Synergistic Effect of Combinatorial Treatment with Curcumin and Mitomycin C on the Induction of Apoptosis of Breast Cancer Cells: A cDNA Microarray Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of Combinatorial Treatment Using Curcumin and Mitomycin C (MMC) on Tumor Growth
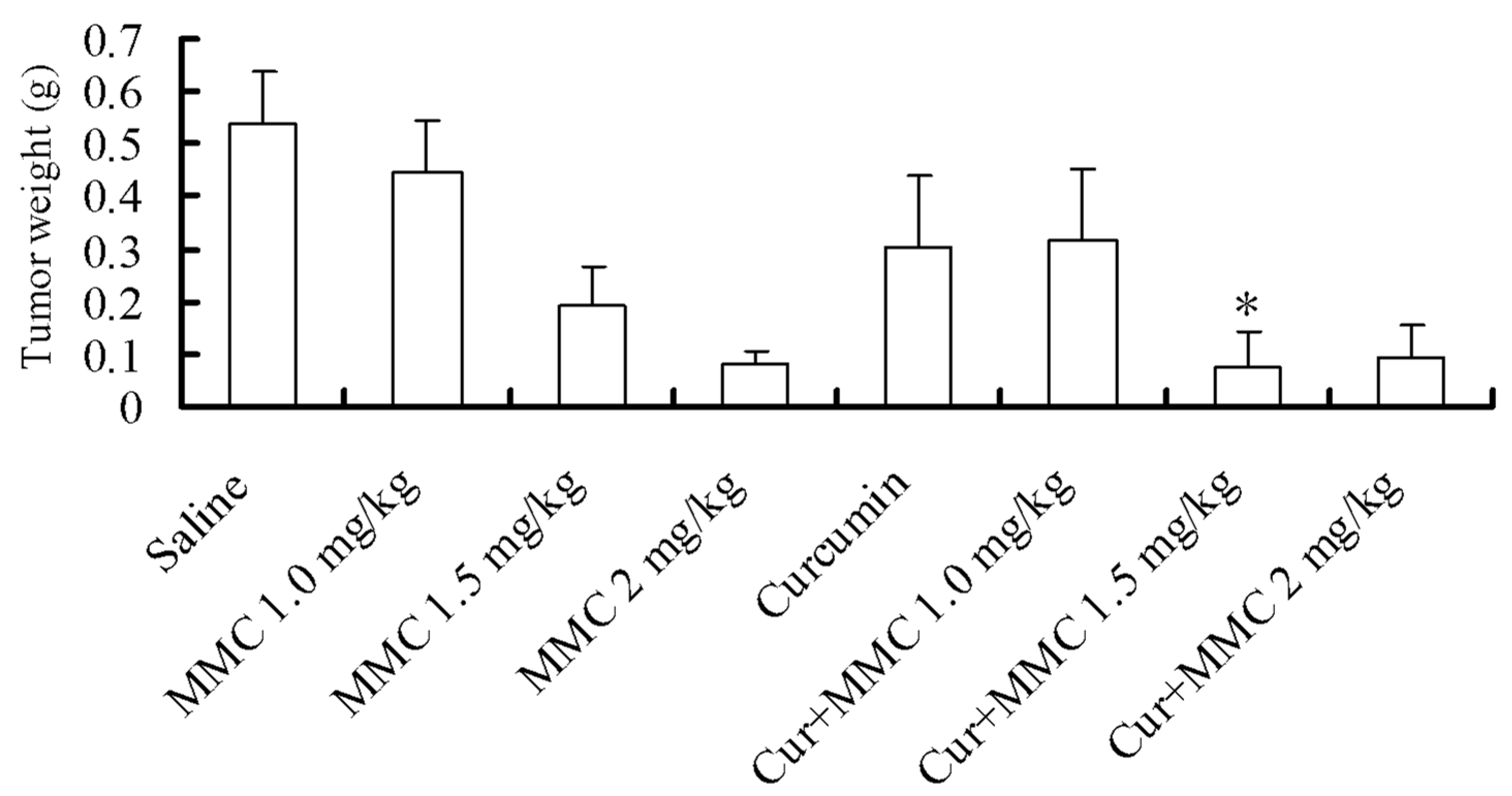
2.2. Gene Expression Profiles by cDNA Microarray Analysis
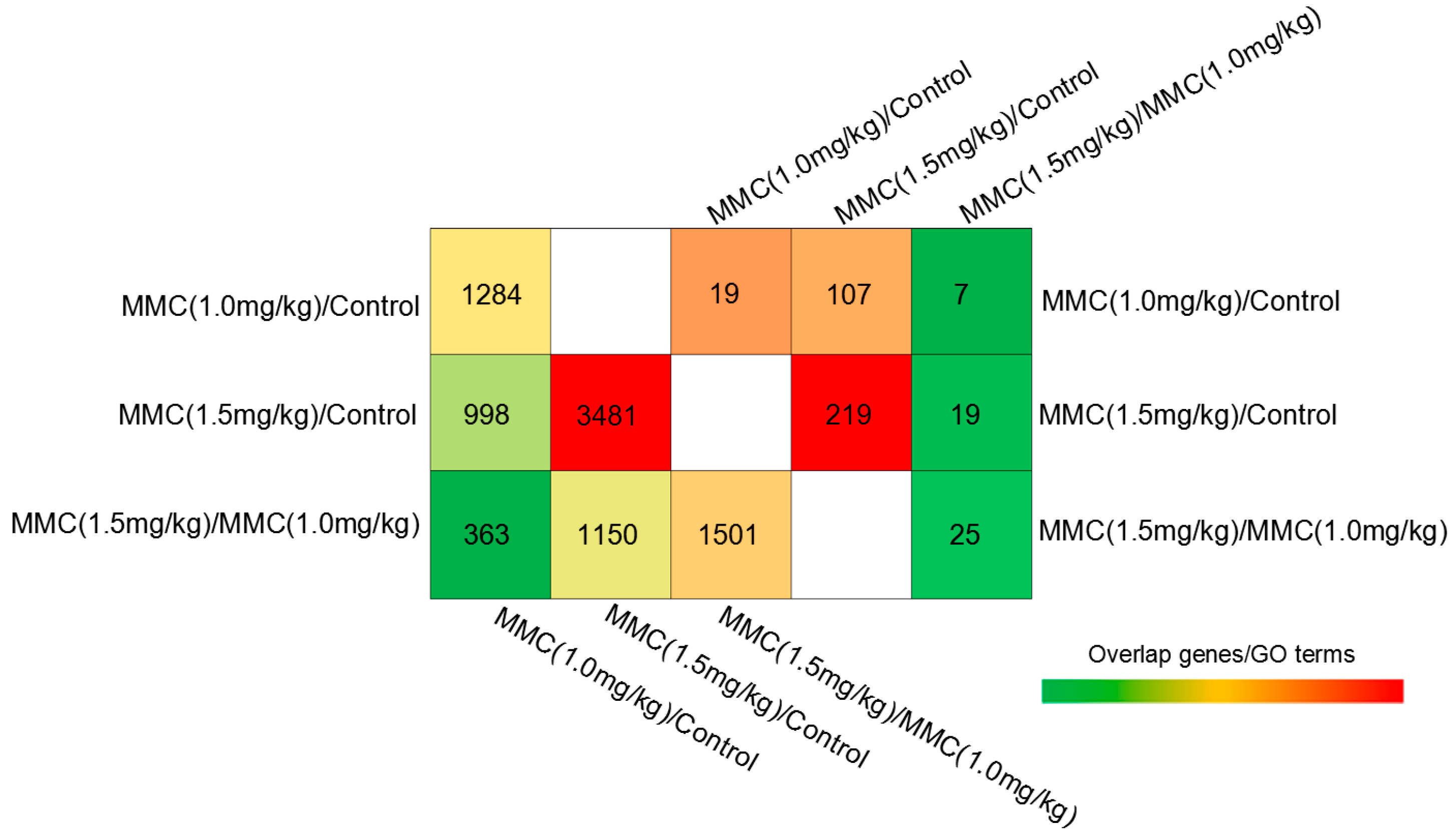
2.3. Modulation of Signaling Pathways Following Combinatorial Treatment of Curcumin/MMC
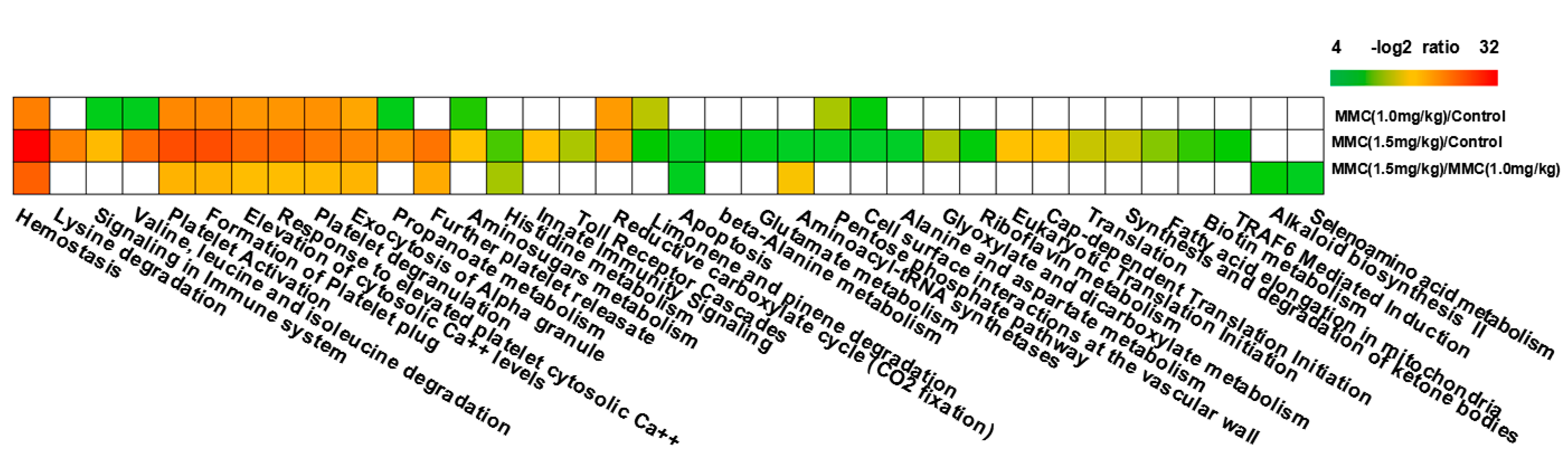
2.4. Relationships among Genes by Signal–Net Analysis
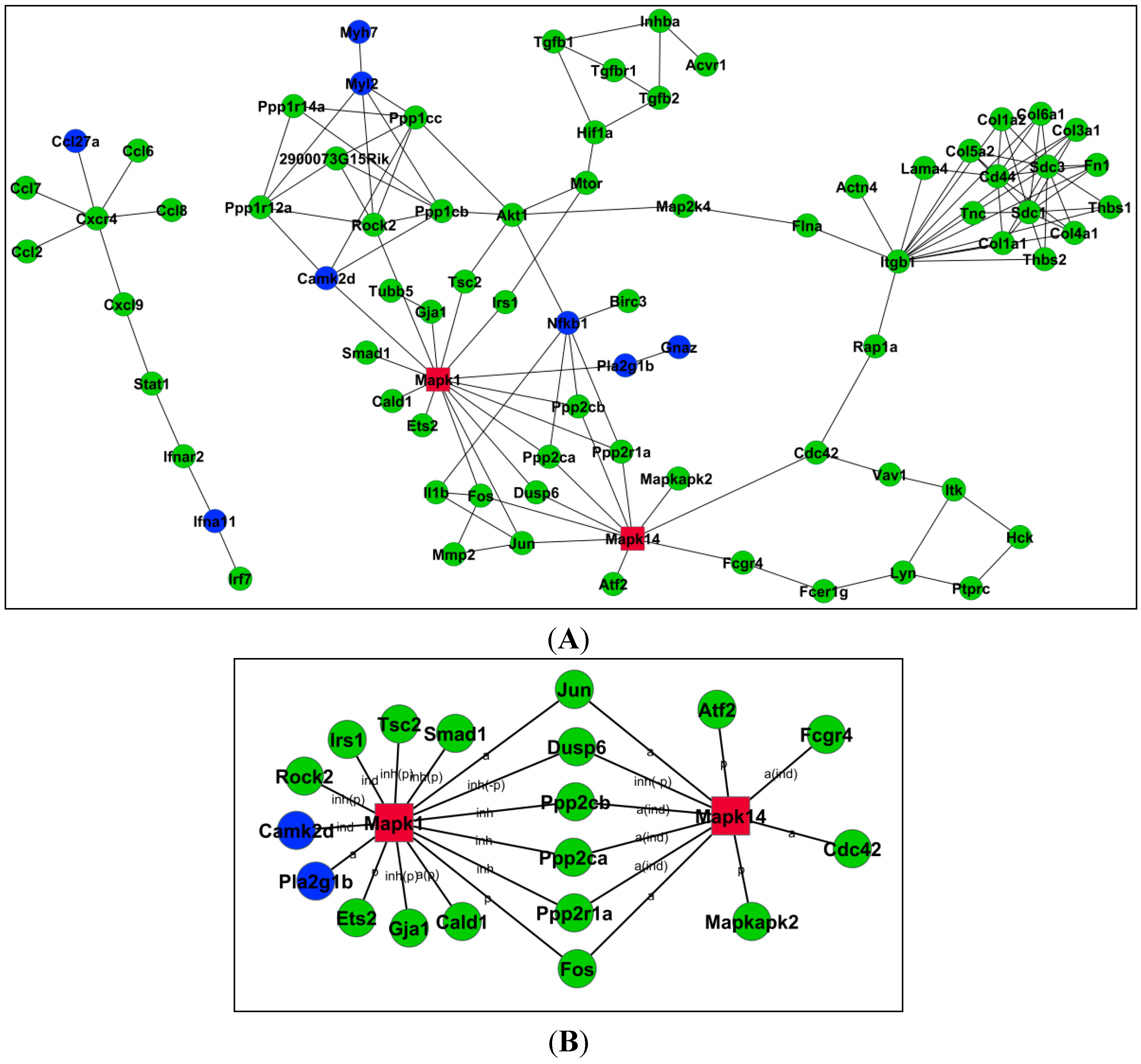
2.5. Validation of DE Genes by Quantitative RT-PCR (qRT-PCR)
| Gene Symbol | Betweenness Centrality | Degree | Indegree | Outdegree | Style |
|---|---|---|---|---|---|
| Mapk1 | 3.43 × 10−2 | 20 | 5 | 15 | up |
| Rock2 | 1.17 × 10−2 | 13 | 1 | 12 | up |
| Mpak14 | 2.25 × 10−2 | 11 | 3 | 8 | up |
| Akt1 | 2.70 × 10−3 | 7 | 1 | 6 | up |
| Nfkb1 | 2.27 × 10−3 | 6 | 1 | 5 | down |
| Mtor | 4.66 × 10−4 | 5 | 1 | 4 | up |
| Ppp1cb | 2.31 × 10−3 | 7 | 4 | 3 | up |
| Ppp1cc | 2.31 × 10−3 | 7 | 4 | 3 | up |
| Ppp1r12a | 1.49 × 10−3 | 6 | 3 | 3 | up |
| Itgb1 | 3.91 × 10−3 | 14 | 14 | 2 | up |
| Fos | 2.10 × 10−3 | 6 | 4 | 2 | up |
| Jun | 2.10 × 10−3 | 6 | 4 | 2 | up |
| Myl2 | 2.24 × 10−3 | 7 | 7 | 1 | up |
| Sdc1 | 0 | 10 | 10 | 0 | up |
| Sdc3 | 0 | 10 | 10 | 0 | up |
| Cd44 | 0 | 8 | 8 | 0 | up |
| 2900073G15Rik | 0 | 6 | 6 | 0 | up |
| Cxcr4 | 0 | 6 | 6 | 0 | up |
