Circulating MicroRNAs as Biomarkers for Sepsis
Abstract
:1. Introduction
2. Biomarkers in Sepsis
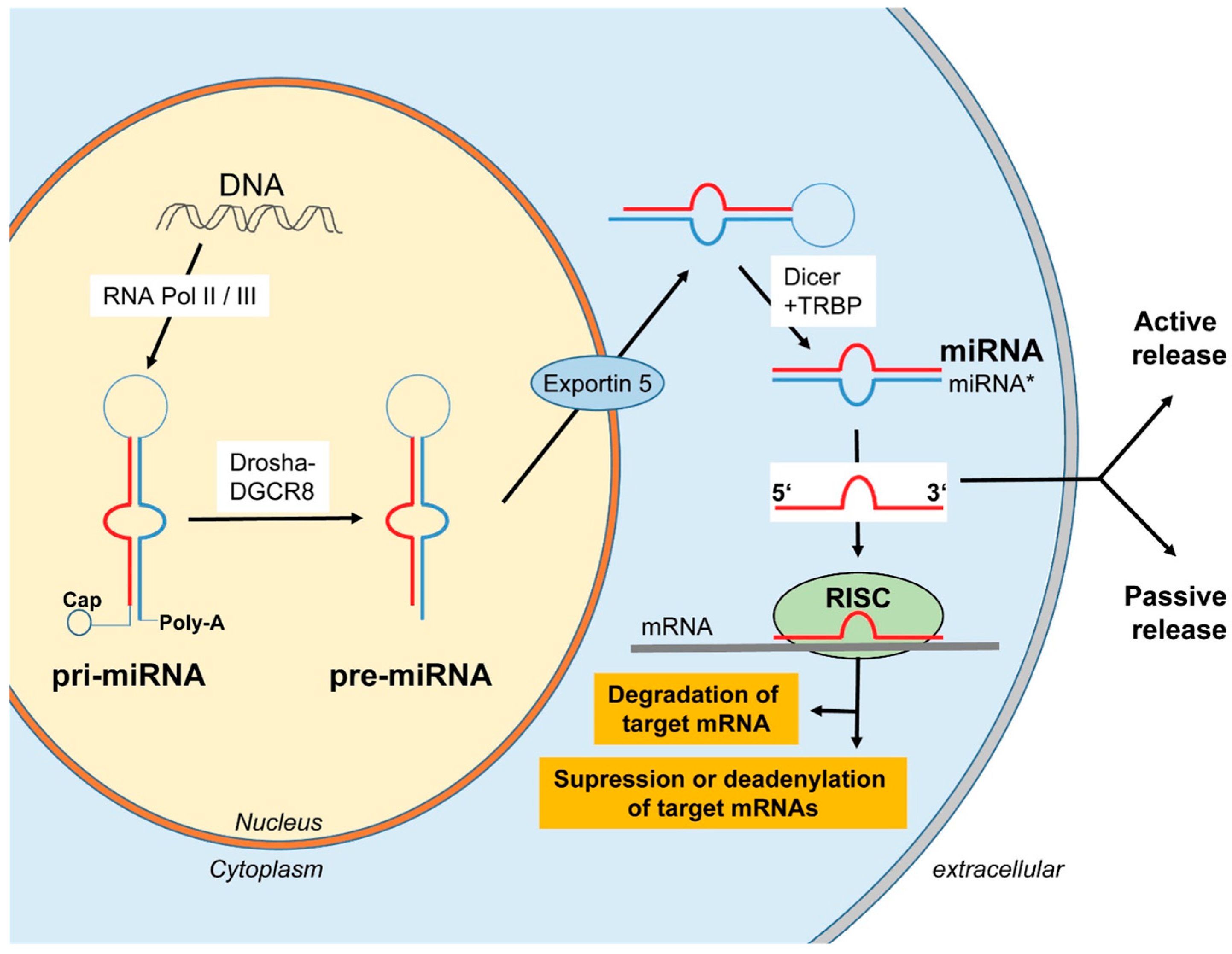
3. Biogenesis and Release of miRNAs
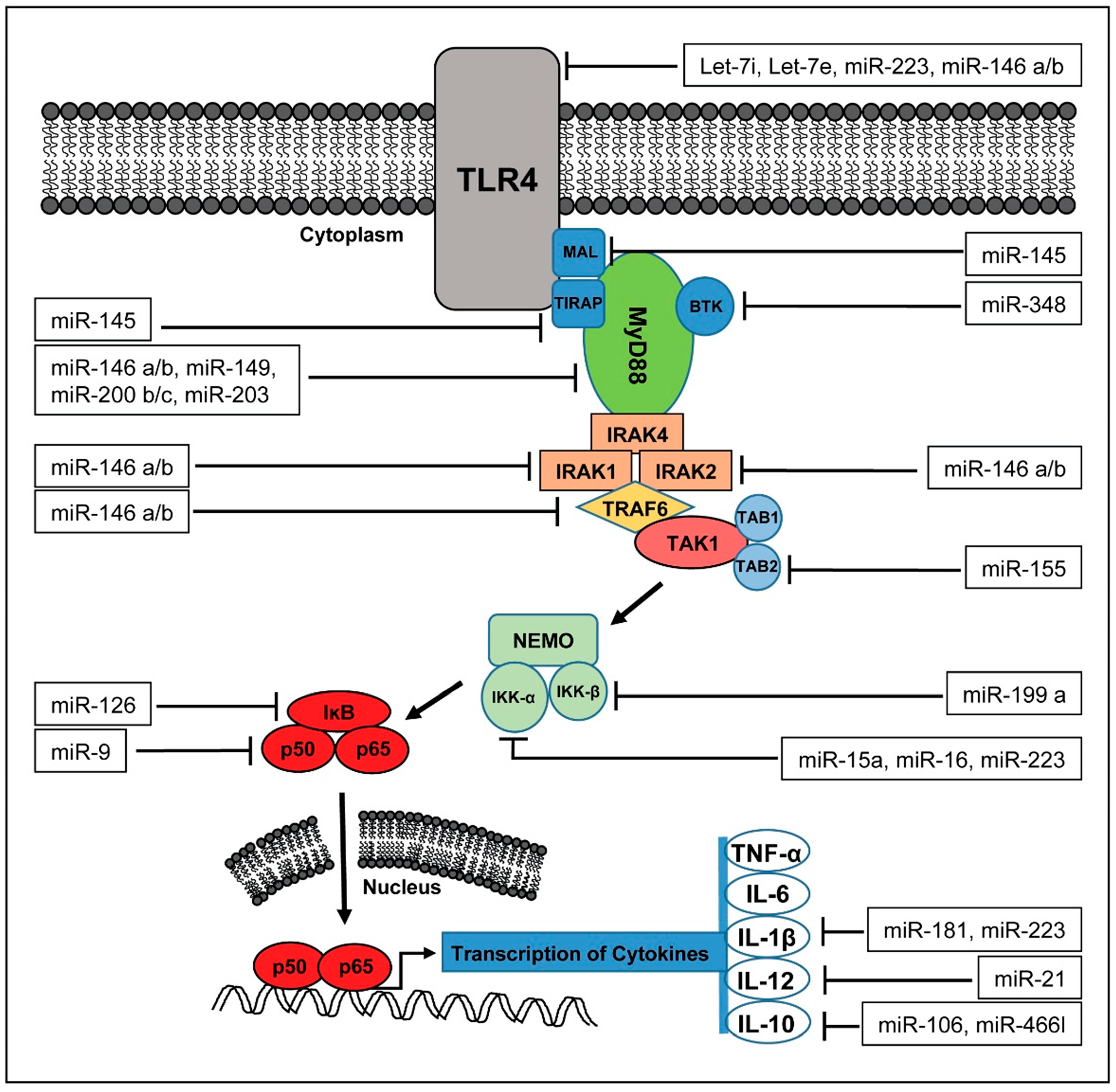
4. miRNAs in the Pathophysiology of Sepsis
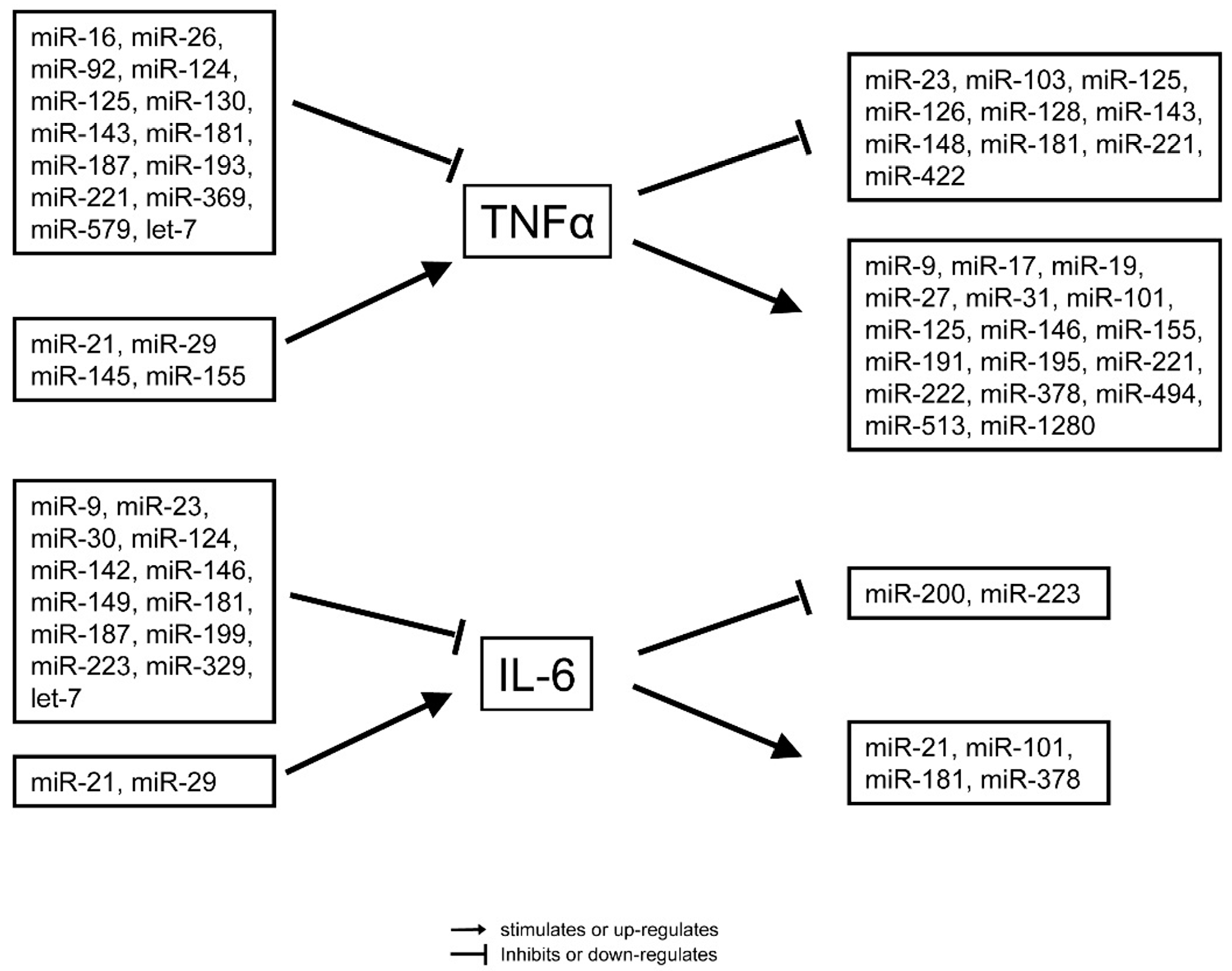
5. Circulating miRNAs as Biomarkers for Sepsis
| Reference | Year | Study Population | Analyzed miRNAs | Results |
|---|---|---|---|---|
| [58] | 2009 | 24 Sepsis, 32 healthy controls | miR-150, | ↓ (in Sepsis) |
| miR-182, | ↑ | |||
| miR-342-5p, | ↓ | |||
| miR-486 | ↑ | |||
| [59] | 2010 | 50 Sepsis, 30 SIRS, 20 healthy controls | miR-146a, | ↓ (and Sepsis vs. SIRS) |
| miR-223, | ↓ (and Sepsis vs. SIRS) | |||
| miR-126, | ↓ | |||
| miR-132, | ↔ | |||
| miR-155, | ↔ | |||
| let-7i | ↔ | |||
| [60] | 2012 | 12 Sepsis-Survivor und 12 Sepsis-Non-Survivor for Microarray-Scan, 66 Sepsis-Survivor und 52 Sepsis-Non-Survivor for Validation | miR-574-5p, | ↑ (in Survivor) |
| miR-297 | ↓ (in Survivor) | |||
| [61] | 2012 | 117 Sepsis-Survivor, 97 Sepsis-Non-Survivor | miR-223, | ↓ (in Non-Survivor), |
| miR-16, | ↓ | |||
| miR-15a, | ↑ | |||
| miR-15b, | ↔ | |||
| miR-122, | ↑ | |||
| miR-193b *, | ↑ | |||
| miR-483-5p, | ↑ | |||
| miR-451, | ↔ | |||
| miR-486-5p, | ↔ | |||
| miR-378, | ↔ | |||
| miR-499-5p, | ↔ | |||
| miR-206 | ↔ | |||
| [62] | 2012 | 166 Sepsis, 32 SIRS, 24 healthy controls | miR-15a, | ↑ (and Sepsis < SIRS) |
| miR-16 | ↑ (and Sepsis = SIRS) | |||
| [63] | 2012 | 123 severe Sepsis, 43 mild Sepsis, 24 healthy controls | miR-223, | ↑ (mild > severe Sepsis) |
| miR-15b, | ↑ (mild > severe) | |||
| miR-483-5p, | ↑ (mild > severe) | |||
| miR-499-5p, | ↓ (severe < mild) | |||
| miR-122, | ↓ (severe = mild) | |||
| miR-193b * | ↓ (severe = mild) | |||
| [64] | 2012 | 17 ICU patients without Sepsis, 36 with Sepsis | miR-181b | ↓ (in Sepsis) |
| [11] | 2013 | 138 ICU with Sepsis, 85 ICU without Sepsis, 76 healthy controls | miR-150 | ↓ |
| [65] | 2013 | 14 SIRS, 14 Sepsis | miR-146a | ↓ (in Sepsis) |
| [66] | 2013 | 22 Sepsis, 22 SIRS, 17 healthy controls | miR-342-3p, | ↓ (Sepsis < SIRS) |
| miR-3173-5p, | ↓ (Sepsis < SIRS) | |||
| miR-191 iso, | ↓ (SIRS = Kontrolle) | |||
| miR-150, | ↓ (Sepsis < SIRS) | |||
| miR-4772-3p, | ↑ (Sepsis = SIRS) | |||
| miR-4772-5p iso, | ↑ (Sepsis > SIRS) | |||
| miR-4772-5p | ↑ (Sepsis = SIRS) | |||
| [67] | 2013 | 3 Sepsis-Survivor, 3 Sepsis-Non-Survivor, 3 ICU-Nonsepsis | miR-466I | ↑ |
| [10] | 2014 | 138 ICU with Sepsis, 85 ICU without Sepsis, 76 healthy controls | miR-133a | ↑ |
| [68] | 2014 | 223 ICU-Patienten (138 Sepsis, 85 Non-Sepsis), 76 healthy controls | miR-122 | |
| [69] | 2014 | 232 Sepsis (106 Non-Survivor = Non-S, 126 Survivor = S), 24 healthy controls | miR-122, | ↑ (Sepsis > C, Non-S > S) |
| miR-193b *, | ↑ (Sepsis = C, Non-S > S) | |||
| miR-483-5p, | ↑ (Sepsis > C, Non-S > S) | |||
| miR-574-5p | ↑ (Sepsis > C, Non-S = S) | |||
| [70] | 2014 | 123 Sepsis (54 with coagulation disorders) (=CA) vs. 69 without coagulation disorders) (=CN) | miR-122, | ↑ (in CA) |
| miR-223, | ↔ | |||
| miR-15a, | ↔ | |||
| miR-16, | ↔ | |||
| miR-193b *, | ↔ | |||
| miR-483-5p | ↔ | |||
| [71] | 2014 | 40 children with sepsis, 20 SIRS, 15 healthy controls | miR-21, | ↔ |
