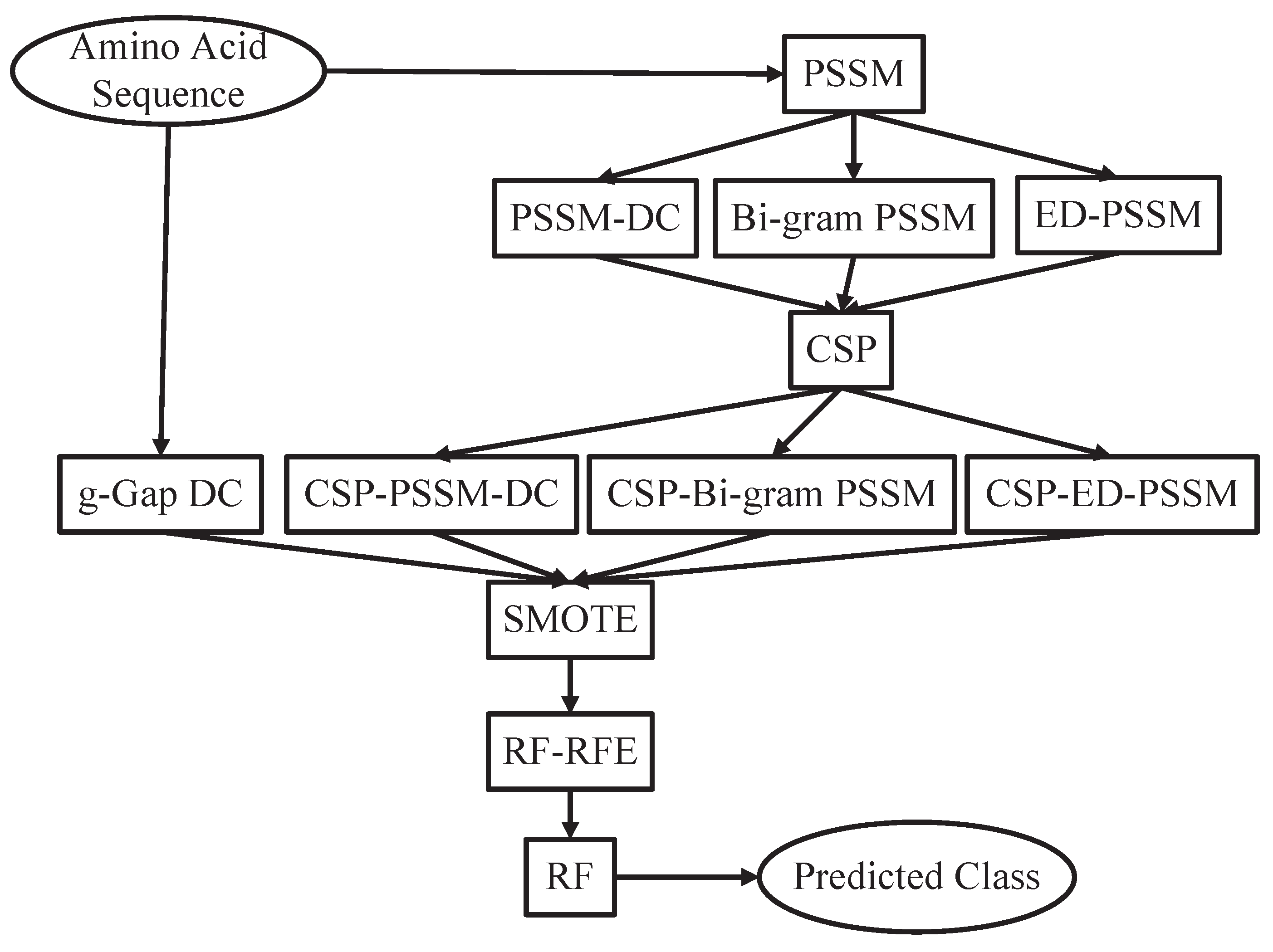
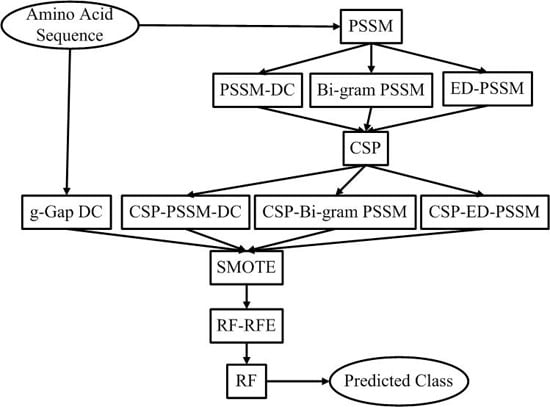
27]. The proposed prediction system is constructed based on an updated benchmark dataset. A CSP based feature extraction strategy is adopted to extract evolutionary information from protein sequences. The perdition performance of CSP based feature extraction method is comparable to that of traditional feature extraction methods. However, the feature number of the CSP based feature extraction method is only 1/20 of traditional feature extraction methods. Therefore, less computational and space cost is needed for the CSP based feature extraction method. CSP reduces computational complexity of our pipeline and effectively explore potential evolutionary information of protein sequences. In order to deal with this imbalanced data problem, we consider the SMOTE (Synthetic Minority Over-sampling Technique) to achieve balance. The Random Forest classifier is used to get an unbiased prediction. The system architecture of the proposed method is shown in
Figure 1. In the 10-fold cross-validation, our method achieves an overall accuracy of 0.908 for the prediction of
cis-Golgi proteins and an overall accuracy of 0.894 for the prediction of
trans-Golgi proteins. To further demonstrate its advantages, the proposed method is tested on the independent dataset given by the existing method [
4]. The results demonstrate that the proposed method is superior to the existing methods. Therefore, our method can be an effective predictor for large-scale determination of Golgi-resident protein types.
Figure 1.
The system architecture of the proposed method. PSSM: Position Specific Scoring Matrix, DC: Dipeptide Composition, ED: Evolutionary Difference, CSP: Common Spatial Patterns, SMOTE: Synthetic Minority Over-sampling Technique, RF: Random Forest, RFE: Recursive Feature Elimination.
Figure 1.
The system architecture of the proposed method. PSSM: Position Specific Scoring Matrix, DC: Dipeptide Composition, ED: Evolutionary Difference, CSP: Common Spatial Patterns, SMOTE: Synthetic Minority Over-sampling Technique, RF: Random Forest, RFE: Recursive Feature Elimination.