Eosinophil-Derived Neurotoxin (EDN/RNase 2) and the Mouse Eosinophil-Associated RNases (mEars): Expanding Roles in Promoting Host Defense
Abstract
:1. Introduction
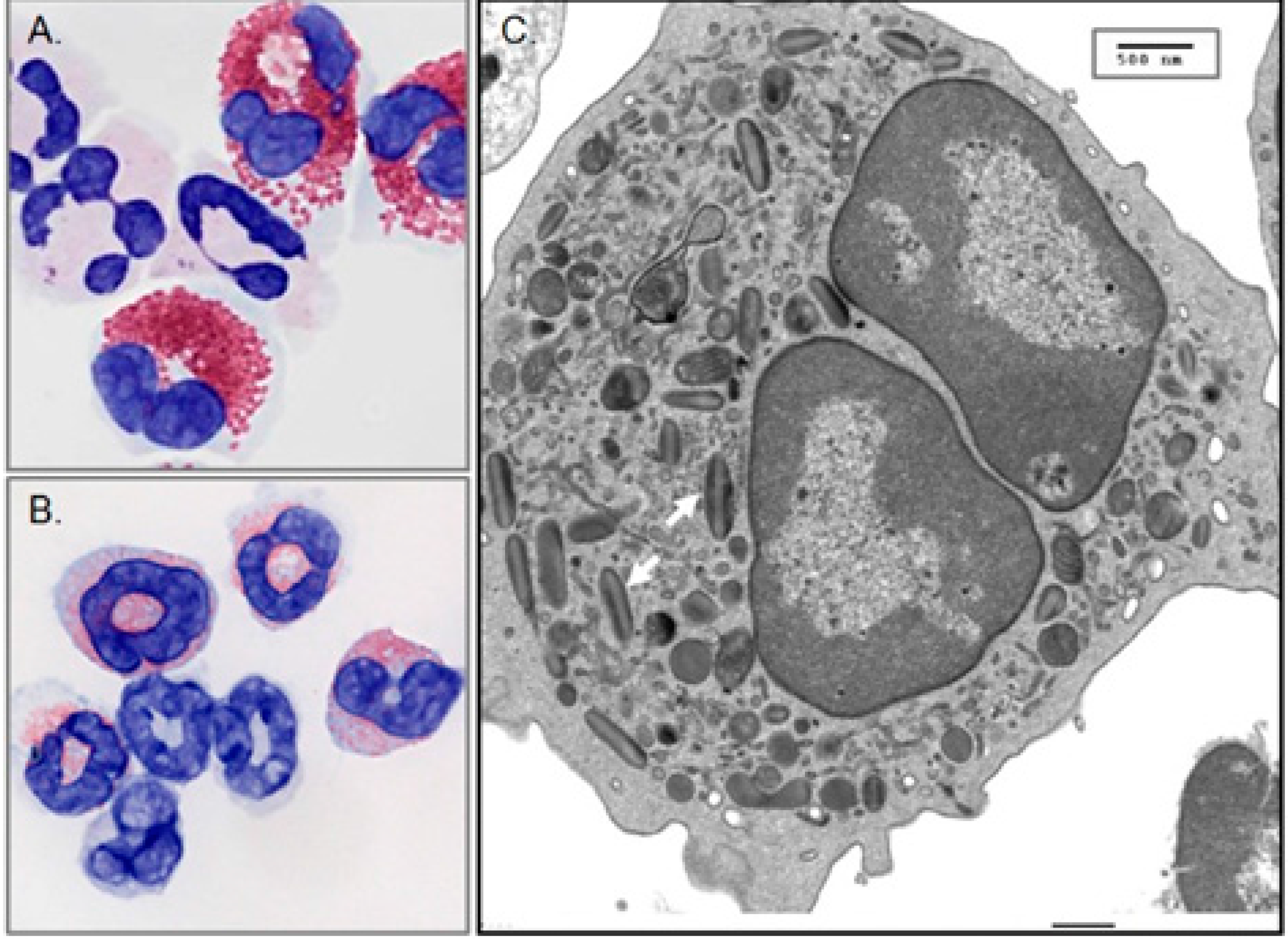
2. Discovery, Isolation and Characterization of EDN
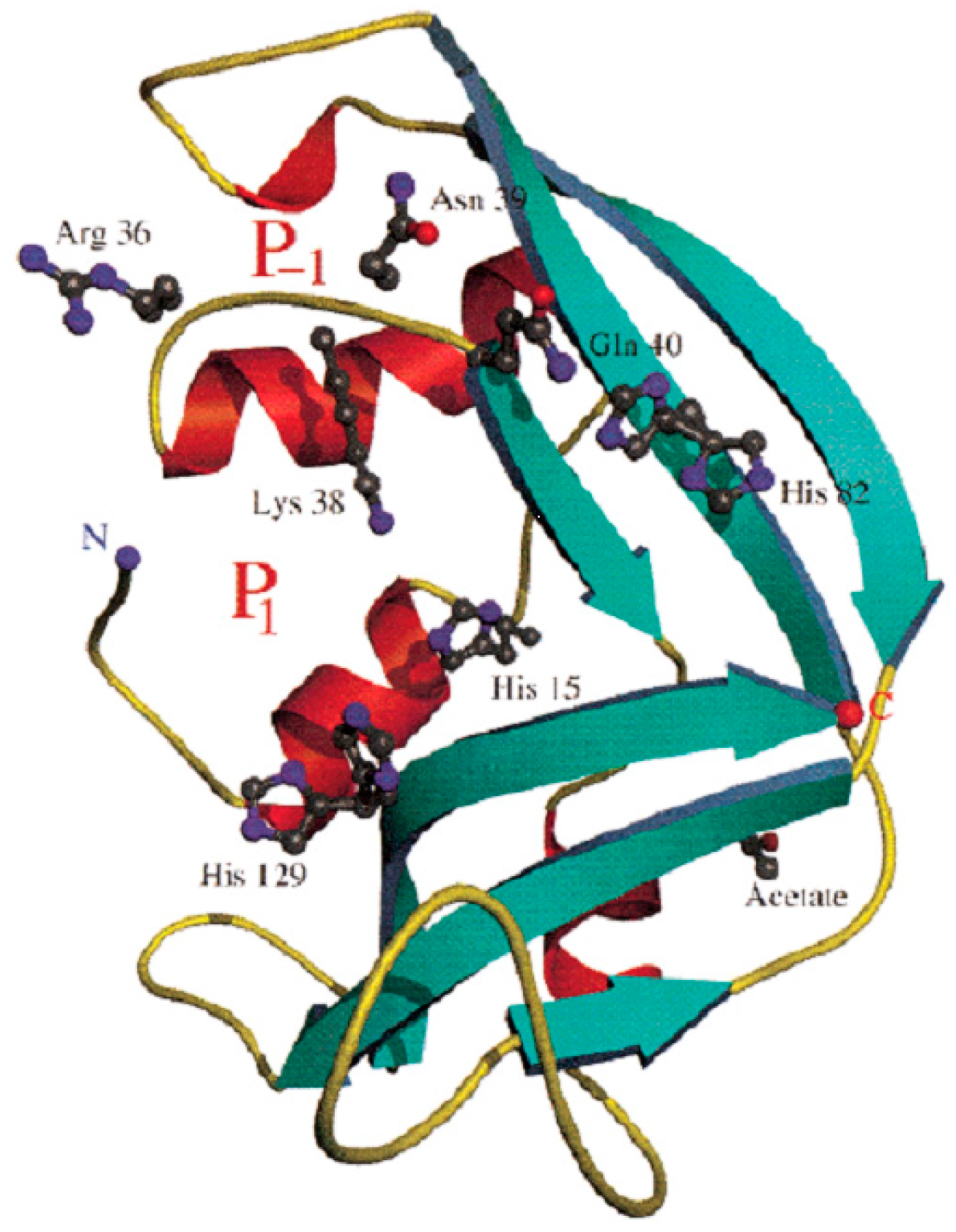
3. EDN Is Member of the RNase A Family of Secretory Ribonucleases
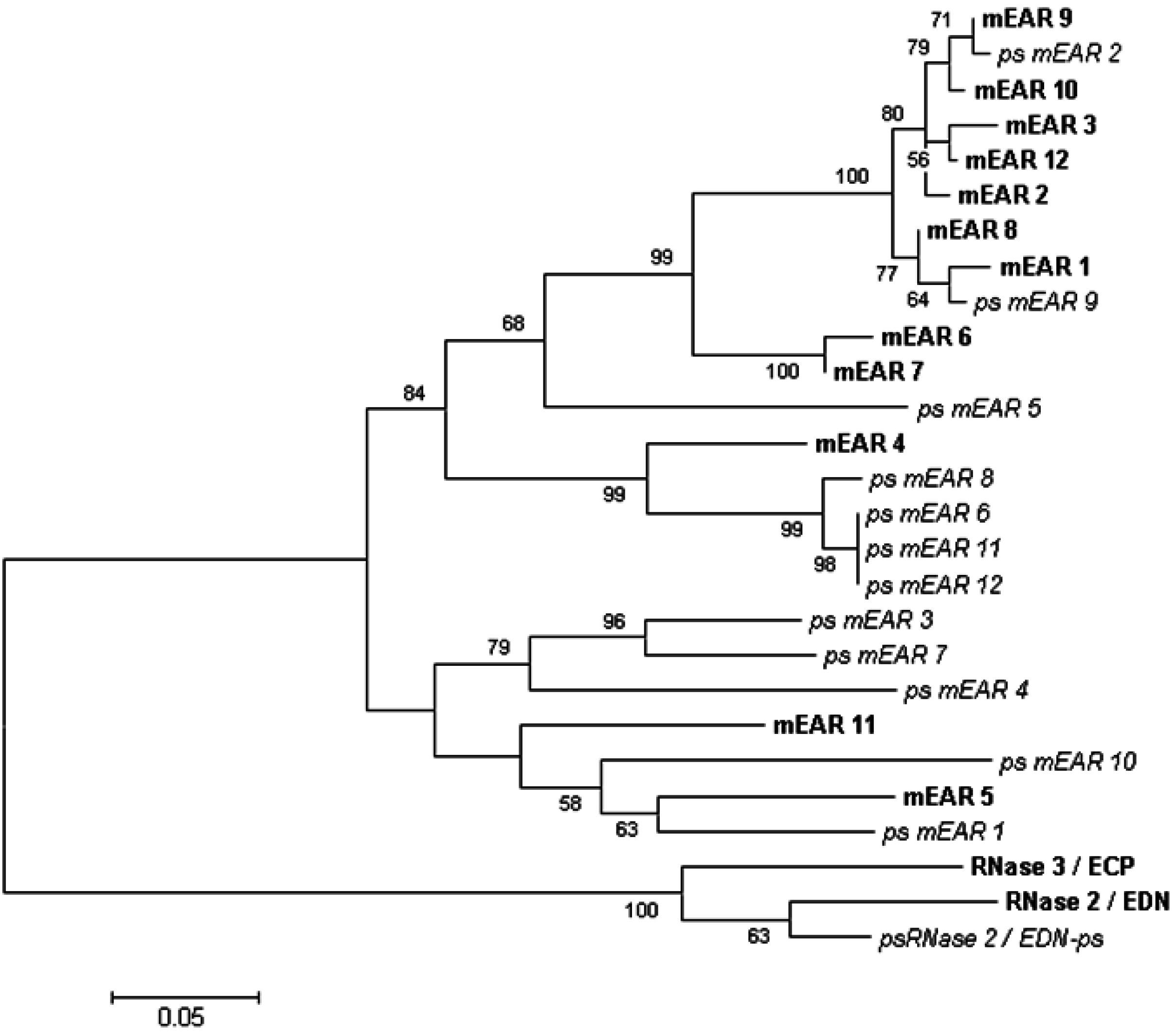
4. Genetics and Diversity of Human EDN and the Mouse Eosinophil-Associated Ribonucleases
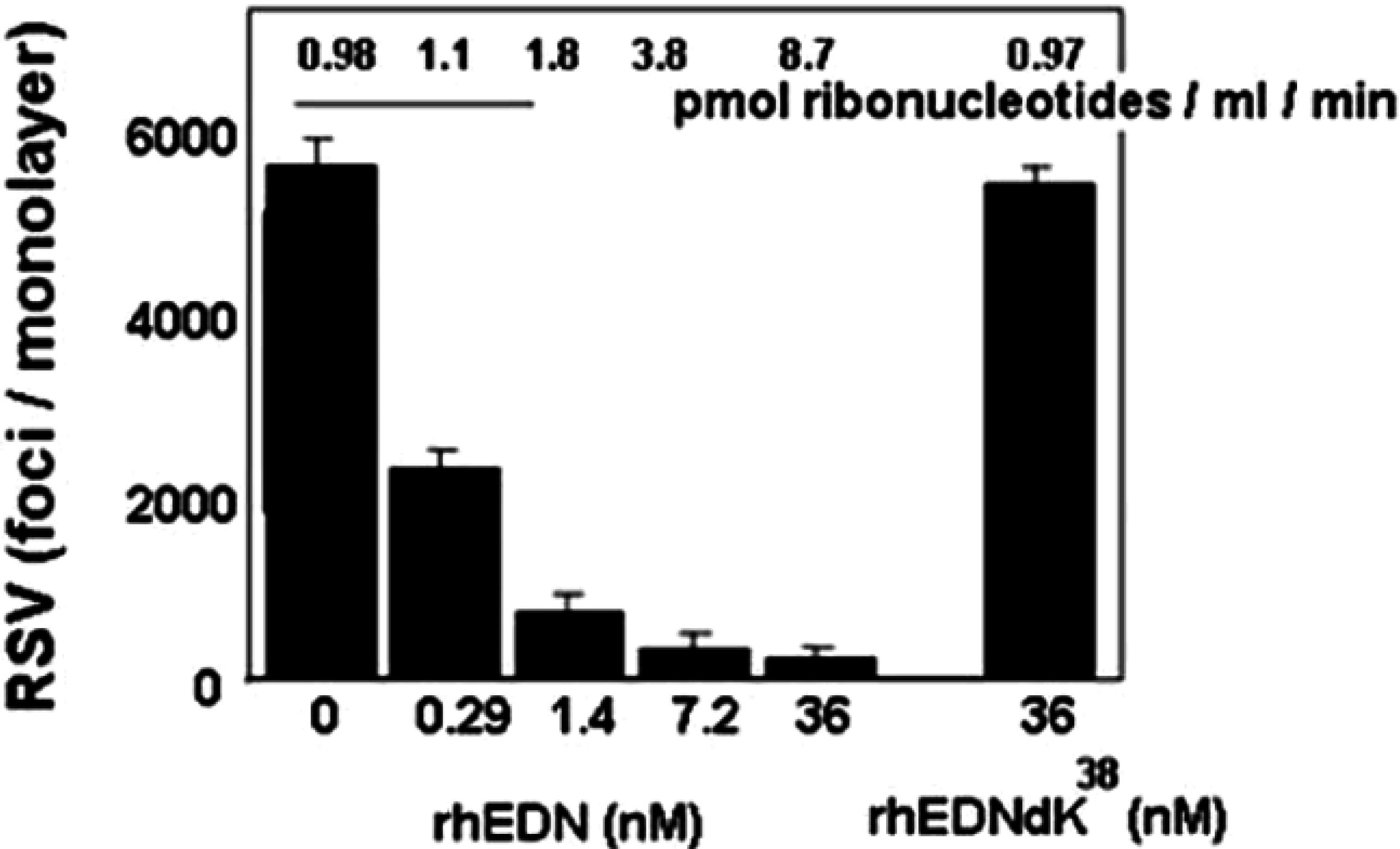
5. EDN and mEars Target Virus Infectivity
6. EDN as a Fusion RNase to Target Hepatitis B Virus (HBV)
7. Eosinophils Interact with Bacteria and Release EDN
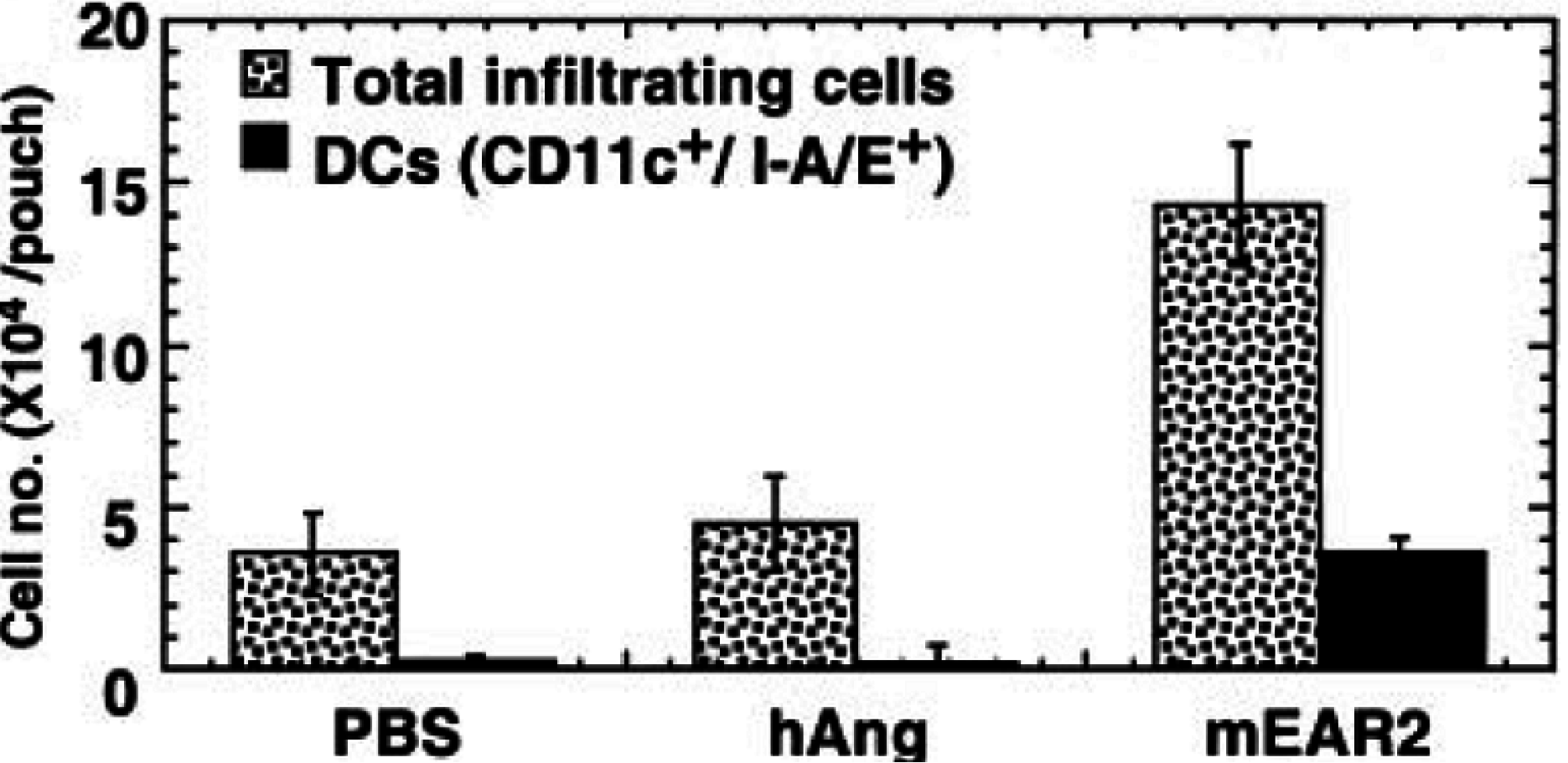
8. EDN and mEar 2 Interact with Dendritic Cells
9. Mouse Ear 6 and Schistosoma mansoni Infection
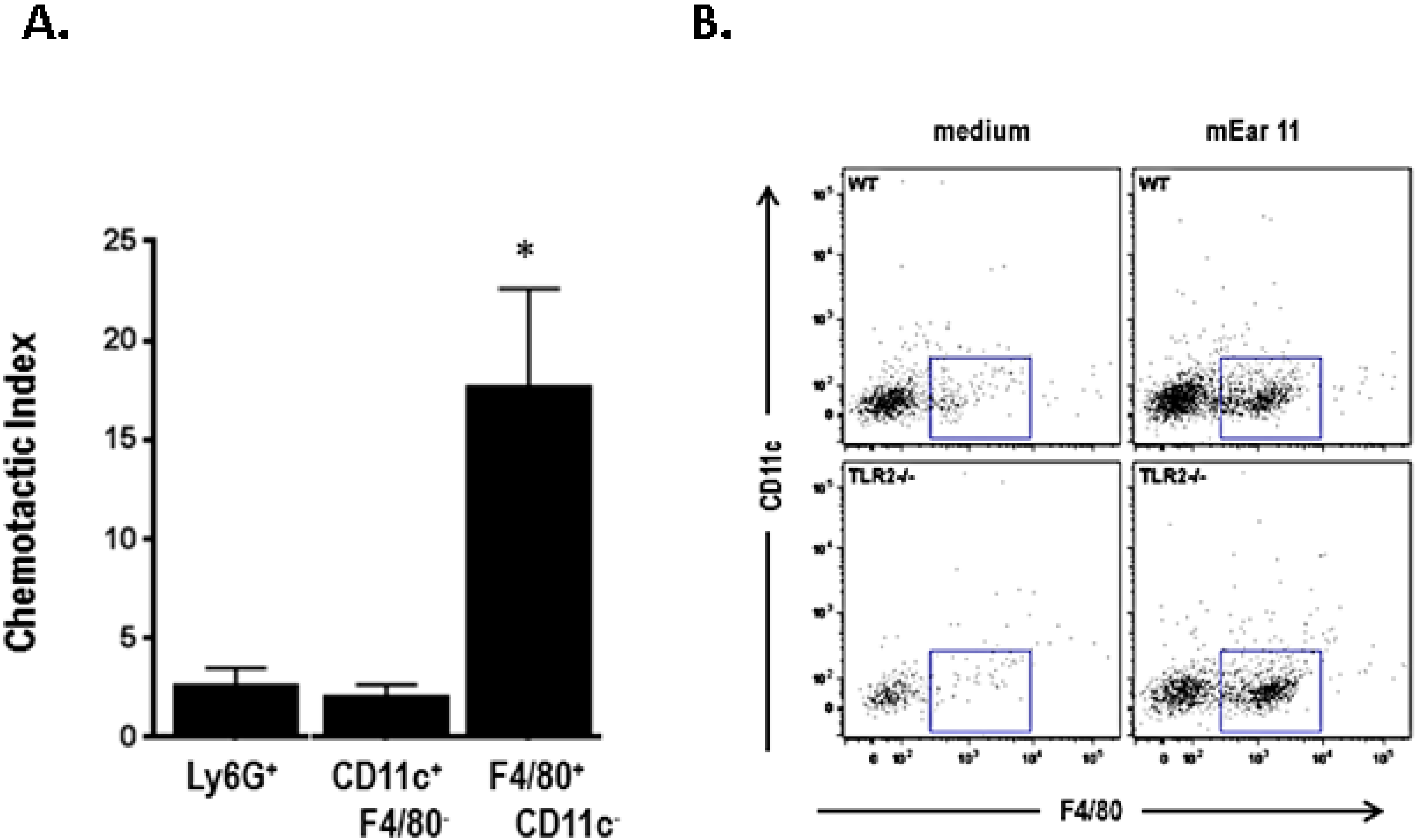
10. Mouse Ear 11 Is Expressed in Response to IL-4, IL-13, and IL-33 and Is a Macrophage Chemoattractant
11. Future Directions
Acknowledgments
Conflicts of Interest
References
- Rosenberg, H.F.; Dyer, K.D.; Foster, P.S. Eosinophils: Changing perspectives in health and disease. Nat. Rev. Immunol. 2013, 13, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Lacy, P.; Moqbel, R. Chapter 7.6: Signaling and degranulation. In Eosinophils in Health and Disease; Lee, J.J., Rosenberg, H.F., Eds.; Elsevier Press: Waltham, MA, USA, 2013; pp. 206–219. [Google Scholar]
- Abu-Ghazaleh, R.I.; Dunnette, S.L.; Loegering, D.A.; Checkel, J.L.; Kita, H.; Thomas, L.L.; Gleich, G.J. Eosinophil granule proteins in peripheral blood granulocytes. J. Leukoc. Biol. 1992, 52, 611–618. [Google Scholar] [PubMed]
- Shin, S.W.; Park, J.S.; Park, C.S. Elevation of Eosinophil-Derived neurotoxin in plasma of subjects with aspirin-exacerbated respiratory disease: A possible peripheral blood protein biomarker. PLoS ONE 2013, 8, e66644. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.K. Eosinophil-derived neurotoxin: A novel biomarker for diagnosis and monitoring of asthma. Korean J. Pediatr. 2013, 56, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.K.; Callaway, Z.; Fletcher, R.; Koh, Y.Y. Eosinophil-derived neurotoxin in childhood asthma: Correlation with disease severity. J. Asthma 2010, 47, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Badar, A.; Hussain, M.M.; Saeed, W.; Aslam, M. Correlation of eosinophil-derived neurotoxin with airway resistance in asthmatics. J. Pak. Med. Assoc. 2010, 60, 97–101. [Google Scholar] [PubMed]
