Proteomic Analysis of Immature Fraxinus mandshurica Cotyledon Tissues during Somatic Embryogenesis: Effects of Explant Browning on Somatic Embryogenesis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Separation of Proteins
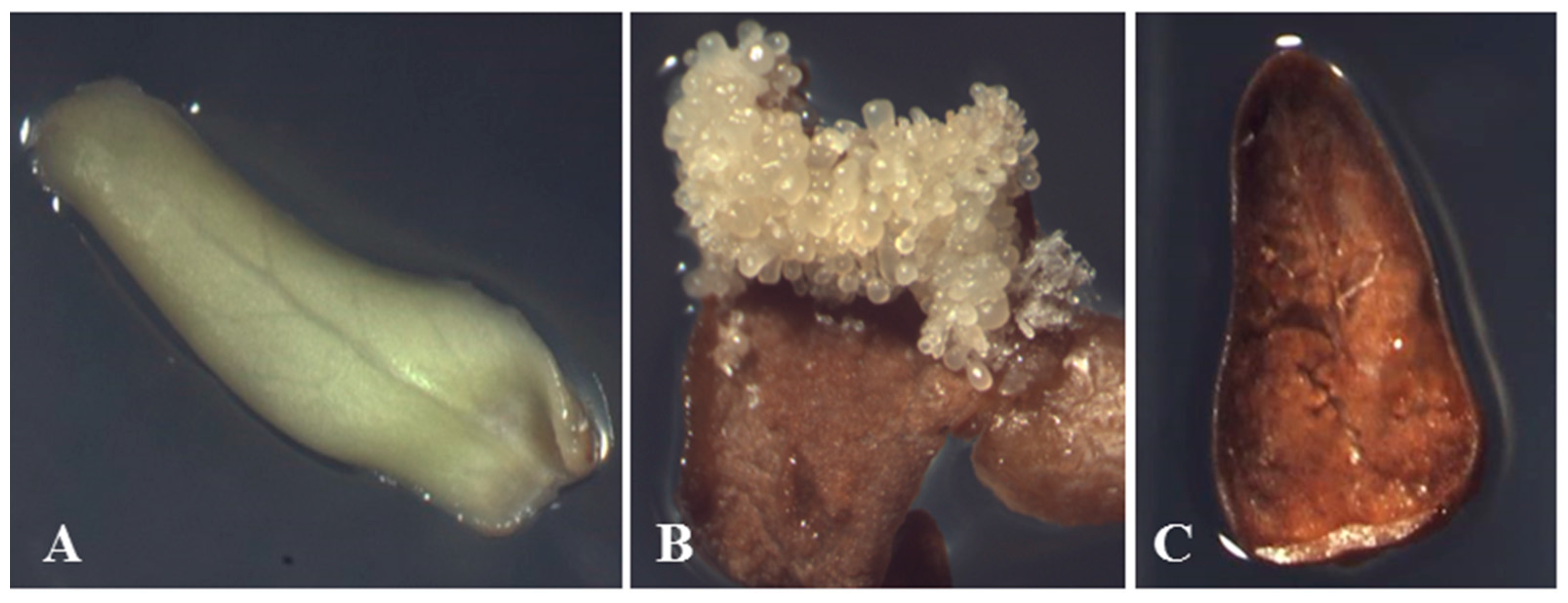

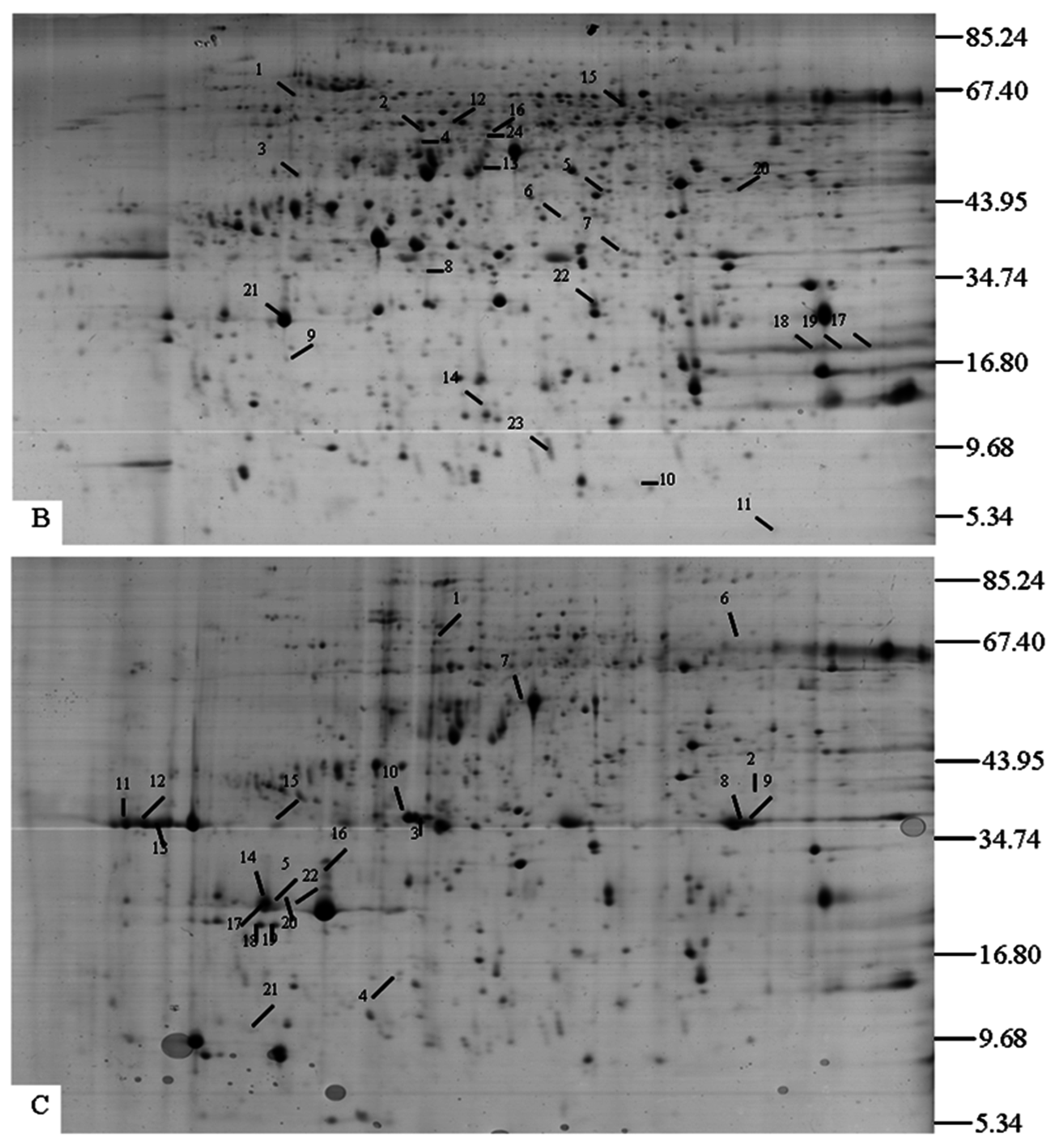
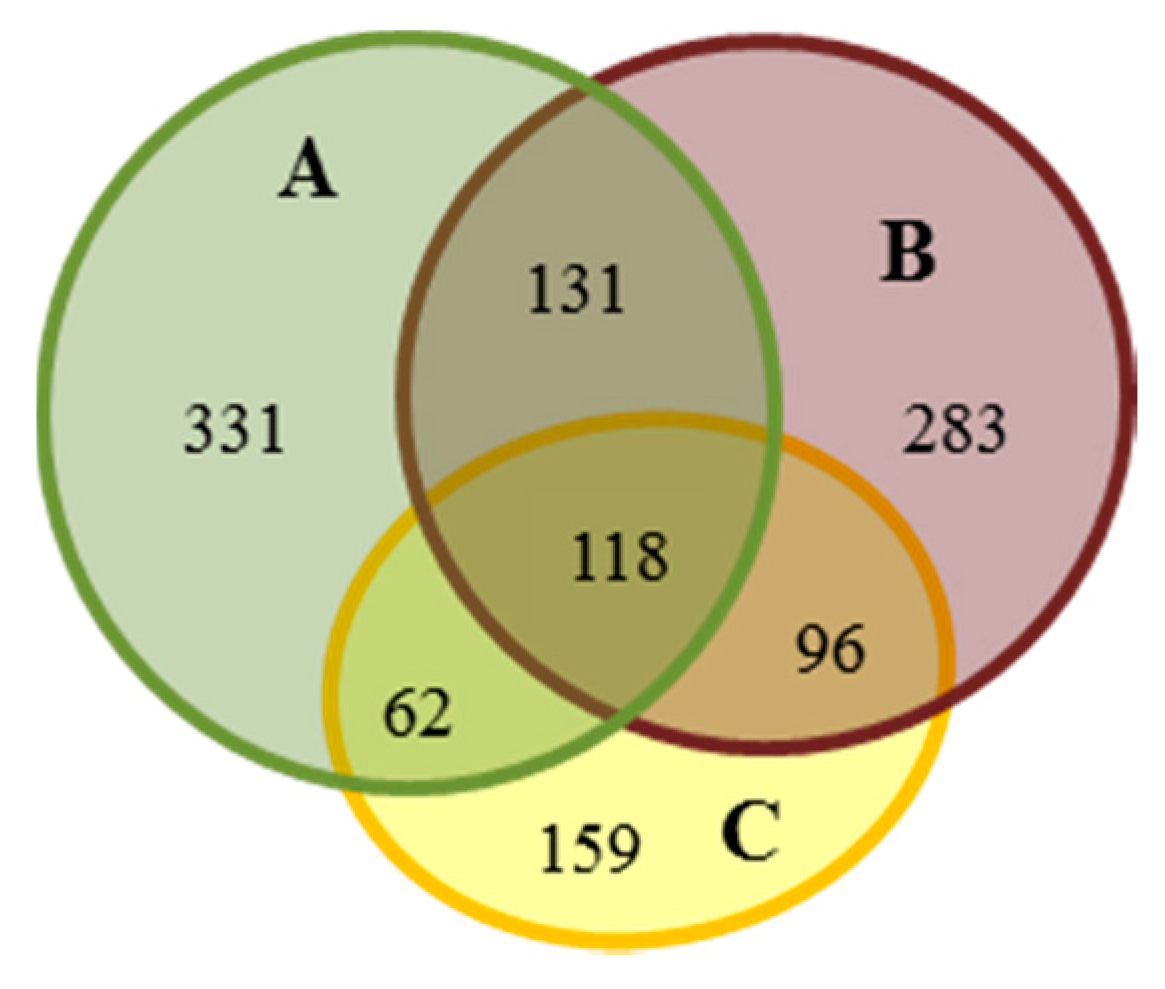
| Compared Gels | A | B | C | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Specially | Differentially | Specially | Differentially | Specially | Differentially | |||||||
| up | mid | down | up | mid | down | up | mid | down | ||||
| A-B | 393 | 29 | 181 | 39 | 379 | 39 | 181 | 29 | - | |||
| B-C | - | 414 | 9 | 172 | 33 | 221 | 33 | 172 | 9 | |||
| A-C | 462 | 14 | 131 | 35 | - | 255 | 35 | 131 | 14 | |||
| ALL (A-B-C) | specially | differentially (A-B) | specially | differentially (B-C) | specially | differentially (A-C) | ||||||
| 331 | 15 | 86 | 17 | 283 | 8 | 93 | 17 | 159 | 19 | 88 | 11 | |
| Spots | Differentially Expression (V%) | ||
|---|---|---|---|
| A | B | C | |
| A24 | 0.735 | - | 0.088 |
| A25 | 0.304 | - | 0.077 |
| A26 | 0.062 | - | 0.240 |
| A27 | 0.040 | - | 0.084 |
| A28 | 0.065 | - | 0.254 |
| A29 | 0.076 | - | 0.500 |
| A30 | 0.025 | - | 0.103 |
| B16 | - | 0.050 | 0.013 |
| B17 | - | 0.024 | 0.110 |
| B18 | - | 0.050 | 0.217 |
| B19 | - | 0.108 | 0.031 |
| B20 | 0.009 | 0.044 | - |
| B21 | 0.171 | 1.148 | - |
| B22 | 0.016 | 0.058 | - |
| B23 | 0.133 | 0.019 | - |
| B24 | 0.794 | 0.182 | - |
| C8 | - | 0.168 | 0.305 |
| C9 | - | 0.079 | 0.277 |
| C17 | - | 0.119 | 0.333 |
| C18 | - | 0.089 | 0.217 |
| C19 | - | 0.055 | 0.223 |
| C22 | - | 0.026 | 0.156 |
2.2. Functional Classifications of Identified Proteins
| Spot a | Matched Protein | gi Number b | Organism | Mass/pI | Score c | Queries Matched d (%) | Sequence Coverage e (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| Stress response and defense (18) | |||||||||
| A2 | Molecular chaperone Hsp90-1 | 38154489 | Lycopersicon esculentum | 80.1/4.98 | 100 | 31 | 35 | ||
| A24 | Dehydrin | 18964 | Zea mays | 16.9/8.04 | 49 | 1 | 5 | ||
| A25 | Dehydrin | 18964 | Zea mays | 16.9/8.04 | 64 | 1 | 5 | ||
| A26 | Peroxidase | 1781326 | Spinacia oleracea | 38.2/6.15 | 212 | 3 | 7 | ||
| A27 | Heat shock protein, putative | 223542544 | Ricinus communis | 71.1/6.10 | 684 | 17 | 23 | ||
| B21 | Osmotin-like protein | 33340043 | Gossypium hirsutum | 26.6/7.41 | 63 | 1 | 4 | ||
| C6 | Peroxidase | 1781326 | Spinacia oleracea | 38.2/6.15 | 177 | 3 | 20 | ||
| C8 | Chitinase precursor | 5880843 | Petroselinum crispum | 29.0/6.3 | 94 | 5 | 30 | ||
| C9 | Chitinase precursor | 5880843 | Petroselinum crispum | 31.3/4.88 | 117 | 6 | 32 | ||
| C15 | Chitinase precursor | 5880843 | Petroselinum crispum | 29.0/6.3 | 109 | 5 | 30 | ||
| C11 | RecName: Full= Acidic endochitinase; Flags: Precursor | 116332 | Solanum tuberosum | 31.3/4.88 | 63 | 5 | 10 | ||
| C12 | RecName: Full= Acidic endochitinase; Flags: Precursor | 116332 | - | 29.0/6.3 | 68 | 9 | 38 | ||
