Characterization of Chemically-Induced Bacterial Ghosts (BGs) Using Sodium Hydroxide-Induced Vibrio parahaemolyticus Ghosts (VPGs)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effects of Chemicals on Bacterial Cell Envelopes
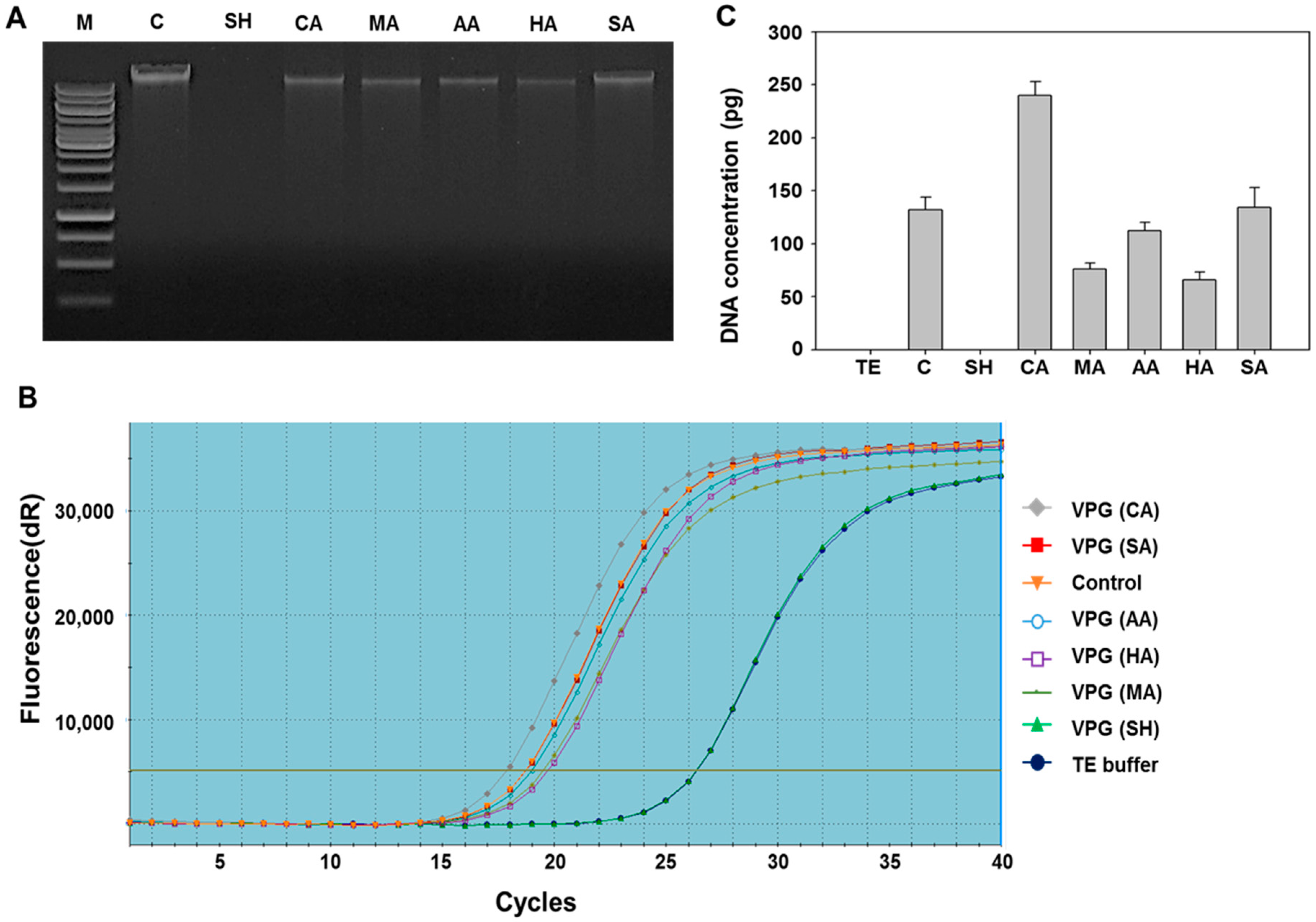
2.2. Determination of DNA-Free Vibrio parahaemolyticus Ghosts (VPGs) by Agarose Gel Electrophoresis and qPCR
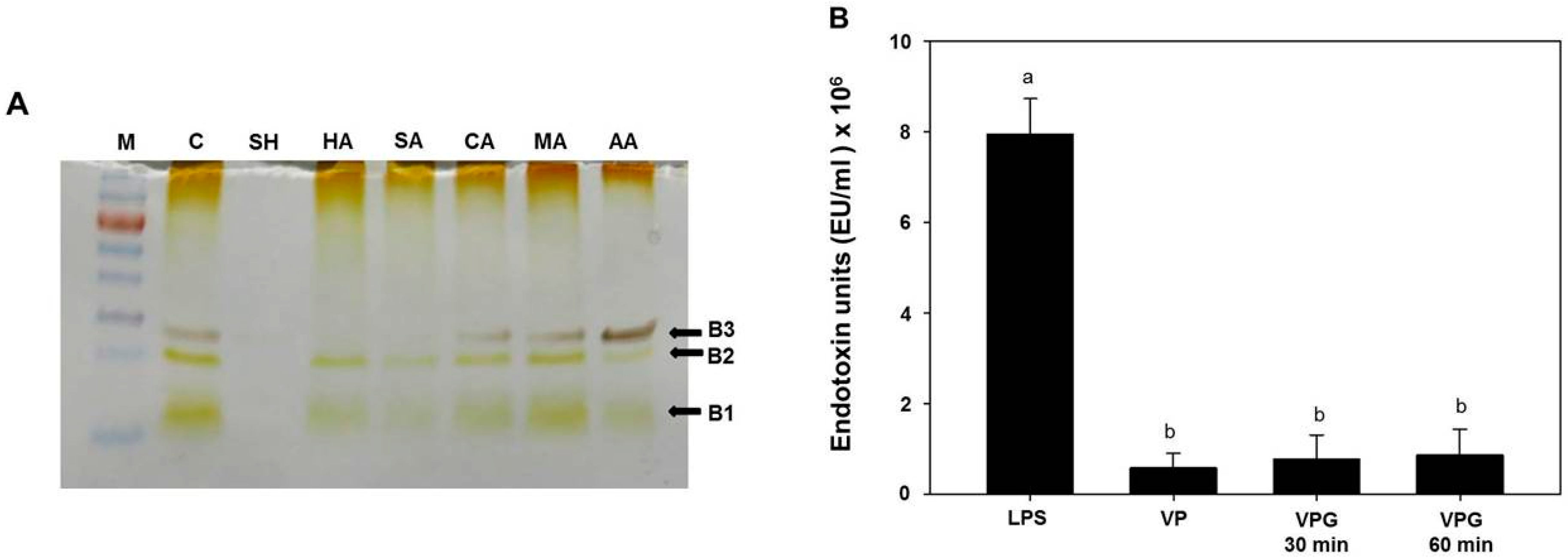
2.3. Analysis of Lipopolysaccharides (LPS) Profile in Chemically-Induced VPGs
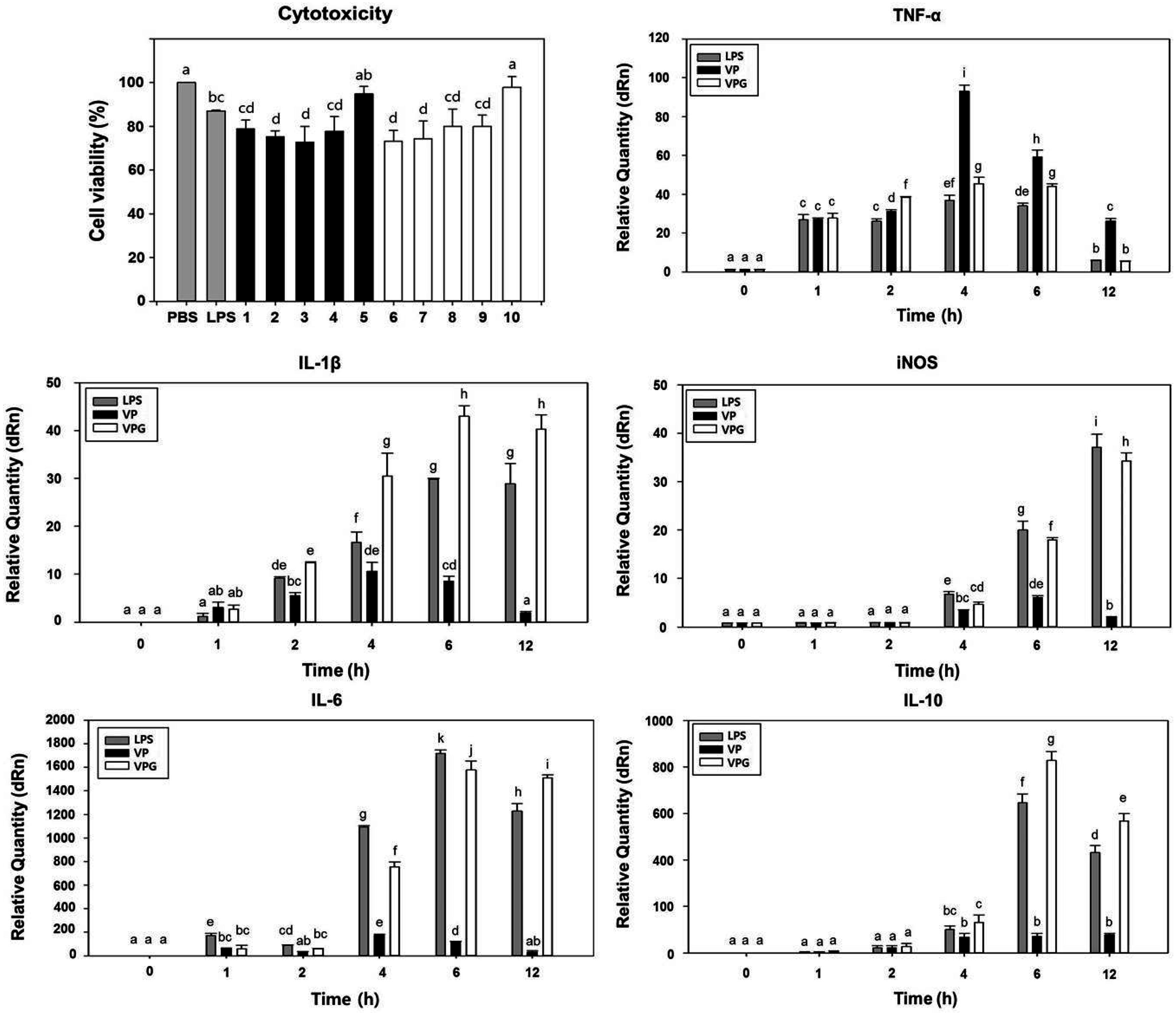
2.4. Comparison of Cytotoxicity Tests for VPGs
2.5. Induction of Cytokine Gene Expression in Murine Macrophages-Exposed VPGs
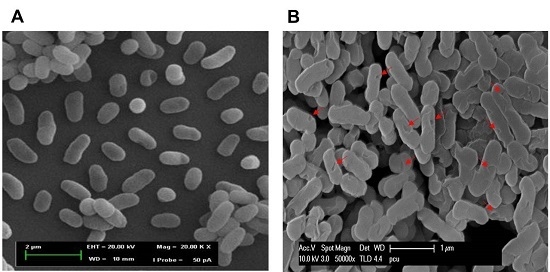
2.6. Morphological Observation of NaOH-Induced VPGs by Scanning Electron Microscopy (SEM)
2.7. Analysis of Protein and DNA Profiles in Sodium Hydroxide-Induced VPGs
3. Materials and Methods
3.1. Bacterial Strain and Culture Condition
3.2. Chemical Agents and Determination of Their MICs (Minimum Inhibitory Concentration)
3.3. Production of VPGs
3.4. Analysis of DNA-Free VPGs by Real-Time PCR
3.5. LPS Extraction, SDS-PAGE, and LAL Assay
3.6. Assessment of Macrophage-Mediated Cytotoxicity
3.7. Quantitative Analysis of Cytokine mRNA by Reverse Transcription (RT)-qPCR
3.8. SEM, SDS-PAGE, and Agarose Gel Electrophoresis Analyses
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Maione, D.; Margarit, I.; Rinaudo, C.D.; Masignani, V.; Mora, M.; Scarselli, M.; Tettelin, H.; Brettoni, C.; Iacobini, E.T.; Rosini, R.; et al. Identification of a universal group B Streptococcus vaccine by multiple genome screen. Science 2005, 309, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Lubitz, P.; Mayr, U.B.; Lubitz, W. Applications of bacterial ghosts in biomedicine. Adv. Exp. Med. Biol. 2009, 655, 159–170. [Google Scholar] [PubMed]
- Kwon, S.R.; Nam, Y.K.; Kim, S.K.; Kim, D.S.; Kim, K.H. Generation of Edwardsiella tarda ghosts by bacteriophage PhiX174 lysis gene E. Aquaculture 2005, 250, 16–21. [Google Scholar] [CrossRef]