- Kalach, N.; Kapel, N.; Waligora-Dupriet, A.J.; Castelain, M.C.; Cousin, M.O.; Sauvage, C.; Ba, F.; Nicolis, I.; Campeotto, F.; Butel, M.J.; et al. Intestinal permeability and fecal eosinophl-derived neurotoxin are the best diagnosis tools for digestive non-IgE mediated cow’s milk allergy in toddlers. Clin. Chem. Lab. Med. 2013, 51, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Kephart, G.M.; Alexander, J.A.; Arora, A.S.; Romero, Y.; Smyrk, T.C.; Talley, N.J.; Kita, H. Marked deposition of eosinophil-derived neurotoxin in adult patients with eosinophilic esophagitis. Am. J. Gastorenterol. 2010, 105, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Pennings, J.L.; Schuurhof, A.; Hodemaekers, H.M.; Buisman, A.; de Rond, L.C.; Widjojoatmodjo, M.N.; Luytjes, W.; Kimpen, J.L.; Bont, L.; Janssen, R. Systemic signature of the lung response to respiratory syncytial virus infection. PLoS ONE 2011, 6, e21461. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F. Eosinophil-derived neurotoxin/RNase 2: Connecting the past, the present and the future. Curr. Pharm. Biotechnol. 2008, 9, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Batra, J.K. Antimicrobial activity of human eosinophil granule proteins: Involvement in host defence against pathogens. Crit. Rev. Microbiol. 2012, 38, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Acharya, K.R.; Ackerman, S.J. Eosinophil granule proteins: Form and function. J. Biol. Chem. 2014, 289, 17406–17415. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Jacobsen, E.A.; Ochkur, S.I.; McGarry, M.P.; Condjella, R.M.; Doyle, A.D.; Luo, H.; Zellner, K.R.; Prothroe, C.A.; Willetts, L.; et al. Human versus mouse eosinophils: “That which we call an eosinophil, by any other name would stain as red”. J. Allergy Clin. Immunol. 2012, 130, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Haigh, B.J.; Griffin, F.J.; Wheeler, T.T. The mammalian secreted RNases: Mechanisms of action in host defence. Innate Immun. 2013, 19, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.H. Remarks on Hodgkin’s Disease: A pathogenic agent in the glands, and its application in diagnosis. Br. Med. J. 1933, 1, 641–644. [Google Scholar] [CrossRef] [PubMed]
- Durack, D.T.; Sumi, S.M.; Klebanoff, S.J. Neurotoxicity of human eosinophils. Proc. Natl. Acad. Sci. USA 1979, 76, 1443–1447. [Google Scholar] [CrossRef] [PubMed]