| C13 | RecName: Full= Acidic endochitinase; Flags: Precursor | 116332 | - | 31.3/4.88 | 64 | 4 | 10 | ||
| C16 | Putative thaumatin-like protein | 53830843 | Solanum tuberosum | 25.0/5.31 | 79 | 2 | 6 | ||
| C17 | Putative thaumatin-like protein | 53830843 | Solanum tuberosum | 25.0/5.31 | 62 | 3 | 12 | ||
| C18 | Putative thaumatin-like protein | 53830847 | Solanum tuberosum | 27.2/8.32 | 149 | 6 | 22 | ||
| C19 | Osmotin-like protein | 12274936 | Fagus sylvatica | 13.8/5.20 | 139 | 2 | 6 | ||
| C22 | Putative thaumatin-like protein | 53830843 | Solanum tuberosum | 25.0/5.31 | 67 | 3 | 12 | ||
| Carbohydrate and energy metabolism (12) | |||||||||
| A1 | C4-specific pyruvate orthophosphate dikinase | 31322756 | Miscanthus giganteus | 102.3/5.50 | 144 | 21 | 16 | ||
| A8 | GLX22(GLYOXALASE2-2); hydroxyacylglutahione hydrolase | 15228389 | Arabidopsis thaliana | 28.8/5.93 | 66 | 3 | 18 | ||
| A9 | NADH dehydrogenase [ubiquinone]1 α subcomplex subunit 5 AltName: Full= NADH-ubiq | 464258 | Solanum tuberosum | 4.1/9.41 | 72 | 4 | 30 | ||
| A10 | Lactoylglutathione lyase family protein, glyoxalase I family protein | 42571377 | Arabidopsis thaliana | 15.4/6.2 | 71 | 1 | 40 | ||
| A28 | ATP synthase D chain, mitochondrial, putative | 223530804 | Ricinus communis | 19.7/5.33 | 339 | 8 | 31 | ||
| A30 | GADPH (383 AA) | 22240 | Zea mays | 40.9/7.21 | 950 | 18 | 39 | ||
| B8 | RecName: Full= triosephosphate isomerase, chloroplastic; short= TIM; short= triose-phosphate isomerase | 1351271 | - | 34.4/6.45 | 245 | 23 | 37 | ||
| B9 | RecName: Full= ATP synthase subunit δ′, mitochondrial; AltName: Full= F-ATPasedelta’subunit; Fl | 2493046 | - | 21.3/5.93 | 121 | 14 | 41 | ||
| B11 | Acyl-CoA-binding protein | 19352190 | Panax ginseng | 9.90/5.43 | 118 | 4 | 18 | ||
| B14 | Putative NADH dehydrogenase (ubiquinone oxidoreductase) | 21536893 | Arabidopsis thaliana | 56.7/8.87 | 64 | 17 | 37 | ||
| B16 | ATP synthase CF1 β subunit | 11467199 | Zea mays | 54.0/5.31 | 730 | 13 | 34 | ||
| C4 | Mitochondrial F0 ATP synthase D chain | 17939851 | Arabidopsis thaliana | 17.2/4.97 | 68 | 3 | 12 | ||
| protein metabolism (5) | |||||||||
| A5 | Aspartic proteinase | 20800441 | Vigna unguiculata | 55.3/5.63 | 140 | 14 | 31 | ||
| A6 | Orf25 | 13449342 | Arabidopsis thaliana | 21.6/9.53 | 54 | 12 | 45 | ||
| A7 | Aspartic proteinase | 20800441 | Vigna unguiculata | 55.3/5.63 | 140 | 4 | 4 | ||
| A22 | Aspartic proteinase | 20800441 | Vigna unguiculata | 55.3/5.63 | 100 | 6 | 7 | ||
| B7 | Putative α7 proteasome subunit | 14594925 | Nicotiana tabacum | 27.2/6.11 | 123 | 6 | 34 | ||
| Signal transduction (3) | |||||||||
| A12 | ANP3 (Arabidopsis NPK1-related protein kinase 3); kinase | 15230612 | Arabidopsis thaliana | 71.6/8.41 | 48 | 14 | 24 | ||
| A21 | kinase associated protein phosphatase | 3328364 | Oryza sativa | 63.3/6.66 | 64 | 15 | 23 | ||
| B12 | RPT3 (root phototropism 3); ATPase | 15237159 | Arabidopsis thaliana | 45.7/5.42 | 124 | 12 | 24 | ||