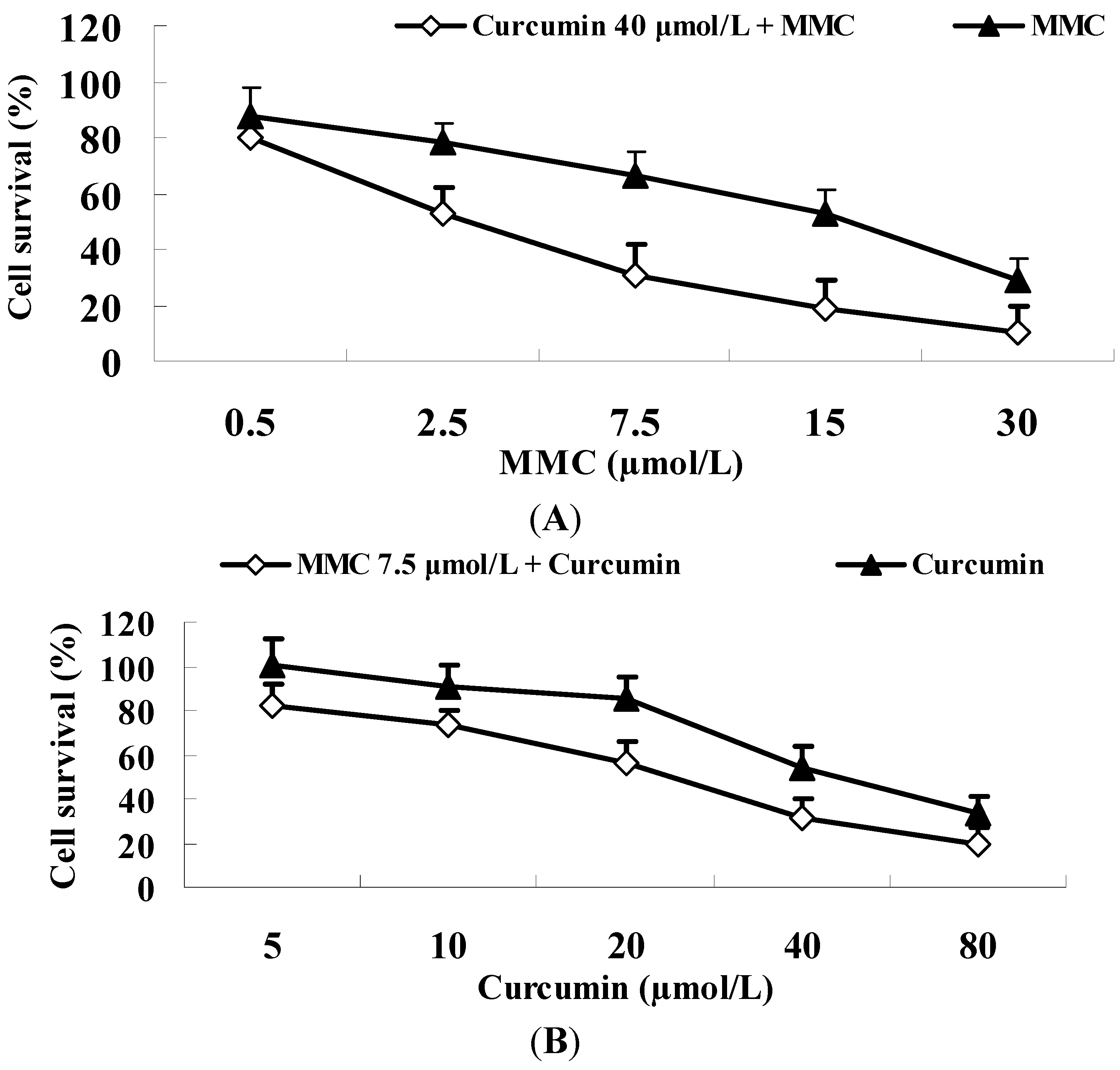
2.6. Curcumin Significantly Improves the Tumoricidal Effect of MMC in MDA-MB-231 Cells

2.7. Apoptosis via ERK Pathway in MCF-7 Cells

3. Experimental Section
3.1. Reagents and Cell Culture
3.2. Tumor Xenograft and Treatment
3.3. Microarray Detection
3.4. Data Analysis
3.5. Regulatory Network Construction of Weighted Genes
3.6. Quantitative RT-PCR (qRT-PCR)
3.7. Western Blot Analysis
3.8. Cell Growth Inhibition Test
3.9. Flow Cytometric Analysis
3.10. Hoechst 33258 Staining
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Curèiæ, M.G.; Stankoviæ, M.S.; Mrkaliæ, E.M.; Matoviæ, Z.D.; Bankoviæ, D.D.; Cvetkoviæ, D.M.; Daèiæ, D.S.; Markoviæ, S.D. Antiproliferative and proapoptotic activities of methanolic extracts from Ligustrum vulgare L. as an individual treatment and in combination with palladium complex. Int. J. Mol. Sci. 2012, 13, 2521–2534. [Google Scholar]
- Critchfield, K.L. Tailoring common treatment principles to fit individual personalities. J. Personal. Disord. 2012, 26, 108–125. [Google Scholar] [CrossRef]
- Montero, A.J.; Diaz-Montero, C.M.; Deutsch, Y.E.; Hurley, J.; Koniaris, L.G.; Rumboldt, T.; Yasir, S.; Jorda, M.; Garret-Mayer, E.; Avisar, E.; et al. Phase II study of neoadjuvant treatment with NOV-002 in combination with doxorubicin and cyclophosphamide followed by docetaxel in patients with HER-2 negative clinical stage II–IIIc breast cancer. Breast Cancer Res. Treat. 2012, 132, 215–223. [Google Scholar] [CrossRef]