| miR-125b, | ↔ | |||
| miR-132, | ↔ | |||
| miR-146a, | ↑ | |||
| miR-155, | ↔ | |||
| miR-223 | ↑ | |||
| [72] | 2015 | 137 Sepsis, 84 Non-Sepsis, 75 healthy controls | miR-223 | ↔ |
| [73] | 2015 | 40 septic shock, 29 Sepsis, 24 healthy controls | miR-150, | ↑ (Sepsis = controls) |
| miR-146a, | ↔ | |||
| miR-223 | ↔ | |||
| [74] | 2015 | 22 Urosepsis, 20 healthy controls | let-7a, | ↓ |
| miR-150, | ↓ | |||
| miR-1249, | ↔ | |||
| miR-199b-5p | ↔ | |||
| [75] | 2015 | 46 with Neonatal-Sepsis, 41 with Neonatal-Pneumonie as controls | miR-15a, | ↑ (in Sepsis) |
| miR-15b, | ↔ | |||
| miR-16, | ↑ | |||
| miR-206, | ↔ | |||
| miR-223, | ↔ | |||
| miR-378, | ↔ | |||
| miR-451 | ↔ | |||
| [76] | 2015 | 70 Sepsis, 30 SIRS | miR-21, | ↔ |
| miR-25, | ↓ (in Sepsis) | |||
| miR-203, | ↔ | |||
| miR-423-5p, | ↔ | |||
| miR-513a-5p, | ↔ | |||
| miR-503 | ↔ |
5.1. miR-25
5.2. miR-122
5.3. miR-133a
5.4. miR-150
5.5. miR-223
5.6. miR-297 and miR-574-5p
5.7. miR-4772
5.8. Other miRNAs
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Danai, P.; Martin, G.S. Epidemiology of sepsis: Recent advances. Curr. Infect. Dis. Rep. 2005, 7, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.L. Management of sepsis in the critically ill patient: Key aspects. Expert Opin. Pharmacother. 2006, 7, 2037–2045. [Google Scholar] [CrossRef] [PubMed]
- Angus, D.C.; van der Poll, T. Severe sepsis and septic shock. N. Engl. J. Med. 2013, 369, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.M.; Artigas, A.; Phillips, G.S.; Rhodes, A.; Beale, R.; Osbor, T.; Vincent, J.-L.; Townsend, S.; Lemeshow, S.; Dellinger, R.P. Outcomes of the Surviving Sepsis Campaign in intensive care units in the USA and Europe: A prospective cohort study. Lancet Infect. Dis. 2012, 12, 919–924. [Google Scholar] [CrossRef]
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving sepsis campaign: International guidelines for management of severe sepsis and septic shock: 2012. Intensive Care Med. 2013, 41, 580–637. [Google Scholar] [CrossRef] [PubMed]
- Rivers, E.; Nguyen, B.; Havstad, S.; Ressler, J.; Muzzin, A.; Knoblich, B.; Peterson, E.; Tomlanovich, M. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N. Engl. J. Med. 2001, 345, 1368–1377. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Bloos, F.; Reinhart, K. Rapid diagnosis of sepsis. Virulence 2014, 5, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Pierrakos, C.; Vincent, J.L. Sepsis biomarkers: A review. Crit. Care 2010, 14, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Tacke, F.; Roderburg, C.; Benz, F.; Cardenas, D.V.; Luedde, M.; Hippe, H.J.; Frey, N.; Vucur, M.; Gautheron, J.; Koch, A.; et al. Levels of circulating miR-133a are elevated in sepsis and predict mortality in critically ill patients. Crit. Care Med. 2014, 42, 1096–1104. [Google Scholar] [CrossRef] [PubMed]
- Roderburg, C.; Luedde, M.; Vargas Cardenas, D.; Vucur, M.; Scholten, D.; Frey, N.; Koch, A.; Trautwein, C.; Tacke, F.; Luedde, T. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS ONE 2013, 8, e54612. [Google Scholar] [CrossRef] [PubMed]
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar]
- Gabay, C.; Kushner, I. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 1999, 340, 448–454. [Google Scholar] [PubMed]