| Nucleotide acid metabolism (2) | |||||||||
| B4 | DEAD box RNA helicase | 25809054 | Pisum sativum | 46.9/5.39 | 270 | 16 | 33 | ||
| B22 | DEAD BOX RNA helicase RH15-like protein | 8953379 | Arabidopsis thaliana | 49.3/5.43 | 273 | 6 | 16 | ||
| Transcriptional regulation (3) | |||||||||
| A15 | WRKY 14 | 34101231 | Theobroma cacao | 4.2/9.50 | 50 | 7 | 50 | ||
| B15 | Pentatricopeptide repeat (PPR)-containing protein | 15228257 | Arabidopsis thaliana | 72.8/6.80 | 66 | 14 | 38 | ||
| C10 | tRNA-splicing endonuclease positive effector-related | 15218807 | Arabidopsis thaliana | 120.2/8.82 | 58 | 3 | 12 | ||
| Storage proteins (2) | |||||||||
| B19 | 7S globulin | 13507023 | Elaeis guineensis | 66.3/6.53 | 74 | 1 | 2 | ||
| B24 | 7S globulin | 13507023 | Elaeis guineensis | 66.3/6.53 | 82 | 1 | 4 | ||
| Unknown function proteins (11) | |||||||||
| A19 | OSJNBb0034G 17.7 | 38605849 | Oryza sativa | 100.1/8.44 | 48 | 12 | 45 | ||
| A13 | Hypothetical protein | 4678216 | Arabidopsis thaliana | 326.1/9.98 | 61 | 13 | 27 | ||
| A23 | Unknown protein | 55168344 | Oryza sativa | 123.8/7.02 | 45 | 20 | 24 | ||
| A29 | Putative protein | 4469015 | Arabidopsis thaliana | 72.1/4.87 | 56 | 1 | 1 | ||
| B3 | Os02g0698000 | 115448091 | Oryza sativa | 44.8/5.68 | 96 | 12 | 36 | ||
| B5 | Os07g0513000 | 115472339 | Oryza sativa | 39.7/8.6 | 64 | 17 | 37 | ||
| B13 | Unknown protein | 42407715 | Oryza sativa | 26.3/11.63 | 104 | 8 | 22 | ||
| B17 | Unknown | 194688752 | Zea mays | 47.2/5.95 | 361 | 8 | 20 | ||
| B23 | Hypothetical protein | 147779485 | Vitis vinifera | 52.9/10.11 | 46 | 3 | 3 | ||
| C2 | Hypothetical protein | 4090293 | Secale cereale | 19.5/8.8 | 64 | 13 | 54 | ||
| C21 | Os08g0500600 | 115477124 | Oryza sativa | 47.6/6.28 | 60 | 6 | 22 | ||
2.3. Proteins Playing Critical Roles in F. mandshurica Explant Browning and SE
2.3.1. Stress Response and Defense Proteins
Chitinase
Aspartic Proteinase
Dehydrins
Peroxidases
Osmotin-Like Protein and Putative Thaumatin-Like Protein
Heat Shock Proteins (HSPs)
2.3.2. Signal Transduction
Arabidopsis NPK1-Related Protein Kinase 3 (ANP3)
Kinase-Associated Protein Phosphatase (KAPP)
2.3.3. Other Proteins
3. Experimental Section
3.1. Materials Preparation
3.2. Protein Extraction
3.3. Two-Dimensional Gel Electrophoresis
3.4. Spot Matching
3.5. In-Gel Digestion of Protein Spots
3.6. Identification of Protein Spots by MALDI-TOF-TOF
3.7. Mascot Database Search
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zimmerman, L.J. Somatic embryogenesis: A model for early development in higher plants. Plant Cell 1993, 5, 1411–1414. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.D.L.; Guzzo, F.; Toonen, M.A.J.; de Vries, S.C. A leucine rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 1997, 124, 2049–2062. [Google Scholar] [PubMed]
- Anjaneyulu, C.; Shyamkumar, B.; Giri, C.C. Somatic embryogenesis from callus cultures of Terminalia chebula Retz: An important medicinal tree. Trees 2004, 18, 547–552. [Google Scholar] [CrossRef]