- Secord, A.A.; Berchuck, A.; Higgins, R.V.; Nycum, L.R.; Kohler, M.F.; Puls, L.E.; Holloway, R.W.; Lewandowski, G.S.; Valea, F.A.; Havrilesky, L.J. A multicenter, randomized, phase II clinical trial to evaluate the efficacy and safety of combination docetaxel and carboplatin and sequential therapy with docetaxel then carboplatin in patients with recurrent platinum-sensitive ovarian cancer. Cancer 2012, 118, 3283–3293. [Google Scholar] [CrossRef]
- Wang, J.Y.; Swami, S.; Krishnan, A.V.; Feldman, D. Combination of calcitriol and dietary soy exhibits enhanced anticancer activity and increased hypercalcemic toxicity in a mouse xenograft model of prostate cancer. Prostate 2012, 72, 1628–1637. [Google Scholar] [CrossRef]
- Urba, S.; van Herpen, C.M.; Sahoo, T.P.; Shin, D.M.; Licitra, L.; Mezei, K.; Reuter, C.; Hitt, R.; Russo, F.; Chang, S.C.; et al. Pemetrexed in combination with cisplatin versus cisplatin monotherapy in patients with recurrent or metastatic head and neck cancer: Final results of a randomized, double-blind, placebo-controlled, phase III study. Cancer 2012, 118, 4694–4705. [Google Scholar] [CrossRef]
- Chou, T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmocol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
- Jia, J.; Zhu, F.; Ma, X.H.; Cao, Z.W.; Li, Y.X.; Chen, Y.Z. Mechanisms of drug combinations: Interaction and network perspecivs. Nat. Rev. 2009, 8, 111–128. [Google Scholar]
