The Influence of PSCA Gene Variation on Its Expression and Gastric Adenocarcinoma Susceptibility in the Northwest Chinese Population
Abstract
:1. Introduction
2. Results
| SNP ID | Position a | Location | Allele A/B b | HWE-p |
|---|---|---|---|---|
| rs2294008 | 143,761,931 | 5'-UTR | T/C | 0.618 |
| rs3736001 | 143,762,807 | Exon 2 | A/G | 0.446 |
| rs2976392 | 143,762,932 | Intron 2 | A/G | 0.939 |
| rs10216533 | 143,763,690 | 3'-UTR | A/G | 0.266 |
| Variable | Case | Control | p | ||
|---|---|---|---|---|---|
| (n = 476) | (n = 481) | ||||
| Age, mean years (SD) | 57.1 | (11.5) | 56.4 | (11.8) | 0.334 a |
| Gender, n (%) | |||||
| Male | 249 | (52.3) | 247 | (51.4) | 0.766 b |
| Female | 227 | (47.7) | 234 | (48.6) | |
| SNP ID | Genotype | No. (Frequency) | OR | (95% CI) | p | |||
|---|---|---|---|---|---|---|---|---|
| Case (n = 476) | Control (n = 481) | |||||||
| rs2294008 | CC | 227 | (47.8) | 261 | (54.4) | 1.00 | ||
| CT | 207 | (43.6) | 183 | (38.1) | 1.30 | (1.00–1.70) | 0.053 | |
| TT | 41 | (8.6) | 36 | (7.5) | 1.31 | (0.81–2.12) | 0.272 | |
| CT + TT | 248 | (52.2) | 219 | (45.6) | 1.30 | (1.01–1.68) | 0.042 * | |
| rs3736001 | GG | 382 | (80.6) | 402 | (83.6) | 1.00 | ||
| GA | 88 | (18.6) | 74 | (15.4) | 1.25 | (0.89–1.76) | 0.195 | |
| AA | 4 | (0.8) | 5 | (1.0) | 0.84 | (0.22–3.16) | 0.936 | |
| GA + AA | 92 | (19.4) | 79 | (16.4) | 1.23 | (0.88–1.71) | 0.229 | |
| rs2976392 | GG | 190 | (43.6) | 231 | (51.2) | 1.00 | ||
| GA | 208 | (47.7) | 184 | (40.8) | 1.37 | (1.04–1.81) | 0.024 * | |
| AA | 38 | (8.7) | 36 | (8.0) | 1.28 | (0.78–2.10) | 0.322 | |
| GA + AA | 246 | (56.4) | 220 | (48.8) | 1.36 | (1.04–1.77) | 0.023 * | |
| rs10216533 | GG | 237 | (53.6) | 256 | (54.4) | 1.00 | ||
| GA | 152 | (34.4) | 176 | (37.4) | 0.93 | (0.71–1.23) | 0.626 | |
| AA | 53 | (12.0) | 39 | (8.3) | 1.47 | (0.94–2.30) | 0.093 | |
| GA + AA | 205 | (46.4) | 215 | (45.7) | 1.03 | (0.79–1.34) | 0.824 | |
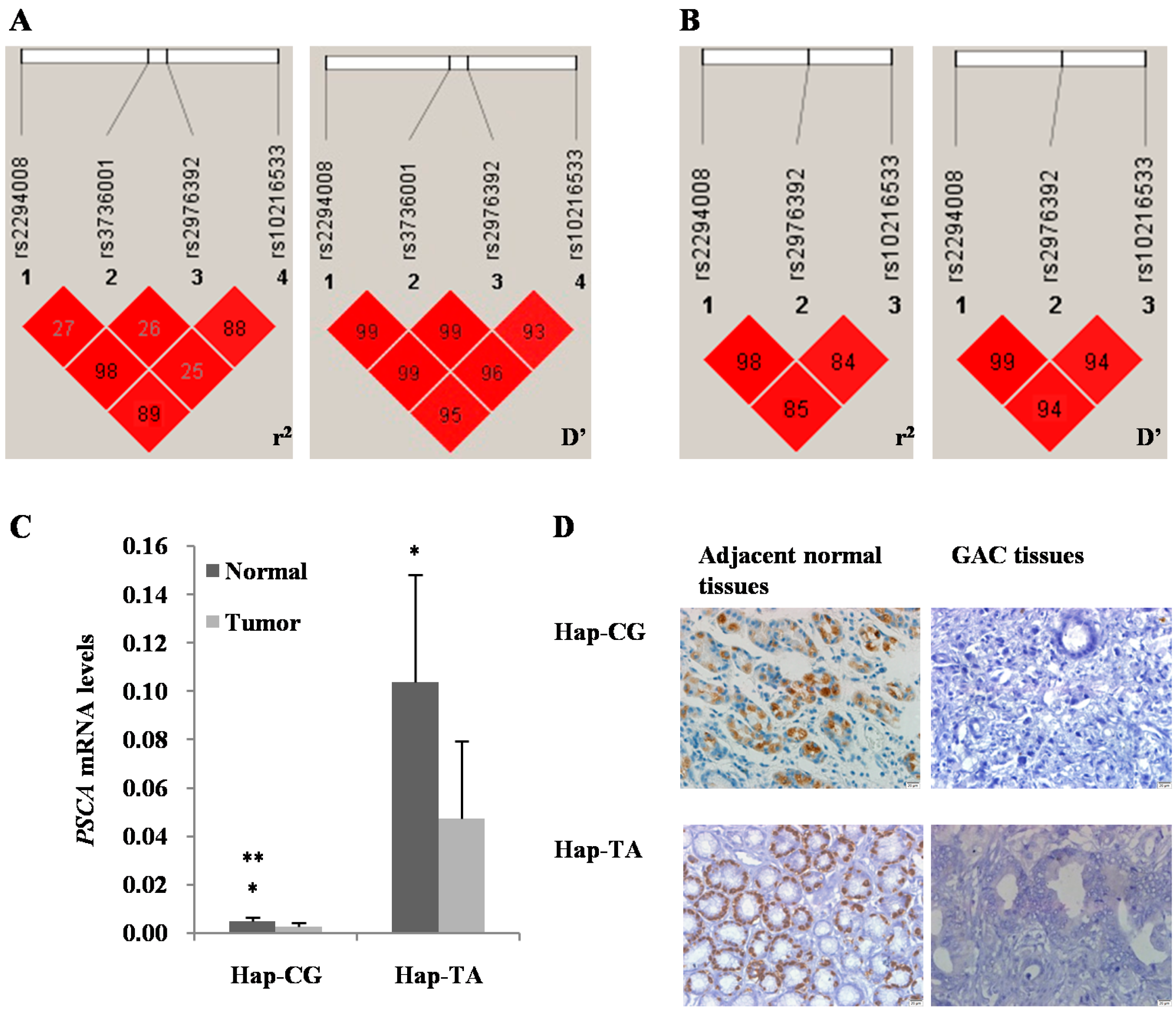
| Haplotype ID | Frequency a | OR (95% CI) | Fisher’s p | Pearson’s p | SNP No. and Haplotype | ||
|---|---|---|---|---|---|---|---|
| Case | Control | ||||||