- Stasolla, C.; Kong, L.; Yeung, E.C.; Thorpe, T.A. Maturation of somatic embryos in conifers: Morphogenesis, physiology, biochemistry, and molecular biology. Vitro Cell Dev. Biol. Plant 2002, 38, 93–105. [Google Scholar] [CrossRef]
- Attree, S.M.; Pomeroy, M.K.; Fowke, L.C. Manipulation of conditions for the culture of somatic embryos of white spruce for improved triacylglycerol biosynthesis and desiccation tolerance. Planta 1992, 187, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Balbuena, T.S.; Silveira, V.; Junqueira, M.; Dias, L.L.C.; Shevchenko, A.; Floh, E.I.S. Changes in the 2-DE protein profile during zygotic embryogenesis in the Brazilian Pine (Araucaria angustifolia). J. Proteome Res. 2009, 72, 337–352. [Google Scholar] [CrossRef]
- Imin, N.; de Jong, F.; Mathesius, U.; van Noorden, G.; Saeed, N.A.; Wang, X.D.; Rose, R.J.; Rolfe, B.G. Proteome reference maps of Medicago truncatula embryogenic cell cultures generated from single protoplasts. Proteomics 2004, 4, 1883–1896. [Google Scholar] [CrossRef] [PubMed]
- Nolan, K.E.; Irwanto, R.R.; Rose, R.J. Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol. 2003, 133, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Zhao, X.Y.; Liu, Y.B.; Zhang, C.L.; O’Neill, S.D.; Zhang, X.S. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009, 59, 448–460. [Google Scholar]
- Bouchabké-Coussa, O.; Obellianne, M.; Linderme, D.; Montes, E.; Maia-Grondard, A.; Vilaine, F.; Pannetier, C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013, 32, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Benson, E.E. Special symposium: In vitro plant recalcitrance. Do free radicals have a role in plant tissue culture recalcitrance? Vitro Cell Dev. Biol. Plant 2000, 36, 163–170. [Google Scholar] [CrossRef]
- Pinto, G.; Silva, S.; Park, Y.S.; Neves, L.; Araújo, C.; Santos, C. Factors influencing somatic embryogenesis induction in Eucalyptus globulus Labill.: Basal medium and anti-browning agents. Plant Cell Tissue Organ Cult. 2008, 95, 79–88. [Google Scholar] [CrossRef]
- Fehér, A.; Pasternak, T.P.; Dudits, D. Transition of somatic plant cells to an embryogenic state. Plant Cell Tissue Organ Cult. 2003, 74, 201–228. [Google Scholar] [CrossRef]
- Gaj, M.D. Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul. 2004, 43, 27–47. [Google Scholar] [CrossRef]
- Quiroz-Figueroa, F.R.; Rojas-Herrera, R.; Galaz-Avalos, R.M.; Loyola-Vargas, V.M. Embryo production through somatic embryogenesis can be used to study cell differentiation in plants. Plant Cell Tissue Organ Cult. 2006, 86, 285–301. [Google Scholar] [CrossRef]