- Eko, F.O.; Mayr, U.B.; Attridge, S.R.; Lubitz, W. Characterization and immunogenicity of Vibrio cholera ghosts expressing toxin-coregulated pili. J. Biotechnol. 2000, 83, 115–123. [Google Scholar] [CrossRef]
- Eko, F.O.; Schukovskaya, T.; Lotzmanova, E.Y.; Firstova, V.V.; Emalyanova, N.V.; Klueva, S.N.; Kravtzov, A.L.; Livanova, L.F.; Kutyrev, V.V.; Igietseme, J.U.; et al. Evaluation of the protective efficacy of Vibrio cholerae ghost (VCG) candidate vaccines in rabbits. Vaccine 2003, 21, 3663–3674. [Google Scholar] [CrossRef]
- Paukner, S.; Kohl, G.; Jalava, K.; Lubitz, W. Sealed bacterial ghosts-novel targeting vehicles for advanced drug delivery of water-soluble substances. J. Drug Target. 2003, 11, 151–161. [Google Scholar] [PubMed]
- Haidinger, W.; Mayr, U.B.; Szostak, M.P.; Resch, S.; Lubitz, W. Escherichia coli ghost production by expression of lysis gene E and Staphylococcal nuclease. Appl. Environ. Microbiol. 2003, 69, 6103–6113. [Google Scholar] [CrossRef]
- Wang, X.; Lu, C. Mice orally vaccinated with Edwardsiella tarda ghosts are significantly protected against infection. Vaccine 2009, 27, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Langemann, T.; Koller, V.J.; Muhammad, A.; Kudela, P.; Mayr, U.B.; Lubitz, W. The bacterial ghost platform system: Production and applications. Bioeng. Bugs 2010, 1, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Champeimont, J.; Mayr, U.B.; Lubitz, W.; Kudela, P. Bacterial ghosts as carriers of protein subunit and DNA-encoded antigens for vaccine applications. Expert Rev. Vaccines 2012, 11, 97–116. [Google Scholar] [CrossRef] [PubMed]
- Jawale, C.V.; Chaudhari, A.A.; Jeon, B.W.; Nandre, R.M.; Lee, J.H. Characterization of a novel inactivated Salmonella enterica serovar Enteritidis vaccine candidate generated using a modified cI857/λ PR/gene E expression system. Infect. Immun. 2012, 80, 1502–1509. [Google Scholar] [CrossRef] [PubMed]