- Durack, D.T.; Ackerman, S.J.; Loegering, D.A.; Gleich, G.J. Purification of human eosinophil-derived neurotoxin. Proc. Natl. Acad. Sci. USA 1981, 78, 5165–5169. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.G.; Venge, P. Purification and characterization of a new cationic protein—Eosinophil protein-X (EPX)-from granules of human eosinophils. Immunology 1983, 50, 19–26. [Google Scholar] [PubMed]
- Sorrentino, S.; Glitz, D.G.; Hamann, K.J.; Loegering, D.A.; Checkel, J.L.; Gleich, G.J. Eosinophil-derived neurotoxin and human liver ribonuclease. Identity of structure and linkage of neurotoxicity to nuclease activity. J. Biol. Chem. 1992, 267, 14859–14865. [Google Scholar] [PubMed]
- Gleich, G.J.; Loegering, D.A.; Bell, M.P.; Checkel, J.L.; Ackerman, S.J.; McKean, D.J. Biochemical and functional similarities between eosinophil-derived neurotoxin and eosinophil cationic protein: Homology with ribonuclease. Proc. Natl. Acad. Sci. USA 1986, 83, 3146–3150. [Google Scholar] [CrossRef] [PubMed]
- Slifman, N.R.; Loegering, D.A.; McKean, D.J.; Gleich, G.J. Ribonuclease activity associated with human eosinophil-derived neurotoxin and eosinophil cationic protein. J. Immunol. 1986, 137, 2913–2917. [Google Scholar] [PubMed]
- Rosenberg, H.F.; Tenen, D.G.; Ackerman, S.J. Molecular cloning of the human eosinophil-derived neurotoxin: A member of the ribonuclease gene family. Proc. Natl. Acad. Sci. USA 1989, 86, 4460–4464. [Google Scholar] [CrossRef] [PubMed]
- Hamann, K.J.; Barker, R.L.; Loegering, D.A.; Pease, L.R.; Gleich, G.J. Sequence of human eosinophil-derived neurotoxin cDNA: Identity of deduced amino acid sequence with human nonsecretory ribonucleases. Gene 1989, 83, 161–167. [Google Scholar] [CrossRef]
- Simanski, M.; Koten, B.; Schroder, J.M.; Glaser, R.; Harder, J. Antimicrobial RNases in cutaneous defense. J. Innate Immun. 2012, 4, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F. RNase A ribonucleases and host defense: An evolving story. J. Leukoc. Biol. 2008, 83, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Dyer, K.D.; Rosenberg, H.F. The RNase A superfamily: Generation of diversity and innate host defense. Mol. Divers. 2006, 10, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Beintema, J.J.; Zhang, J. The ribonuclease A superfamily of mammals and birds: Identifying new members and tracing evolutionary histories. Genomics 2005, 85, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Goo, S.M.; Cho, S. The expansion and functional diversification of the mammalian ribonuclease A superfamily epitomizes the efficiency of multigene families at generating biological novelty. Genome Biol. Evol. 2013, 5, 2124–2140. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, G.J.; Holloway, D.E.; Veluraja, K.; Acharya, K.R. Atomic resolution (0.98A) structure of eosinophil-derived neurotoxin. Biochemistry 2002, 41, 3341–3352. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D.; Tiffany, H.L.; Gonzalez, M. Rapid evolution of a unique family of primate ribonuclease genes. Nat. Genet. 1995, 10, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Rosenberg, H.F.; Nei, M. Positive Darwinian selection after gene duplicaton in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 1998, 95, 3708–3713. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D. Eosinophil cationic protein and eosinophil-derived neurotoxin. Evolution of novel function in a primate ribonuclease gene family. J. Biol. Chem. 1995, 270, 21539–21544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Rosenberg, H.F. Complementary advantageous substitutions in the evolution of an antiviral RNase of higher primates. Proc. Natl. Acad. Sci. USA 2002, 99, 5486–5491. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D. Diversity among the primate eosinophil-derived neurotoxin genes: A specific C-terminal sequence is necessary for enhanced ribonuclease activity. Nucleic Acids Res. 1997, 25, 3532–3536. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; D’Alessio, G. The success of the RNase scaffold in the advance of biosciences and in evolution. Gene 2007, 406, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Benito, A.; Laurents, D.V.; Ribo, M.; Vilanova, M. The structural determinants that lead to the formation of particular oligomeric structures in the pancreatic-type ribonuclease family. Curr. Protein Pept. Sci. 2008, 9, 370–393. [Google Scholar] [CrossRef] [PubMed]
- Podlaha, O.; Zhang, J. Processed pseudogenes: The fossilized footprings of past gene expression. Trends Genet. 2009, 25, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Trabesinger-Ruef, N.; Jermann, T.; Zankel, T.; Durrant, B.; Frank, G.; Benner, S.A. Pseudogenes in ribonuclease evolution: A source of new biomacromolecular function? FEBS Lett. 1996, 382, 319–322. [Google Scholar] [CrossRef]