- Somasundaram, S.; Edmund, N.A.; Moore, D.T.; Small, G.W.; Shi, Y.Y.; Orlowski, R.Z. Dietary curcumin inhibits chemotherapyinduced apoptosis in models of human breast cancer. Cancer Res. 2002, 62, 3868–3875. [Google Scholar]
- Du, B.; Jiang, L.; Xia, Q.; Zhong, L. Synergistic inhibitory effects of curcumin and 5-fluorouracil on the growth of the human colon cancer cell line HT-29. Chemotherapy 2006, 52, 23–28. [Google Scholar] [CrossRef]
- Zhou, Q.M.; Wang, X.F.; Liu, X.J.; Zhang, H.; Lu, Y.Y.; Huang, S.; Su, S.B. Curcumin improves MMC-based chemotherapy by simultaneously sensitising cancer cells to MMC and reducing MMC-associated side-effects. Eur. J. Cancer 2011, 47, 2240–2247. [Google Scholar] [CrossRef]
- Zhou, Q.M.; Wang, X.F.; Liu, X.J.; Zhang, H.; Lu, Y.Y.; Su, S.B. Curcumin cooperates with mitomycin C to inhibit cell proliferation and induce cell cycle arrest in breast cancer models in vitro and in vivo. Acta Pharmacol. Sin. 2011, 32, 1402–1410. [Google Scholar] [CrossRef]
- Zhou, Q.M.; Zhang, H.; Lu, Y.Y.; Wang, X.F.; Su, S.B. Curcumin reduced the side effects of mitomycin C by inhibiting GRP58-mediated DNA cross-linking in MCF-7 breast cancer xenografts. Cancer Sci. 2009, 100, 2040–2045. [Google Scholar] [CrossRef]
- Tamm, I.; Schriever, F.; Dorken, B. Apoptosis: Implications of basic research for clinical oncology. Lancet Oncol. 2001, 2, 33–42. [Google Scholar] [CrossRef]
- Simstein, R.; Burow, M.; Parker, A.; Weldon, C.; Beckman, B. Apoptosis, chemoresistance, and breast cancer: Insights from the mcf-7 cell model system. Exp. Biol. Med. 2003, 228, 995–1003. [Google Scholar]