- Barati, M.; Alinejad, F.; Bahar, M.A.; Tabrisi, M.S.; Shamshiri, A.R.; Bodouhi, N.O.; Karimi, H. Comparison of WBC, ESR, CRP and PCT serum levels in septic and non-septic burn cases. Burns 2008, 34, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Povoa, P.; Coelho, L.; Almeida, E.; Fernandes, A.; Mealha, R.; Moreira, P.; Sabino, H. C-reactive protein as a marker of infection in critically ill patients. Clin. Microbiol. Infect. 2005, 11, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Schmit, X.; Vincent, J.L. The time course of blood C-reactive protein concentrations in relation to the response to initial antimicrobial therapy in patients with sepsis. Infection 2008, 36, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Muller, B.; White, J.C.; Nylen, E.S.; Snider, R.H.; Becker, K.L.; Habener, J.F. Ubiquitous expression of the calcitonin-i gene in multiple tissues in response to sepsis. J. Clin. Endocrinol. Metab. 2001, 86, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Gauvin, F.; Amre, D.K.; Saint-Louis, P.; Lacroix, J. Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: A systematic review and meta-analysis. Clin. Infect. Dis. 2004, 39, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Kopterides, P.; Siempos, II.; Tsangaris, I.; Tsantes, A.; Armaganidis, A. Procalcitonin-guided algorithms of antibiotic therapy in the intensive care unit: A systematic review and meta-analysis of randomized controlled trials. Crit. Care Med. 2010, 38, 2229–2241. [Google Scholar] [CrossRef] [PubMed]
- Selberg, O.; Hecker, H.; Martin, M.; Klos, A.; Bautsch, W.; Kohl, J. Discrimination of sepsis and systemic inflammatory response syndrome by determination of circulating plasma concentrations of procalcitonin, protein complement 3a, and interleukin-6. Crit. Care Med. 2000, 28, 2793–2798. [Google Scholar] [CrossRef] [PubMed]
- Harbarth, S.; Harbarth, S.; Holeckova, K.; Froidevaux, C.; Pittet, D.; Ricou, B.; Grau, G.E.; Vadas, L.; Pugin, J. Diagnostic value of procalcitonin, interleukin-6, and interleukin-8 in critically ill patients admitted with suspected sepsis. Am. J. Respir. Crit. Care Med. 2001, 164, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Pathan, N.; Hemingway, C.A.; Alizadeh, A.A.; Stephens, A.C.; Boldrick, J.C.; Oragui, E.E.; McCabe, C.; Welch, S.B.; Whitney, A.; O’Gara, P.; et al. Role of interleukin 6 in myocardial dysfunction of meningococcal septic shock. Lancet 2004, 363, 203–209. [Google Scholar] [CrossRef]
- Jawa, R.S.; Anillo, S.; Huntoon, K.; Baumann, H.; Kulaylat, M. Analytic review: Interleukin-6 in surgery, trauma, and critical care: Part I: Basic science. J. Intensive Care Med. 2011, 26, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Jawa, R.S.; Anillo, S.; Huntoon, K.; Baumann, H.; Kulaylat, M. Interleukin-6 in surgery, trauma, and critical care part II: Clinical implications. J. Intensive Care Med. 2011, 26, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Samraj, R.S.; Zingarelli, B.; Wong, H.R. Role of biomarkers in sepsis care. Shock 2013, 40, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, N.I.; Trzeciak, S.; Hollander, J.E.; Birkhahn, R.; Otero, R.; Osborn, T.M.; Moretti, E.; Nguyen, H.B.; Gunnerson, K.J.; Milzman, D.; et al. A prospective, multicenter derivation of a biomarker panel to assess risk of organ dysfunction, shock, and death in emergency department patients with suspected sepsis. Crit. Care Med. 2009, 37, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- miRBase: The microRNA Database. Available online: http://www.mirbase.org/ (accessed on 23 November 2015).