- Panthel, K.; Jechlinger, W.; Matis, A.; Rohde, M.; Szostak, M.; Lubitz, W.; Haas, R. Generation of Helicobacter pylori ghosts by PhiX protein E-mediated inactivation and their evaluation as vaccine candidates. Infect. Immun. 2003, 71, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Mayr, U.B.; Haller, C.; Haidinger, W.; Masignani, V.; Mora, M.; Scarselli, M.; Tettelin, H.; Brettoni, C.; Iacobini, E.T.; Rosini, R.; et al. Bacterial ghosts as an oral vaccine: A single dose of Escherichia coli O157:H7 bacterial ghosts protects mice against lethal challenge. Infect. Immun. 2005, 73, 4810–4817. [Google Scholar] [CrossRef] [PubMed]
- Tu, F.P.; Chu, W.H.; Zhuang, X.Y.; Lu, C.P. Effect of oral immunization with Aeromonas hydrophila ghosts on protection against experimental fish infection. Lett. Appl. Microbiol. 2010, 50, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Amara, A.A.; Salem-Bekhit, M.M.; Alanazi, F.K. Sponge-like: A new protocol for preparing bacterial ghosts. Sci. World J. 2013, 2013, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vinod, N.; Oh, S.; Kim, S.; Choi, C.W.; Kim, S.C.; Jung, C.H. Chemically induced Salmonella enteritidis ghosts as a novel vaccine candidate against virulent challenge in a rat model. Vaccine 2014, 32, 3249–3255. [Google Scholar] [CrossRef] [PubMed]
- Vinod, N.; Oh, S.; Park, H.J.; Koo, J.M.; Choi, C.W.; Kim, S.C. Generation of a novel Staphylococcus aureus ghost vaccine and examination of its immunogenicity against virulent challenge in rats. Infect. Immun. 2015, 83, 2957–2965. [Google Scholar] [CrossRef] [PubMed]
- Sandle, T. A paractical approach to depyrogenation studies using bacterial endotoxin. J. GXP Compliance 2011, 15, 90–96. [Google Scholar]
- Newton, A.; Kendall, M.; Vugia, D.J.; Henao, O.L.; Mahon, B.E. Increasing rates of vibriosis in the United States, 1996–2010: Review of surveillance data from 2 systems. Clin. Infect. Dis. 2012, 54, S391–S395. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.C.; Liu, C. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007, 24, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Nair, G.B.; Ramamurthy, T.; Bhattacharya, S.K.; Dutta, B.; Takeda, Y.; Sack, D.A. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 2007, 20, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, D.; Hasan, N.A.; Huq, A.; Colwell, R.R. Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.R.; Kang, Y.J.; Lee, D.J.; Lee, E.H.; Nam, Y.K.; Kim, S.K.; Kim, K.H. Generation of Vibrio anguillarum ghost by coexpression of PhiX 174 lysis E gene and staphylococcal nuclease A gene. Mol. Biotechnol. 2009, 42, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.R. Comparison of the immunogenicity between bacterial ghost and formalin-killed bacteria for Vibrio vulnificus. J. Fish Pathol. 2012, 25, 159–164. [Google Scholar] [CrossRef]
- Midolo, P.D.; Lambert, J.R.; Hull, R.; Luo, F.; Grayson, M.L. In vitro inhibition of Helicobacter pylori NCTC 11637 by organic acids and lactic acid bacteria. J. Appl. Bacteriol. 1995, 79, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Fujita, T.; Yamamoto, A. Role of cyclooxygenase-2 in Helicobacter pylori-induced gastritis in Mongolian gerbils. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G791–G798. [Google Scholar] [PubMed]
- Hu, M.; Zhang, Y.; Xie, F.; Li, G.; Li, J.; Si, W.; Liu, S.; Hu, S.; Zhang, Z.; Shen, N.; et al. Protection of piglets by a Haemophilus parasuis ghost vaccine against homologous challenge. Clin. Vaccine Immunol. 2012, 20, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Yang, G.; Zhang, Y.; Yuan, J.; An, L. Generation of biotechnology-derived Flavobacterium columnare ghosts by PhiX174 gene E-mediated inactivation and the potential as vaccine candidates against infection in grass carp. J. Biomed. Biotechnol. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Nishibuchi, M.; Fasano, A.; Russell, R.G.; Kaper, J.B. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direc themolysin. Infect. Immun. 1992, 60, 3539–3545. [Google Scholar] [PubMed]
- Honda, T.; Ni, Y.X.; Miwatani, T. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 1988, 56, 961–965. [Google Scholar] [PubMed]