- Makin, G.; Dive, C. Modulating sensitivity to drug-induced apoptosis: The future for chemotherapy? Breast Cancer Res. 2001, 3, 150–153. [Google Scholar] [CrossRef]
- Roy, S.K.; Srivastava, R.K.; Shankar, S. Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of FOXO transcription factor, leading to cell cycle arrest and apoptosis in pancreatic cancer. J. Mol. Signal. 2010, 19, 5–10. [Google Scholar]
- Azzam, D.G.; Tay, J.W.; Greeve, M.A.; Harvey, J.M.; Bentel, J.M. ERK/MAPK regulation of the androgen responsiveness of breast cancer cells. Adv. Exp. Med. Biol. 2008, 617, 429–435. [Google Scholar]
- Chen, J.; Sun, L. Formononetin-induced apoptosis by activation of Ras/p38 mitogen-activated protein kinase in estrogen receptor-positive human breast cancer cells. Horm. Metab. Res. 2012, 44, 943–948. [Google Scholar] [CrossRef]
- Zhang, J.D.; Wiemann, S. KEGGgraph: A graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics 2009, 25, 1470–1471. [Google Scholar] [CrossRef]
- Pang, W.; Leng, X.; Lu, H.; Yang, H.; Song, N.; Tan, L.; Jiang, Y.; Guo, C. Depletion of intracellular zinc induces apoptosis of cultured hippocampal neurons through suppression of ERK signaling pathway and activation of caspase-3. Neurosci. Lett. 2013, 552, 140–145. [Google Scholar] [CrossRef]
- Lee, H.J.; Auh, Q.S.; Lee, Y.M.; Kang, S.K.; Chang, S.W.; Lee, D.S.; Kim, Y.C.; Kim, E.C. Growth inhibition and apoptosis-inducing effects of cudraflavone B in human oral cancer cells via MAPK, NF-κB, and SIRT1 signaling pathway. Planta Med. 2013, 79, 1298–1306. [Google Scholar] [CrossRef]