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, S.; Pfeffer, S.; Baumert, T.F.; Zeisel, M.B. miR-122—A key factor and therapeutic target in liver disease. J. Hepatol. 2015, 62, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, S.; Ding, J.; Zhao, Y.; Liang, L.; Liu, T.; Zhan, R.; He, X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene 2010, 29, 2302–2308. [Google Scholar] [CrossRef] [PubMed]
- Roderburg, C.; Luedde, T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J. Hepatol. 2014, 61, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Remaley, A.T. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 2012, 23, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. MicroRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.H.; Liang, Y.B.; Tang, H.; Chen, Z.B.; Li, Z.Y.; Hu, X.C.; Ma, Z.F. Dexamethasone down-regulates the expression of microRNA-155 in the livers of septic mice. PLoS ONE 2013, 8, e80547. [Google Scholar] [CrossRef] [PubMed]
- Puimege, L.; van Hauwermeiren, F.; Steeland, S.; Van Ryckeghem, S.; Vandewalle, J.; Lodens, S.; Dejager, L.; Vandevyver, S.; Staelens, J.; Timmermans, S.; et al. Glucocorticoid-induced microRNA-511 protects against TNF by down-regulating TNFR1. EMBO Mol. Med. 2015, 7, 1004–1017. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, H.; Ono, S.; Efron, P.A.; Scumpia, P.O.; Moldawer, L.L.; Mochizuki, H. Role of Toll-like receptors in the development of sepsis. Shock 2008, 29, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K.; Roth, J.; Oppenheim, J.J.; Tracey, K.J.; Vogl, T.; Feldmann, M.; Horwood, N.; Nanchahal, J. Alarmins: Awaiting a clinical response. J. Clin. Investig. 2012, 122, 2711–2719. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Juarez, G.A.; de Haan, B.J.; Faas, M.M.; de Vos, P. The role of pathogen-associated molecular patterns in inflammatory responses against alginate based microcapsules. J. Control. Release 2013, 172, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Avlas, O.; Fallach, R.; Shainberg, A.; Porat, E.; Hochhauser, E. Toll-like receptor 4 stimulation initiates an inflammatory response that decreases cardiomyocyte contractility. Antioxid. Redox Signal. 2011, 15, 1895–1909. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.G.; Yang, J.; Zheng, Y.; Jin, Y. MiR-15a/16 regulates macrophage phagocytosis after bacterial infection. J. Immunol. 2014, 193, 4558–4567. [Google Scholar] [CrossRef] [PubMed]
- El Gazzar, M.; Church, A.; Liu, T.; McCall, C.E. MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-α during TLR4-induced gene reprogramming. J. Leukoc. Biol. 2011, 90, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, T.; Wu, B.; Liu, H.; Luo, J.; Feng, D.; Shi, Y. STAT1 regulates MD-2 expression in monocytes of sepsis via miR-30a. Inflammation 2014, 37, 1903–1911. [Google Scholar] [CrossRef] [PubMed]