- Joseph, S.; Colwell, R.; Kaper, J. Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 1982, 10, 77–124. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.N.; Chaudhuri, K. Lipopolysaccharides of Vibrio cholerae: III. Biological functions. Biochim. Biophys. Acta 2006, 1762, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mader, H.J.; Szostak, M.P.; Hensel, A.; Lubitz, W.; Haslberger, A.G. Endotoxicity does not limit the use of bacterial ghosts as candidate vaccines. Vaccine 1997, 15, 195–202. [Google Scholar] [CrossRef]
- Han, T.J.; Chai, T.J. Electrophoretic and chemical characterization of lipopolysaccharides of Vibrio parahaemolyticus. J. Bacteriol. 1992, 174, 3140–3146. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.M.; Frasch, C.E.; Rivera, E.; Hochstein, H.D. Measurements of lipopolysaccharide (endotoxin) in meningococcal protein and polysaccharide preparations for vaccine usage. J. Biol. Stand. 1989, 17, 249–258. [Google Scholar] [CrossRef]
- Bandekar, J.R.; Nerkar, D.P. Antitumor activity of lipopolysaccharide and radio-detoxified lipopolysaccharide of Vibrio parahaemolyticus. Microbiol. Immunol. 1987, 31, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Seid, R.C.; Sadoff, J.C. Preparation and characterization of detoxified lipopolysaccharide-protein conjugates. J. Biol. Chem. 1981, 256, 7305–7310. [Google Scholar] [PubMed]
- Von Eschen, K.B.; Rudbach, J.A. Antibody responses of mice to alkaline detoxified lipopolysaccharide. J. Immunol. 1976, 116, 8–11. [Google Scholar] [PubMed]
- ČiŽnár, I.; Shands, J.W. Effect of alkali on the immunological reactivity of lipopolysaccharide from Salmonella typhimurium. Infect. Immun. 1970, 2, 549–555. [Google Scholar] [PubMed]
- Waters, S.; Luther, S.; Joerger, T.; Richards, G.P.; Fidelma Boyd, E.; Parent, M.A. Murine macrophage inflammatory cytokine production and immune activation in response to Vibrio parahaemolyticus infection. Microbiol. Immunol. 2013, 57, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Castejon, G.; Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, C.; Rollinghoff, M.; Diefenbach, A. The role of nitric oxide in innate immunity. Immunol. Rev. 2000, 173, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Moncada, S.; Palmer, R.M.; Higgs, E.A. Nitric oxide: Physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 1991, 43, 109–142. [Google Scholar] [PubMed]