- Shi, J.; Wu, X.; Surma, M.; Vemula, S.; Zhang, L.; Yang, Y.; Kapur, R.; Wei, L. Distinct roles for ROCK1 and ROCK2 in the regulation of cell detachment. Cell Death Dis. 2013, 4, e483. [Google Scholar] [CrossRef]
- Green, B.D.; Jabbour, A.M.; Sandow, J.J.; Riffkin, C.D.; Masouras, D.; Daunt, C.P.; Salmanidis, M.; Brumatti, G.; Hemmings, B.A.; Guthridge, M.A.; et al. Akt1 is the principal Akt isoform regulating apoptosis in limiting cytokine concentrations. Cell Death Differ. 2013, 20, 1341–1349. [Google Scholar] [CrossRef]
- Yeh, C.T.; Yen, G.C. Induction of apoptosis by the anthocyanidins through regulation of Bcl-2 gene and activation of c-Jun N-terminal kinase cascade in hepatoma cells. J. Agric. Food Chem. 2005, 53, 1740–1749. [Google Scholar] [CrossRef]
- Volpato, M.; Seargent, J.; Loadman, P.M.; Phillips, R.M. Formation of DNA interstrand cross-links as a marker of mitomycin C bioreductive activation and chemosensitivity. Eur. J. Cancer 2005, 41, 1331–1338. [Google Scholar] [CrossRef]
- Chalasani, P.; Kurtin, S.; Dragovich, T. Response to a third-line mitomycin C (MMC)-based chemotherapy in a patient with metastatic pancreatic adenocarcinoma carrying germline BRCA2 mutation. JOP 2008, 9, 305–308. [Google Scholar]
- Wu, H.I.; Brown, J.A.; Dorie, M.J.; Lazzeroni, L.; Brown, J.M. Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C. Cancer Res. 2004, 64, 3940–3948. [Google Scholar] [CrossRef]