- Bosmann, M.; Ward, P.A. The inflammatory response in sepsis. Trends Immunol. 2013, 34, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, C.; Rossi, S.; Shimizu, M.; Tudor, S.; Veronese, A.; Ferracin, M.; Nicoloso, M.S.; Barbarotto, E.; Popa, M.; Stanciulea, O.; et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS ONE 2009, 4, e7405. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Yu, M.L.; Yu, G.; Bian, J.J.; Deng, X.M.; Wan, X.J.; Zhu, K.M. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 2010, 394, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, K.; Chen, W.; Feng, D.; Jia, Y.; Xie, L. Serum miR-574-5p: a prognostic predictor of sepsis patients. Shock 2012, 37, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, P.; Chen, W.; Feng, D.; Jia, Y.; Xie, L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients' mortality: A prospective observational study. PLoS ONE 2012, 7, e38885. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, P.; Chen, W.; Feng, D.; Jia, Y.; Xie, L.X. Evidence for serum miR-15a and miR-16 levels as biomarkers that distinguish sepsis from systemic inflammatory response syndrome in human subjects. Clin. Chem. Lab. Med. 2012, 50, 1423–1428. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Zhang, P.J.; Chen, W.J.; Feng, D.; Jia, Y.H.; Xie, L.X. Four serum microRNAs identified as diagnostic biomarkers of sepsis. J. Trauma Acute Care Surg. 2012, 73, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Icli, B.; Wara, A.K.; Belkin, N.; He, S.; Kobzik, L.; Hunninghake, G.M.; Vera, M.P.; Blackwell, T.S.; Baron, R.M.; et al. MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J. Clin. Investig. 2012, 122, 1973–1990. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, H.C.; Chen, C.; Zeng, J.; Wang, Q.; Zheng, L.; Yu, H.D. Differential expression of plasma miR-146a in sepsis patients compared with non-sepsis-SIRS patients. Exp. Ther. Med. 2013, 5, 1101–1104. [Google Scholar] [PubMed]
- Ma, Y.; Vilanova, D.; Atalar, K.; Delfour, O.; Edgeworth, J.; Ostermann, M.; Hernandez-Fuentes, M.; Razafimahatratra, S.; Michot, B.; Persing, D.H.; et al. Genome-wide sequencing of cellular microRNAs identifies a combinatorial expression signature diagnostic of sepsis. PLoS ONE 2013, 8, e75918. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dalli, J.; Chiang, N.; Baron, R.M.; Quintana, C.; Serhan, C.N. Plasticity of leukocytic exudates in resolving acute inflammation is regulated by MicroRNA and proresolving mediators. Immunity 2013, 39, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Roderburg, C.; Benz, F.; Vargas Cardenas, D.; Koch, A.; Janssen, J.; Vucur, M.; Gautheron, J.; Schneider, A.T.; Koppe, C.; Kreggenwinkel, K.; et al. Elevated miR-122 serum levels are an independent marker of liver injury in inflammatory diseases. Liver Int. 2015, 35, 1172–1184. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yu, B.; Deng, J.; Jin, Y.; Xie, L. Serum miR-122 correlates with short-term mortality in sepsis patients. Trauma 2014, 25, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Deng, J.; Wang, J.Y.; Zhang, P.J.; Xin, Z.; Xiao, K.; Feng, D.; Jia, Y.H.; Liu, Y.N.; Xie, L.X. Serum miR-122 levels are related to coagulation disorders in sepsis patients. Clin. Chem. Lab. Med. 2014, 52, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, C.; He, Y.; Li, Q.; Wang, G.; Wen, P.; Yang, W.; Yang, Y. [Relationship between expression of microRNA and inflammatory cytokines plasma level in pediatric patients with sepsis]. Zhonghua Er Ke Za Zhi 2014, 52, 28–33. (In Chinese) [Google Scholar] [PubMed]