- O’Shea, J.J.; Ma, A.; Lipsky, P. Cytokines and autoimmunity. Nat. Rev. Immunol. 2002, 2, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-B.; Lee, Y.-J.; Park, S.K.; Kim, H.-C.; Bae, H.; Kim, H.M.; Ko, S.-G.; Choi, H.Y.; Oh, M.S.; Park, W. Anti-inflamatory effects of Scutellaria baicalensis water extract on LPS-activated RAW 264.7 macrophages. J. Ethnopharm. 2009, 125, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, J.E.; Santos, M.J.; Canhão, H.; Choy, E. Interleukin-6 as a key player in systemic inflammation and joint destruction. Autoimmun. Rev. 2009, 8, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. BBA-Mol. Cell Res. 2011, 1813, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, S.; Marincola, F.; Rossi, C.R.; Nitti, D.; Lise, M. The multifaceted relationship between IL-10 and adaptive immunity: Putting together the pieces of a puzzle. Cytokine Growth Factor Rev. 2004, 15, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Pfaffl, M.W. Relative quantification. In Real-Time PCR; Dorak, T., Ed.; International University Line: La Jolla, CA, USA, 2006; pp. 63–82. [Google Scholar]
- Fomsgaard, A.; Freudenberg, M.A.; Galanos, C. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J. Clin. Microbiol. 1990, 28, 2627–2631. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]


| Chemical | MIC (mg/mL) | Medium pH |
|---|---|---|
| Sodium hydroxide (NaOH) | 3.125 | 9.99 |
| Acetic acid (CH3COOH) | 6.25 | 4.28 |
| Boric acid (BH3O3) | <50 | - |
| Citric acid (C6H8O7) | 25 | 3.11 |
| Hydrochloric acid (HCl) | 1.56 | 3.95 |
| Maleic acid (C4H4O4) | 6.25 | 3.56 |
| Sulfuric acid (H2SO4) | 0.781 | 5.83 |
| Gene | Orientation | Primer Sequences (5′–3′) |
|---|---|---|
| 16s rRNA | forward | ATATGCCTAGGTGGGATTAGCTAGT |