- Verwey, J.; de Vries, J.; Pinedo, H.M. Mitomycin C-induced renal toxicity, a dose-dependent side effect? Eur. J Cancer Clin. Oncol. 1987, 23, 195–199. [Google Scholar] [CrossRef]
- Campbell, F.C.; Collett, G.P. Chemopreventive properties of curcumin. Future Oncol. 2005, 1, 405–414. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Kumar, A.; Bharti, A.C. Anticancer potential of curcumin: Preclinical and clinical studies. Anticancer Res. 2003, 23, 363–398. [Google Scholar]
- Xia, Z.; Dickens, M.; Raingeaud, J.; Davis, R.J.; Greenberg, M.E. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 1995, 270, 1326–1331. [Google Scholar] [CrossRef]
- Lee, J.C.; Laydon, J.T.; McDonnell, P.C.; Gallagher, T.F.; Kumar, S.; Green, D.; McNulty, D.; Blumenthal, M.J.; Heys, J.R.; Landvatter, S.W.; et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 1994, 372, 739–746. [Google Scholar] [CrossRef]
- Han, J.; Lee, J.D.; Bibbs, L.; Ulevitch, R.J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994, 265, 808–811. [Google Scholar] [CrossRef]
- Lee, K.H.; Hyun, M.S.; Kim, J.R. Growth factor-dependent activation of the MAPK pathway in human pancreatic cancer: MEK/ERK and p38 MAP kinase interaction in uPA synthesis. Clin. Exp. Metastasis 2003, 20, 499–505. [Google Scholar] [CrossRef]
- Gao, J.; Tian, J.; Lv, Y.; Shi, F.; Kong, F.; Shi, H.; Zhao, L. Leptin induces functional activation of cyclooxygenase-2 through JAK2/STAT3, MAPK/ERK, and PI3K/AKT pathways in human endometrial cancer cells. Cancer Sci. 2009, 100, 389–395. [Google Scholar] [CrossRef]