- Benz, F.; Tacke, F.; Luedde, M.; Trautwein, C.; Luedde, T.; Koch, A.; Roderburg, C. Circulating microRNA-223 serum levels do not predict sepsis or survival in patients with critical illness. Dis. Markers. 2015, 2015, 384208. [Google Scholar] [CrossRef] [PubMed]
- Puskarich, M.A.; Nandi, U.; Shapiro, N.I.; Trzeciak, S.; Kline, J.A.; Jones, A.E. Detection of microRNAs in patients with sepsis. J. Acute Dis. 2015, 4, 101–106. [Google Scholar] [CrossRef]
- How, C.K.; Hou, S.K.; Shih, H.C.; Huang, M.S.; Chiou, S.H.; Lee, C.H.; Juan, C.C. Expression profile of MicroRNAs in gram-negative bacterial sepsis. Shock 2015, 43, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, X.; Liu, X.; Wang, X.; Xu, J.; Hou, S.; Zhang, X.; Ding, Y. miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. Int. J. Clin. Exp. Med. 2015, 8, 5683–5690. [Google Scholar] [PubMed]
- Yao, L.; Liu, Z.; Zhu, J.; Li, B.; Chai, C.; Tian, Y. Clinical evaluation of circulating microRNA-25 level change in sepsis and its potential relationship with oxidative stress. Int. J. Clin. Exp. Pathol. 2015, 8, 7675–7684. [Google Scholar] [PubMed]
- Fry, R.C.; Rager, J.E.; Bauer, R.; Sebastian, E.; Peden, D.B.; Jaspers, I.; Alexis, N.E. Air toxics and epigenetic effects: ozone altered microRNAs in the sputum of human subjects. Am. J. Physiol. Lung Cell Mol. Physiol. 2014, 306, L1129–L1137. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.; Akerstrom, T.; Rinnov, A.; Yfanti, C.; Scheele, C.; Pedersen, B.K.; Laye, M.J. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS ONE 2014, 9, e87308. [Google Scholar] [CrossRef] [PubMed]
- Bihrer, V.; Friedrich-Rust, M.; Kronenberger, B.; Forestier, N.; Haupenthal, J.; Shi, Y.; Peveling-Oberhag, J.; Radeke, H.H.; Sarrazin, C.; Herrmann, E.; et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am. J. Gastroenterol. 2011, 106, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Waidmann, O.; Bihrer, V.; Pleli, T.; Farnik, H.; Berger, A.; Zeuzem, S.; Kronenberger, B.; Piiper, A. Serum microRNA-122 levels in different groups of patients with chronic hepatitis B virus infection. J. Viral Hepat. 2012, 19, 1365–2893. [Google Scholar] [CrossRef] [PubMed]
- Koberle, V.; Kronenberger, B.; Pleli, T.; Trojan, J.; Imelmann, E.; Peveling-Oberhag, J.; Welker, M.W.; Elhendawy, M.; Zeuzem, S.; Piiper, A.; et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur. J. Cancer 2013, 49, 3442–3449. [Google Scholar] [CrossRef] [PubMed]
- Waidmann, O.; Koberle, V.; Brunner, F.; Zeuzem, S.; Piiper, A.; Kronenberger, B. Serum microRNA-122 predicts survival in patients with liver cirrhosis. PLoS ONE 2012, 7, e45652. [Google Scholar] [CrossRef] [PubMed]