- Zhang, J.; Rosenberg, H.F. Sequence variation at two eosinophil-associated ribonuclease loci in humans. Genetics 2000, 156, 1949–1958. [Google Scholar] [PubMed]
- Blom, K.; Rubin, J.; Halfvarson, J.; Torkvist, L.; Ronnblom, A.; Sangfelt, P.; Lordal, M.; Jonsson, U.-B.; Sjoqvist, U.; Hakansson, L.D.; et al. Eosinophil associated genes in the inflammatory bowel disease 4 region: Correlation to inflammatory bowel disease revealed. World J. Gastroenterol. 2012, 18, 6409–6419. [Google Scholar] [CrossRef] [PubMed]
- Larson, K.A.; Olson, E.V.; Madden, B.J.; Gleich, G.J.; Lee, N.A.; Lee, J.J. Two highly homologous ribonuclease genes expressed in mouse eosinophils identify a larger subgroup of the mammalian ribonuclease superfamily. Proc. Natl. Acad. Sci. USA 1996, 93, 12370–12375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dyer, K.D.; Rosenberg, H.F. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl. Acad. Sci. USA 2000, 97, 4701–4706. [Google Scholar] [CrossRef] [PubMed]
- Singhania, N.A.; Dyer, K.D.; Zhang, J.; Deming, M.S.; Bonville, C.A.; Domachowske, J.B.; Rosenberg, H.F. Rapid evolution of the ribonuclease A superfamily: Adaptive expansion of independent gene clusters in rats and mice. J. Mol. Evol. 1999, 49, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Shamri, R.; Melo, R.C.; Young, K.M.; Bivas-Benita, M.; Xenakis, J.J.; Spencer, L.A.; Weller, P.F. CCL11 elicits secretion of RNases from mouse eosinophils and their cell-free granules. FASEB J. 2012, 26, 2084–2093. [Google Scholar] [CrossRef] [PubMed]
- Shamri, R.; Young, K.M.; Weller, P.F. PI3K, ERK, p38 MAPK and integrins regulate CCR3-mediated secretion of mouse and human eosinophil-associated RNaes. Allergy 2013, 68, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Pulido, D.; Moussaoui, M.; Nogues, M.V.; Torrent, M.; Boix, E. Towards the rational design of antimicrobial proteins: Single points mutations can switch on bactericidal and agglutinating activities on the RNase A superfamily lineages. FEBS J. 2013, 280, 5841–5852. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.J.; Johnston, S.L. The role of viruses in acute exacerbations of asthma. J. Allergy Clin. Immunol. 2010, 125, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Green, R.M.; Custovic, A.; Sanderson, G.; Hunter, J.; Johnston, S.L.; Woodcock, A. Synergism between allergens and viruses and risk of hospital admission with asthma: Case control study. BMJ 2002, 324, 763. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.K.; Foster, P.S.; Rosenberg, H.F. Respiratory viral infection, epithelial cytokines and innate lymphoid cells in asthma exacerbations. J. Leukoc. Biol. 2014, 96, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Phipps, S.; Lam, C.E.; Mahalingam, S.; Newhouse, M.; Ramirez, R.; Rosenberg, H.F.; Foster, P.S.; Matthaei, K.I. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood 2007, 110, 1578–1586. [Google Scholar] [CrossRef] [PubMed]
- Percopo, C.M.; Dyer, K.D.; Ochkur, S.I.; Luo, J.L.; Fischer, E.R.; Lee, J.J.; Lee, N.A.; Domachowske, J.B.; Rosenberg, H.F. Activated mouse eosinophils protect against lethal respiratory virus infection. Blood 2014, 123, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Domachowske, J.B.; Dyer, K.D.; Bonville, C.A.; Rosenberg, H.F. Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. J. Infect. Dis. 1998, 177, 1458–1464. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D.; Domachowske, J.B. Respiratory viruses and eosinophils: Exploring the connections. Antivir. Res. 2009, 83, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rugeles, M.T.; Trubey, C.M.; Bedoya, V.I.; Pinto, L.A.; Oppenheim, J.J.; Rybak, S.M.; Shearer, G.M. Ribonuclease is partly repsonsible for the HIV-1 inhibitory effect activated by HLA alloantigen recognition. AIDS 2003, 17, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Bedoya, V.I.; Boasso, A.; Hardy, A.W.; Rybak, S.; Shearer, G.M.; Rugeles, M.T. Ribonucleases in HIV type I inhibition: Effect of recombinant RNases on infection of primary T cells and immune activation-induced RNase gene and protein expression. AIDS Res. Hum. Retroviruses 2006, 22, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Moreau, J.M.; Dyer, K.D.; Bonville, C.A.; Nitto, T.; Vasquez, N.L.; Easton, A.J.; Domachowske, J.B.; Rosenberg, H.F. Diminished expression of an antiviral ribonuclease in response to pneumovirus infection in vivo. Antivir. Res. 2003, 59, 181–191. [Google Scholar] [CrossRef]
- Gaudreault, E.; Gosselin, J. Leukotriene B4-mediated release of antimicrobial peptides against cytomegalovirus is BLT1 dependent. Viral Immunol. 2007, 20, 407–420. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, M.A.; Yee, M.; Buczynski, B.W.; Vitiello, P.F.; Keng, P.C.; Welle, S.L.; Finkelstein, J.N.; Dean, D.A.; Lawrence, B.P. Neonatala oxygen increases sensitivity to influenza A virus infection in adult mice by suppressing epithelial expression of Ear 1. Am. J. Pathol. 2012, 181, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.H.; Xue, C.F.; Ding, J.; Gong, W.D.; Zhao, Y.; Huang, Y.X. Targeted ribonuclease can inhibit replication of hepatitis B virus. World J. Gastroenterol. 2003, 9, 295–299. [Google Scholar] [PubMed]