| reverse | TGTCTCAGTTCCAGTGTGGCTG | |
| TNF-α | forward | ATGAGCACAGAA AGCATGATCCG |
| reverse | GCTGAGACATAGGCACCGC | |
| Il-1β | forward | ATGGCAACTGTTCCTGAACTCAACT |
| reverse | AGTAGCCCTTCATCTTTTGGGG | |
| IL-6 | forward | ATGAAGTTCCTCTCTGCAAGAGACT |
| reverse | GTCTCCTCTCCGGACTTGTGA | |
| IL-10 | forward | ATGCCTGGCTCAGCACTGCTA |
| reverse | CTGGGAAGTGGGTGCAGTTATTG | |
| IL-12 | forward | ATGTGTCAATCACGCTACCTCCT |
| reverse | GACTGGCTAAGACACCTGGC | |
| iNOS | forward | ATGAACCCCAAGAGTTTGACCAGA |
| reverse | GGAGCCATAATACTGGTTGATGAAC | |
| GAPDH | forward | ATGGTGAAGGTCGGTGTGAACG |
| reverse | CAATGAAGGGGTCGTTGATGGC |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.J.; Oh, S.; Vinod, N.; Ji, S.; Noh, H.B.; Koo, J.M.; Lee, S.H.; Kim, S.C.; Lee, K.-S.; Choi, C.W. Characterization of Chemically-Induced Bacterial Ghosts (BGs) Using Sodium Hydroxide-Induced Vibrio parahaemolyticus Ghosts (VPGs). Int. J. Mol. Sci. 2016, 17, 1904. https://doi.org/10.3390/ijms17111904
Park HJ, Oh S, Vinod N, Ji S, Noh HB, Koo JM, Lee SH, Kim SC, Lee K-S, Choi CW. Characterization of Chemically-Induced Bacterial Ghosts (BGs) Using Sodium Hydroxide-Induced Vibrio parahaemolyticus Ghosts (VPGs). International Journal of Molecular Sciences. 2016; 17(11):1904. https://doi.org/10.3390/ijms17111904
Chicago/Turabian StylePark, Hyun Jung, Sung Oh, Nagarajan Vinod, Seongmi Ji, Han Byul Noh, Jung Mo Koo, Su Hyeong Lee, Sei Chang Kim, Ki-Sung Lee, and Chang Won Choi. 2016. "Characterization of Chemically-Induced Bacterial Ghosts (BGs) Using Sodium Hydroxide-Induced Vibrio parahaemolyticus Ghosts (VPGs)" International Journal of Molecular Sciences 17, no. 11: 1904. https://doi.org/10.3390/ijms17111904
APA StylePark, H. J., Oh, S., Vinod, N., Ji, S., Noh, H. B., Koo, J. M., Lee, S. H., Kim, S. C., Lee, K.-S., & Choi, C. W. (2016). Characterization of Chemically-Induced Bacterial Ghosts (BGs) Using Sodium Hydroxide-Induced Vibrio parahaemolyticus Ghosts (VPGs). International Journal of Molecular Sciences, 17(11), 1904. https://doi.org/10.3390/ijms17111904