- Trebicka, J.; Anadol, E.; Elfimova, N.; Strack, I.; Roggendorf, M.; Viazov, S.; Wedemeyer, I.; Drebber, U.; Rockstroh, J.; Sauerbruch, T.; et al. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J. Hepatol. 2013, 58, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Mitchelson, K.R.; Qin, W.Y. Roles of the canonical myomiRs miR-1, -133 and -206 in cell development and disease. World J. Biol. Chem. 2015, 6, 162–208. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lu, Y.; Li, Z.; Wang, Q. MicroRNA-133: Expression, function and therapeutic potential in muscle diseases and cancer. Curr. Drug Targets 2014, 15, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Davidson-Moncada, J.; Papavasiliou, F.N.; Tam, W. MicroRNAs of the immune system: Roles in inflammation and cancer. Ann. N. Y. Acad. Sci. 2010, 2010, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, W.M.; Spiel, A.O.; Jilma, B.; Wolzt, M.; Muller, M. In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem. Biophys. Res. Commun. 2009, 380, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chaudhry, H.; Zhong, Y.; Ali, M.M.; Perkins, L.A.; Owens, W.B.; Morales, J.E.; McGuire, F.R.; Zumbrun, E.E.; Zhang, J.; et al. Dysregulation in microRNA expression in peripheral blood mononuclear cells of sepsis patients is associated with immunopathology. Cytokine 2015, 71, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Haneklaus, M.; Gerlic, M.; O'Neill, L.A.; Masters, S.L. MiR-223: Infection, inflammation and cancer. J. Intern. Med. 2013, 274, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Fasseu, M.; Treton, X.; Guichard, C.; Pedruzzi, E.; Cazals-Hatem, D.; Richard, C.; Aparicio, T.; Daniel, F.; Soule, J.C.; Moreau, R.; et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS ONE 2010, 5, e13160. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.T.; Chen, S.Y.; Wang, C.R.; Liu, M.F.; Lin, C.C.; Jou, I.M.; Shiau, A.L.; Wu, C.L. Brief report: Amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis Rheum. 2013, 64, 3240–3245. [Google Scholar] [CrossRef] [PubMed]
- Essandoh, K.; Fan, G.C. Role of extracellular and intracellular microRNAs in sepsis. Biochim. Biophys. Acta 2014, 1842, 2155–2162. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, W.; Yang, Y.; Wang, Y.; Peng, T.; Chang, J.; Caldwell, C.C.; Zingarelli, B.; Fan, G.C. Loss of duplexmiR-223 (5p and 3p) aggravates myocardial depression and mortality in polymicrobial sepsis. Biochim. Biophys. Acta 2014, 2014, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Benz, F.; Roderburg, C.; Vargas Cardenas, D.; Vucur, M.; Gautheron, J.; Koch, A.; Zimmermann, H.; Janssen, J.; Nieuwenhuijsen, L.; Luedde, M.; et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp. Mol. Med. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Yang, J.C.; Rau, C.S.; Chen, Y.C.; Lu, T.H.; Lin, M.W.; Tzeng, S.L.; Wu, Y.C.; Wu, C.J.; Hsieh, C.H. Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. PLoS ONE 2013, 8, e77936. [Google Scholar] [CrossRef] [PubMed]
- Petrocca, F.; Visone, R.; Onelli, M.R.; Shah, M.H.; Nicoloso, M.S.; de Martino, I.; Iliopoulos, D.; Pilozzi, E.; Liu, C.G.; Negrini, M.; et al. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008, 13, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Cossart, P.; Lebreton, A. A trip in the “New Microbiology” with the bacterial pathogen Listeria monocytogenes. FEBS Lett. 2014, 588, 2437–2445. [Google Scholar] [CrossRef] [PubMed]