| 1–3 | |||||||
| 1 | 0.67 | 0.72 | 0.83 | (0.67–1.01) | 0.065 | 0.065 | C–G |
| 2 | 0.32 | 0.28 | 1.21 | (0.99–1.48) | 0.065 | 0.065 | T–A |
| 1–3–4 | |||||||
| 3 | 0.64 | 0.71 | 0.78 | (0.64–0.96) | 0.020 * | 0.020 * | C–G–G |
| 4 | 0.29 | 0.28 | 1.08 | (0.87–1.33) | 0.478 | 0.478 | T–A–A |
| 5 | 0.04 | 0.00 | 12.28 | (3.75–40.27) | 0.00 * | 0.00 * | T–A–G |
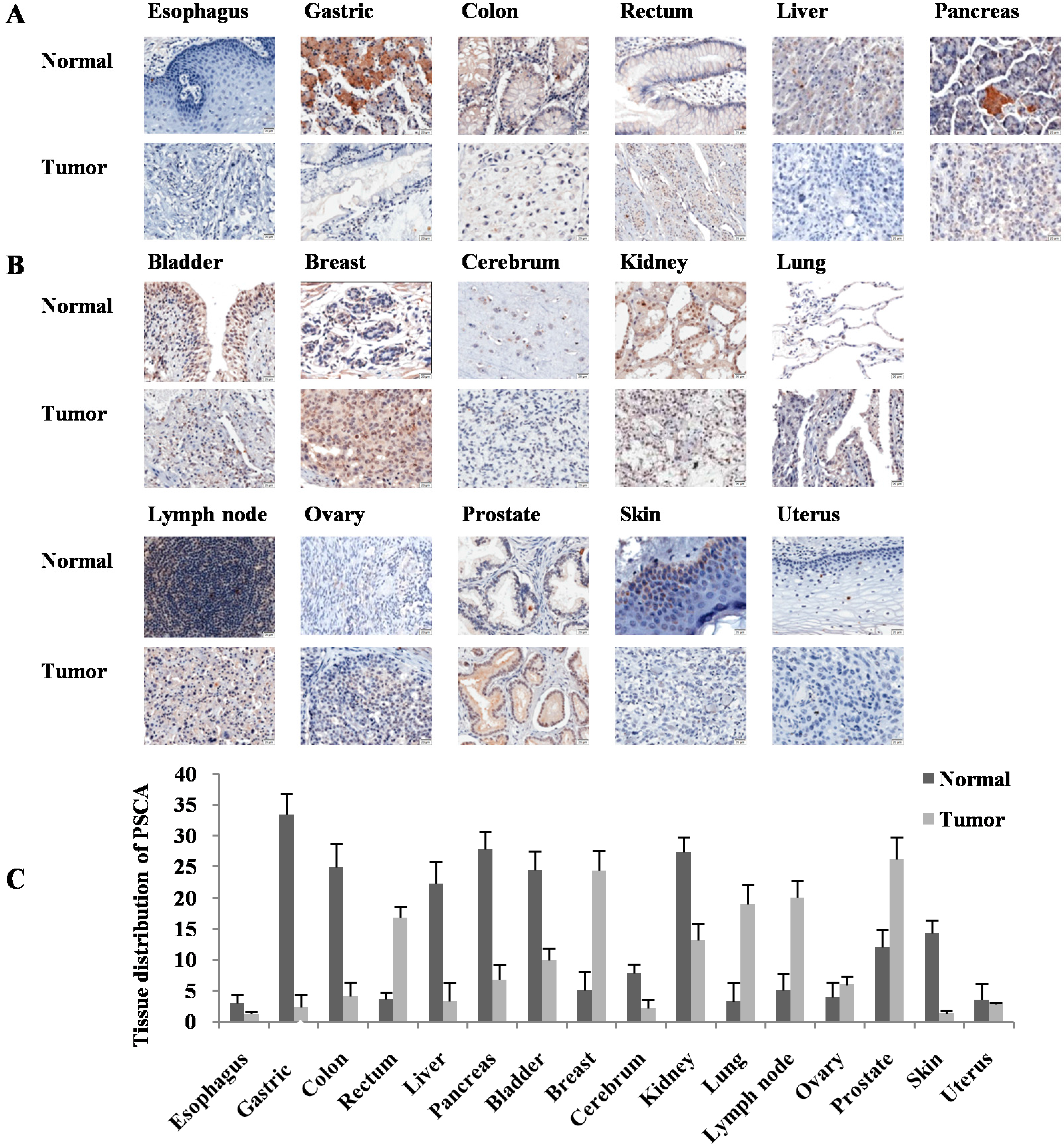
3. Discussion
4. Experimental Section
4.1. Patients and Samples
4.2. SNP Selection and Genotyping
4.3. Quantitative Real-Time PCR
4.4. Immunohistochemistry and Tissue Microarray
4.5. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA. Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Kelley, J.R.; Duggan, J.M. Gastric cancer epidemiology and risk factors. J. Clin. Epidemiol. 2003, 56, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, L. Incidence and mortality of gastric cancer in China. World J. Gastroenterol. 2006, 12, 17–20. [Google Scholar] [PubMed]
- Wadhwa, R.; Song, S.; Lee, J.S.; Yao, Y.; Wei, Q.; Ajani, J.A. Gastric cancer-molecular and clinical dimensions. Nat. Rev. Clin. Oncol. 2013, 10, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Duraes, C.; Almeida, G.M.; Seruca, R.; Oliveira, C.; Carneiro, F. Biomarkers for gastric cancer: Prognostic, predictive or targets of therapy? Virchows Arch. 2014, 464, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Pharoah, P.D.; Dunning, A.M.; Ponder, B.A.; Easton, D.F. Association studies for finding cancer-susceptibility genetic variants. Nat. Rev. Cancer 2004, 4, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.E.; Gu, Z.; Watabe, T.; Thomas, G.; Szigeti, K.; Davis, E.; Wahl, M.; Nisitani, S.; Yamashiro, J.; LeBeau, M.M.; et al. Prostate stem cell antigen: A cell surface marker overexpressed in prostate cancer. Proc. Natl. Acad. Sci. USA 1998, 95, 1735–1740. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Thomas, G.; Yamashiro, J.; Shintaku, I.P.; Dorey, F.; Raitano, A.; Witte, O.N.; Said, J.W.; Loda, M.; Reiter, R.E. Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene 2000, 19, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Saffran, D.C.; Raitano, A.B.; Hubert, R.S.; Witte, O.N.; Reiter, R.E.; Jakobovits, A. Anti-PSCA mAbs inhibit tumor growth and metastasis formation and prolong the survival of mice bearing human prostate cancer xenografts. Proc. Natl. Acad. Sci. USA 2001, 98, 2658–2663. [Google Scholar] [CrossRef] [PubMed]
- Tran, C.P.; Lin, C.; Yamashiro, J.; Reiter, R.E. Prostate stem cell antigen is a marker of late intermediate prostate epithelial cells. Mol. Cancer Res. 2002, 1, 113–121. [Google Scholar] [PubMed]
- Gu, Z.; Yamashiro, J.; Kono, E.; Reiter, R.E. Anti-prostate stem cell antigen monoclonal antibody 1G8 induces cell death in vitro and inhibits tumor growth in vivo via a Fc-independent mechanism. Cancer Res. 2005, 65, 9495–9500. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Yoshimura, K.; Saeki, N.; Katai, H.; Shimoda, T.; Matsuno, Y.; Saito, D.; Sugimura, H.; Tanioka, F.; Kato, S.; et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat. Genet. 2008, 40, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zhang, W.; Xu, L.; Cai, D. Quantitative assessment of the influence of prostate stem cell antigen polymorphisms on gastric cancer risk. Tumour Biol. 2014, 35, 2167–2174. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, S.; Verdi, D.; Pooley, K.A.; Nitti, D. Genetic variation and gastric cancer risk: A field synopsis and meta-analysis. Gut 2015. [Google Scholar] [CrossRef]
- Hu, Z.; Ajani, J.A.; Wei, Q. Molecular epidemiology of gastric cancer: Current status and future prospects. Gastrointest. Cancer Res. 2007, 1, 12–19. [Google Scholar] [PubMed]
- Hirschhorn, J.N.; Daly, M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005, 6, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Yoo, J.Y.; Shin, A.; Kim, Y.; Lee, E.S.; Lee, Y.S. Prostate stem cell antigen single nucleotide polymorphisms influence risk of estrogen receptor negative breast cancer in Korean females. Asian Pac. J. Cancer Prev. 2012, 13, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ye, Y.; Kiemeney, L.A.; Sulem, P.; Rafnar, T.; Matullo, G.; Seminara, D.; Yoshida, T.; Saeki, N.; Andrew, A.S.; et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 2009, 41, 991–995. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tang, J.; Wang, M.; Yuan, L.; Zhang, Z. Genetic variation in PSCA and bladder cancer susceptibility in a Chinese population. Carcinogenesis 2010, 31, 621–624. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.P.; Kohaar, I.; Rothman, N.; Earl, J.; Figueroa, J.D.; Ye, Y.; Malats, N.; Tang, W.; Liu, L.; Garcia-Closas, M.; et al. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc. Natl. Acad. Sci. USA 2012, 109, 4974–4979. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, G.; Jeon, E.; Sohn, E.J.; Lee, Y.; Kang, H.; Lee, D.W.; Kim, D.H.; Hwang, I. The immediate upstream region of the 5'-UTR from the AUG start codon has a pronounced effect on the translational efficiency in Arabidopsis thaliana. Nucleic Acids Res. 2014, 42, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Lunghi, M.; Galizi, R.; Magini, A.; Carruthers, V.B.; DiCristina, M. Expression of the glycolytic enzymes enolase and lactate dehydrogenase during the early phase of Toxoplasma differentiation is regulated by an intron retention mechanism. Mol. Microbiol. 2015. [Google Scholar] [CrossRef]
- Maiti, G.P.; Ghosh, A.; Mondal, P.; Baral, A.; Datta, S.; Samadder, S.; Nayak, S.P.; Chakrabarti, J.; Biswas, J.; Sikdar, N.; et al. SNP rs1049430 in the 3'-UTR of SH3GL2 regulates its expression: Clinical and prognostic implications in head and neck squamous cell carcinoma. Biochim. Biophys. Acta 2015, 1852, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Dai, N.; Zheng, M.; Wang, C.; Ji, Y.; Du, J.; Zhu, C.; He, Y.; Zhu, M.; Zhu, X.; Sun, M.; et al. Genetic variants at 8q24 are associated with risk of esophageal squamous cell carcinoma in a Chinese population. Cancer Sci. 2014, 105, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Tanikawa, C.; Urabe, Y.; Matsuo, K.; Kubo, M.; Takahashi, A.; Ito, H.; Tajima, K.; Kamatani, N.; Nakamura, Y.; Matsuda, K. A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat. Genet. 2012, 44, 430–434, S1–S2. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.Y.; Lee, Y.S.; Park, S.; Yoon, H.; Lee, S.J.; Park, W.S.; Seo, H.K.; Chung, J.; Kim, S.Y.; Hong, S.H.; et al. Haplotype analysis of prostate stem cell antigen and association with prostate cancer risk. J. Urol. 2011, 185, 2112–2118. [Google Scholar] [CrossRef] [PubMed]