- Li, Y.; Zhao, Y.; Liu, J.; Huang, Y.; Liu, Z.; Xue, C. A promising alternative anti-HBV agent: The targeted ribonuclease. Int. J. Mol. Med. 2010, 26, 51–56. [Google Scholar] [PubMed]
- Kvarnhammar, A.M.; Cardell, L.O. Pattern recognition receptors in human eosinophils. Immunology 2012, 136, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Szklarek, D.; Barton, A.; Ganz, T.; Hamann, K.J.; Gleich, G.J. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J. Immunol. 1989, 142, 4428–4434. [Google Scholar] [PubMed]
- Ackerman, S.J.; Gleich, G.J.; Logering, D.A.; Richardson, B.A.; Butterworth, A.E. Comparative toxicity of purified human eosinophil granule cationic proteins for schistosomula of Schistosoma mansoni. Am. J. Trop. Med. Hyg. 1985, 34, 735–745. [Google Scholar] [PubMed]
- Hosoki, K.; Nakamura, A.; Nagao, M.; Hiraguchi, Y.; Tokuda, R.; Wada, H.; Nobori, T.; Fujisawa, T. Differential activation of eosinophils by “probiotic” Bifidiobacterium bifidum and “pathogenic” Clostridium difficle. Int. Arch. Allergy Immunol. 2010, 152 (Suppl. 1), 83–89. [Google Scholar] [CrossRef] [PubMed]
- Hosoki, K.; Nakamura, A.; Kainuma, K.; Sugimoto, M.; Nagao, M.; Hiraguchi, Y.; Tanida, H.; Tokuda, R.; Wada, H.; Nobori, T.; et al. Differential activation of eosinophils by bacteria associated with asthma. Int. Arch. Allergy Immunol. 2013, 161 (Suppl. 2), 16–22. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Rosenberg, H.F.; Chen, Q.; Dyer, K.D.; Kurosaka, K.; Oppenheim, J.J. Eosinophil-derived neurotoxin (EDN), an antimicrobial protein with chemotactic activities for dendritic cells. Blood 2003, 102, 3396–3403. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, Q.; Rosenberg, H.F.; Rybak, S.M.; Newton, D.L.; Wang, Z.Y.; Fu, Q.; Tchernev, V.T.; Wang, M.; Schweitzer, B.; et al. Human ribonuclease A superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induced dendritic cell maturation and activation. J. Immunol. 2004, 173, 6134–6142. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, Q.; Su, S.B.; Zhang, P.; Kurosaka, K.; Caspi, R.R.; Michalek, S.M.; Rosenberg, H.F.; Zhang, N.; Oppenheim, J.J. Eosinophil-derived neurotixin acts as an alarmin to activated the TLR2-MyD88 signal pathway in dendritic cells and enhances the Th2 immune responses. J. Exp. Med. 2008, 205, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.E. DAMPs, PAMPs, and alarmins: All we need to know about danger. J. Leukoc. Biol. 2007, 81, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Botos, I.; Segal, D.M.; Davies, D.R. The structural biology of Toll-like receptors. Structure 2011, 19, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.J.; Barker, T.; Dyer, K.D.; Rice, T.A.; Percopo, C.M.; Garcia-Crespo, K.E.; Cho, S.; Lee, J.J.; Druey, K.M.; Rosenberg, H.F. Eosinophil-associated ribonuclease 11 is a macrophage chemoattractant. J. Biol. Chem. 2015, 290, 8863–8875. [Google Scholar] [CrossRef] [PubMed]
- Nitto, T.; Dyer, K.D.; Mejia, R.A.; Bystrom, J.; Wynn, T.A.; Rosenberg, H.F. Characterization of the divergent eosinophil ribonuclease, mEar 6, and its expression in response to Schistosoma mansoni infection in vivo. Genes Immun. 2004, 5, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Cormier, S.A.; Yuan, S.; Crosby, J.R.; Protheroe, C.A.; Dimina, D.M.; Hines, E.M.; Lee, N.A.; Lee, J.J. T(H)2-mediated pulmonary inflammation leads to the differential expression of ribonuclease genes by alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 2002, 27, 678–687. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosenberg, H.F. Eosinophil-Derived Neurotoxin (EDN/RNase 2) and the Mouse Eosinophil-Associated RNases (mEars): Expanding Roles in Promoting Host Defense. Int. J. Mol. Sci. 2015, 16, 15442-15455. https://doi.org/10.3390/ijms160715442
Rosenberg HF. Eosinophil-Derived Neurotoxin (EDN/RNase 2) and the Mouse Eosinophil-Associated RNases (mEars): Expanding Roles in Promoting Host Defense. International Journal of Molecular Sciences. 2015; 16(7):15442-15455. https://doi.org/10.3390/ijms160715442
Chicago/Turabian StyleRosenberg, Helene F. 2015. "Eosinophil-Derived Neurotoxin (EDN/RNase 2) and the Mouse Eosinophil-Associated RNases (mEars): Expanding Roles in Promoting Host Defense" International Journal of Molecular Sciences 16, no. 7: 15442-15455. https://doi.org/10.3390/ijms160715442
APA StyleRosenberg, H. F. (2015). Eosinophil-Derived Neurotoxin (EDN/RNase 2) and the Mouse Eosinophil-Associated RNases (mEars): Expanding Roles in Promoting Host Defense. International Journal of Molecular Sciences, 16(7), 15442-15455. https://doi.org/10.3390/ijms160715442