- Furci, L.; Schena, E.; Miotto, P.; Cirillo, D.M. Alteration of human macrophages microRNA expression profile upon infection with Mycobacterium tuberculosis. Int. J. Mycobacteriol. 2013, 2, 128–134. [Google Scholar] [CrossRef]
- Davis, M.A.; Lim, J.Y.; Soyer, Y.; Harbottle, H.; Chang, Y.F.; New, D.; Orfe, L.H.; Besser, T.E.; Call, D.R. Development and validation of a resistance and virulence gene microarray targeting Escherichia coli and Salmonella enterica. J. Microbiol. Methods 2010, 82, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Ibeagha-Awemu, E.M.; Liang, G.; Beaudoin, F.; Zhao, X.; Guan le, L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Chen, D.S.; Wu, Y.Q.; Xu, X.J.; Zhang, H.; Chen, C.F.; Chen, H.C.; Liu, Z.F. MicroRNA expression profile in RAW264.7 cells in response to Brucella melitensis infection. Int. J. Biol. Sci. 2012, 8, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, X.; Ye, Y.; Zhao, K.; Zhuang, Y.; Li, Y.; Wei, Y.; Wu, M. MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.L.; Gao, J.X.; Zou, C.G.; Ma, Y.C.; Zhang, K.Q. miR-233 modulates the unfolded protein response in C. elegans during Pseudomonas aeruginosa infection. PLoS Pathog. 2015, 11, e1004606. [Google Scholar] [CrossRef] [PubMed]
- Gong, A.Y.; Gong, A.Y.; Zhou, R.; Hu, G.; Liu, J.; Sosnowska, D.; Drescher, K.M.; Dong, H.; Chen, X.M. Cryptosporidium parvum induces B7-H1 expression in cholangiocytes by down-regulating microRNA-513. J. Infect. Dis. 2010, 201, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; Splinter, P.L.; O'Hara, S.P.; LaRusso, N.F. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J. Biol. Chem. 2007, 282, 28929–28938. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Hu, G.; Zhou, R.; Liu, J.; Gong, A.Y.; Eischeid, A.N.; Dittman, J.W.; Chen, X.M. MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J. Immunol. 2009, 183, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Geraci, N.S.; Tan, J.C.; McDowell, M.A. Characterization of microRNA expression profiles in Leishmania-infected human phagocytes. Parasite Immunol. 2015, 37, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Chamnanchanunt, S.; Kuroki, C.; Desakorn, V.; Enomoto, M.; Thanachartwet, V.; Sahassananda, D.; Sattabongkot, J.; Jenwithisuk, R.; Fucharoen, S.; Svasti, S.; et al. Downregulation of plasma miR-451 and miR-16 in Plasmodium vivax infection. Exp. Parasitol. 2015, 155, 19–25. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benz, F.; Roy, S.; Trautwein, C.; Roderburg, C.; Luedde, T. Circulating MicroRNAs as Biomarkers for Sepsis. Int. J. Mol. Sci. 2016, 17, 78. https://doi.org/10.3390/ijms17010078
Benz F, Roy S, Trautwein C, Roderburg C, Luedde T. Circulating MicroRNAs as Biomarkers for Sepsis. International Journal of Molecular Sciences. 2016; 17(1):78. https://doi.org/10.3390/ijms17010078
Chicago/Turabian StyleBenz, Fabian, Sanchari Roy, Christian Trautwein, Christoph Roderburg, and Tom Luedde. 2016. "Circulating MicroRNAs as Biomarkers for Sepsis" International Journal of Molecular Sciences 17, no. 1: 78. https://doi.org/10.3390/ijms17010078
APA StyleBenz, F., Roy, S., Trautwein, C., Roderburg, C., & Luedde, T. (2016). Circulating MicroRNAs as Biomarkers for Sepsis. International Journal of Molecular Sciences, 17(1), 78. https://doi.org/10.3390/ijms17010078