- Saeki, N.; Gu, J.; Yoshida, T.; Wu, X. Prostate stem cell antigen: A Jekyll and Hyde molecule? Clin. Cancer Res. 2010, 16, 3533–3538. [Google Scholar] [CrossRef] [PubMed]
- Adamec, C. Example of the use of the nonparametric test. Test X2 for comparison of 2 independent examples. Cesk Zdrav 1964, 12, 613–619. [Google Scholar] [PubMed]
- Bland, J.M.; Altman, D.G. Statistics notes. The odds ratio. BMJ 2000, 320, 1468. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Liang, P.; Wang, W.; Dai, P.; Wang, Q.; Yan, W.; Zhao, J.; Sun, J.; Peng, Y.; Cui, D.; et al. The Influence of PSCA Gene Variation on Its Expression and Gastric Adenocarcinoma Susceptibility in the Northwest Chinese Population. Int. J. Mol. Sci. 2015, 16, 11648-11658. https://doi.org/10.3390/ijms160511648
Zhang W, Liang P, Wang W, Dai P, Wang Q, Yan W, Zhao J, Sun J, Peng Y, Cui D, et al. The Influence of PSCA Gene Variation on Its Expression and Gastric Adenocarcinoma Susceptibility in the Northwest Chinese Population. International Journal of Molecular Sciences. 2015; 16(5):11648-11658. https://doi.org/10.3390/ijms160511648
Chicago/Turabian StyleZhang, Wentao, Ping Liang, Weihua Wang, Peng Dai, Qin Wang, Wei Yan, Jinrong Zhao, Jianbin Sun, Yong Peng, Daxiang Cui, and et al. 2015. "The Influence of PSCA Gene Variation on Its Expression and Gastric Adenocarcinoma Susceptibility in the Northwest Chinese Population" International Journal of Molecular Sciences 16, no. 5: 11648-11658. https://doi.org/10.3390/ijms160511648
APA StyleZhang, W., Liang, P., Wang, W., Dai, P., Wang, Q., Yan, W., Zhao, J., Sun, J., Peng, Y., Cui, D., & Yan, Z. (2015). The Influence of PSCA Gene Variation on Its Expression and Gastric Adenocarcinoma Susceptibility in the Northwest Chinese Population. International Journal of Molecular Sciences, 16(5), 11648-11658. https://doi.org/10.3390/ijms160511648