- Elvira, M.I.; Galdeano, M.M.; Gilardi, P.; García-Luque, I.; Serra, M.T. Proteomic analysis of pathogenesis-related proteins (PRs) induced by compatible and incompatible interactions of pepper mild mottle virus (PMMoV) in Capsicum chinense L3 plants. J. Exp. Bot. 2008, 59, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Soosaar, J.L.M.; Burch-Smith, T.M.; Dinesh-Kumar, S.P. Mechanisms of plant resistance to viruses. Nat. Rev. 2005, 3, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Passarinho, P.A.; Hengel, A.J.V.; Fransz, P.F.; de Vries, S. Expression pattern of the Arabidopsis thaliana AtEP3/AtchitIV endochitinase gene. Planta 2001, 212, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Ekramoddoullah, A.K.; Zamani, A. A class IV chitinase is up-regulated by fungal infection and abiotic stresses and associated with slow-canker-growth resistance to Cronartium ribicola in western white pine (Pinus monticola). Phytopathology 2005, 95, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Xu, L.; Fu, Z.; Yang, Y.; Guo, J.; Wang, S.; Que, Y. ScChi, encoding an acidic class III chitinase of sugarcane, confers positive responses to biotic and abiotic stresses in sugarcane. Int. J. Mol. Sci. 2014, 15, 2738–2760. [Google Scholar] [CrossRef] [PubMed]
- Sabater-Jara, A.B.; Almagro, L.; Pedreño, M.A. Induction of extracellular defense-related proteins in suspension cultured-cells of Daucus carota elicited with cyclodextrins and methyl jasmonate. Plant Physiol. Biochem. 2014, 77, 133–139. [Google Scholar] [CrossRef] [PubMed]
- De Jong, A.; Cordewener, J.; LoSchiavo, F.; Terzi, M.; Vandekerckhove, J.; van Kammen, A.; de Vries, S.C. A carrot somatic embryo is rescued by chitinase. Plant Cell 1992, 4, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Van Hengel, A.; Tadesse, Z.; Immerzeel, P.; Schols, H.; van Kammen, A.; de Vries, S.C. N-Acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol. 2001, 125, 1880–1891. [Google Scholar] [CrossRef] [PubMed]
- Maillot, P.; Lebel, S.; Schellenbaum, P.; Jacques, A.; Walter, B. Differential regulation of SERK, LEC1-Like and pathogenesis-related genes during indirect secondary somatic embryogenesis in grapevine. Plant Physiol. Biochem. 2009, 47, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Helleboid, S.; Hendricks, T.; Bauw, G.; Inzé, D.; Vasseur, J.; Hilbert, J.L. Three major somatic embryogenesis related proteins in Cichorium identified as PR proteins. J. Exp. Bot. 2000, 51, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Filonova, L.H.; Bozhkov, P.V.; Brukhin, V.B.; Daniel, G.; Zhivotovsky, B.; von Arnold, S. Two waves of programmed cell death occur during formation anddevelopment of somatic embryos in the gymnosperm, Norway spruce. J. Cell Sci. 2000, 113, 4399–4441. [Google Scholar] [PubMed]
- Ge, X.; Dietrich, C.; Matsuno, M.; Li, G.; Berg, H.; Xia, Y. An Arabidopsis aspartic protease functions as an anti-cell death component in reproduction and embryogenesis. EMBO Rep. 2005, 6, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Tan, S.K.; Kamada, H. Characterization of a dehydrin-like phosphoprotein (ECPP-44) relating to somatic embryogenesis in carrot. Plant Mol. Biol. Rep. 2006, 24, 253a–253j. [Google Scholar] [CrossRef]
- Choi, D.W.; Close, T.J. A newly identified barley gene, Dhn12, encodes a YSK2DHN, is located on chromosome 6H and has embryo-specific expression. Theor. Appl. Genet. 2000, 100, 1274–1278. [Google Scholar] [CrossRef]
- Nylander, M.; Svensson, J.; Palva, E.T.; Welin, B.V. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 2001, 45, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Bernards, M.A.; Fleming, W.D.; Llewellyn, D.B.; Priefer, R.; Yang, X.; Sabatino, A.; Plourde, G.L. Biochemical characterization of the suberization associated anionic peroxidase of potato. Plant Physiol. 1999, 121, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef] [PubMed]
- De Marco, A.; Roubelakis-Angelakis, K.A. The complexity of enzymic control of hydrogen peroxide concentration may affect the regeneration potential of plant protoplasts. Plant Physiol. 1996, 110, 137–145. [Google Scholar] [PubMed]
- Morita, S.; Kaminaka, H.; Masumura, T.; Tanaka, K. Induction of rice cytosolic ascorbate peroxidase mRNA by oxidative stress: The involvement of hydrogen peroxide in oxidative stress signaling. Plant Cell Physiol. 1999, 40, 417–422. [Google Scholar] [CrossRef]
- Brownleader, M.D.; Hopkins, J.; Mobasheri, A.; Dey, P.M.; Jackson, P.; Trevan, M. Role of extensin peroxidase in tomato (Lycopersicon esculentum Mill.) seedling growth. Planta 2000, 210, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Kotake, T.; Nakagawa, N.; Sakurai, N.; Nevins, D.J. Expression and function of cell wall-bound cationic peroxidase in asparagus somatic embryogenesis. Plant Physiol. 2003, 131, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Cordewener, J.; Booij, H.; van der Zandt, H.; van Engelen, F.; van Kammen, A.; de Vries, S. Tunicamycin-inhibited carrot somatic embryogenesis can be restored by secreted cationic peroxidase isoenzymes. Planta 1991, 184, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Lai, Z.; Fang, Z.; He, Y.; Jiang, S. Proteomics analysis of early somatic embryogenesis in Longan (Dimocarpus longan Lour.). Sci. Agric. Sin. 2012, 45, 1775–1790. [Google Scholar]
- Baba, A.I.; Nogueira, F.C.S.; Pinheiro, C.B.; Brasil, J.N.; Jereissati, E.S.; Jucá, T.L.; Soares, A.A.; Santos, M.F.; Domont, G.B.; Campos, F.A.P. Proteome analysis of secondary somatic embryogenesis in cassava (Manihot esculenta). Plant Sci. 2008, 175, 717–723. [Google Scholar] [CrossRef]
- Richard-Forget, F.C.; Gauillard, F.A. Oxidation of chlorogenic acid, catechins and 4-methylcatechol in model solutions by combinations of pear (Pyrus communis cv. Williams) polyphenol oxidase and peroxidase: A possible involvement of peroxidase in enzymatic browning. J. Agric. Food Chem. 1997, 45, 2472–2476. [Google Scholar] [CrossRef]
- Rather, I.A.; Awasthi, P.; Mahajan, V.; Bedi, Y.S.; Vishwakarma, R.A.; Gandhi, S.G. Molecular cloning and functional characterization of an antifungal PR-5 protein from Ocimum basilicum. Gene 2015, 558, 143–151. [Google Scholar] [PubMed]
- Györgyey, J.; Gartner, A.; Németh, K.; Magyar, Z.; Hirt, H.; Heberle-Bors, E.; Dudits, D. Alfalfa heat shock genes are differentially expressed during somatic embryogenesis. Plant Mol. Biol. 1991, 16, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Nishihama, R.; Ishikawa, M.; Araki, S.; Soyano, T.; Asada, T.; Machida, Y. The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev. 2001, 15, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.Z.; Dunstan, D.I. Expression of abundant mRNAs during somatic embryogenesis of white spruce (Picea glauca (Moench) Voss). Planta 1996, 199, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Kovtun, Y.; Chiu, W.L.; Tena, G.; Sheen, J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. PNAS 2000, 97, 2940–2945. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.Y.; Adler, V.; Buschmann, T.; Yin, Z.; Wu, X.; Jones, S.N.; Ronai, Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 1998, 12, 2658–2663. [Google Scholar] [CrossRef] [PubMed]
- Reichheld, J.P.; Vernoux, T.; Lardon, F.; van Montagu, M.; Inzé, D. Specific checkpoints regulate plant cell cycle progression in response to oxidative stress. Plant J. 1999, 17, 647–656. [Google Scholar] [CrossRef]
- Stone, J.M.; Trotochaud, A.E.; Walker, J.C.; Clark, S.E. Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 1998, 117, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Torii, K.U. Receptor kinase activation and signal transduction in plants: An emerging picture. Curr. Opin. Plant Biol. 2000, 3, 361–367. [Google Scholar] [CrossRef]
- Williams, R.W.; Wilson, J.M.; Meyerowitz, E.M. A possible role for kinase associated protein phosphatase in the Arabidopsis CLAVATA1 signalling pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 10467–10472. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Vielle-Calzada, J.P.; Hartog, M.V.; Schmidt, E.D.; Boutilier, K.; Grossniklaus, U.; de Vries, S.C. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 2001, 127, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Russinova, E.; Gadella, T.W.; Willemse, J.; de Vries, S.C. The Arabidopsis kinase associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev. 2002, 16, 1707–1720. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.R.; Fu, Y.R.; Wang, W.J.; Qiu, Z.Y. Quantitative analysis of mitochondrial hydrophobic proteome in HCPT - induced apoptosis of hepatoma cells. Chin. J. Biochem. Mol. Biol. 2008, 24, 373–382. [Google Scholar]
- Abdelhaleem, M. RNA Helicases: Regulators of differentiation. Clin. Biochem. 2005, 38, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, E.A.; Wessel, G.M. DEAD-box helicases: Posttranslational regulation and function. Biochem. Biophys. Res. Commun. 2010, 395, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Dong, C.H.; Lee, H.J.; Zhu, J.H.; Xiong, L.; Gong, D.; Stevenson, B.; Zhu, J.K. A DEAD Box RNA Helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 2005, 17, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Paulus, C.; Köllner, B.; Jacobsen, H.J. Physiological and biochemical characterization of glyoxalase I, a general marker for cell proliferation, from a soybean cell suspension. Planta 1993, 189, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, O.; Pal, S.; Guha-Mukherjee, S.; Sopory, S.K. Correlation of glyoxalase I activity with cell proliferation in Datura callus culture. Plant Cell Rep. 1984, 3, 12l–124. [Google Scholar] [CrossRef] [PubMed]
- Douglas, K.T.; Nadvi, I.N. Inhibition of glyoxalase I: A possible transition-state analogue inhibitor approach to potential antineoplastic agents? FEBS Lett. 1979, 106, 393–396. [Google Scholar] [CrossRef]
- Singla-Pareek, S.L.; Yadav, S.K.; Pareek, A.; Reddy, M.K.; Sopory, S.K. Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol. 2006, 140, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Yadav, S.K.; Pareek, A.; Reddy, M.K.; Sopory, S.K. Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Transgenic Res. 2008, 17, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 2005, 337, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Transgenic tobacco plants overexpressing glyoxalase enzymes resist an increase in methylglyoxal and maintain higher reduced glutathione levels under salinity stress. FEBS Lett. 2005, 579, 6265–6271. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Hasanuzzaman, M.; Fujita, M. Up-regulation of antioxidant and glyoxalase systems by exogenous glycinebetaine and proline in mung bean confer tolerance to cadmium stress. Physiol. Mol. Biol. Plants 2010, 16, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.J.; Zhang, J.B.; Jia, C.H.; Jin, Z.Q.; Xu, B.Y. Enhancement of tolerance to abiotic stress of Saccharomyces cerevisiae transformed by a gene encoding glyoxalase from banana. China Biotechnol. 2010, 30, 22–26. [Google Scholar]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-P.; Yang, L.; Shen, H.-L. Proteomic Analysis of Immature Fraxinus mandshurica Cotyledon Tissues during Somatic Embryogenesis: Effects of Explant Browning on Somatic Embryogenesis. Int. J. Mol. Sci. 2015, 16, 13692-13713. https://doi.org/10.3390/ijms160613692
Liu C-P, Yang L, Shen H-L. Proteomic Analysis of Immature Fraxinus mandshurica Cotyledon Tissues during Somatic Embryogenesis: Effects of Explant Browning on Somatic Embryogenesis. International Journal of Molecular Sciences. 2015; 16(6):13692-13713. https://doi.org/10.3390/ijms160613692
Chicago/Turabian StyleLiu, Chun-Ping, Ling Yang, and Hai-Long Shen. 2015. "Proteomic Analysis of Immature Fraxinus mandshurica Cotyledon Tissues during Somatic Embryogenesis: Effects of Explant Browning on Somatic Embryogenesis" International Journal of Molecular Sciences 16, no. 6: 13692-13713. https://doi.org/10.3390/ijms160613692
APA StyleLiu, C.-P., Yang, L., & Shen, H.-L. (2015). Proteomic Analysis of Immature Fraxinus mandshurica Cotyledon Tissues during Somatic Embryogenesis: Effects of Explant Browning on Somatic Embryogenesis. International Journal of Molecular Sciences, 16(6), 13692-13713. https://doi.org/10.3390/ijms160613692