- Lim, S.C.; Duong, H.Q.; Parajuli, K.R.; Han, S.I. Pro-apoptotic role of the MEK/ERK pathway in ursodeoxycholic acid-induced apoptosis in SNU601 gastric cancer cells. Oncol. Rep. 2012, 4, 1429–1434. [Google Scholar]
- Price, J.E.; Polyzos, A.; Zhang, R.D.; Daniels, L.M. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res. 1990, 50, 717–721. [Google Scholar]
- Chen, J.; Hui, E.; Ip, T.; Thompson, L.U. Dietary flaxseed enhances the inhibitory effect of Tamoxifen on the growth of estrogen-dependent human breast cancer (MCF-7) in nude mice. Clin. Cancer Res. 2004, 10, 7703–7711. [Google Scholar] [CrossRef]
- Jansen, R.; Greenbaum, D.; Gerstein, M. Relating whole-genome expression data with protein–protein interactions. Genome Res. 2002, 12, 37–46. [Google Scholar] [CrossRef]
- Spirin, V.; Mirny, LA. Protein complexes and functional modules in molecular networks. Proc. Natl. Acad. Sci. USA 2003, 100, 12123–12128. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhou, Q.-M.; Chen, Q.-L.; Du, J.; Wang, X.-F.; Lu, Y.-Y.; Zhang, H.; Su, S.-B. Synergistic Effect of Combinatorial Treatment with Curcumin and Mitomycin C on the Induction of Apoptosis of Breast Cancer Cells: A cDNA Microarray Analysis. Int. J. Mol. Sci. 2014, 15, 16284-16301. https://doi.org/10.3390/ijms150916284
Zhou Q-M, Chen Q-L, Du J, Wang X-F, Lu Y-Y, Zhang H, Su S-B. Synergistic Effect of Combinatorial Treatment with Curcumin and Mitomycin C on the Induction of Apoptosis of Breast Cancer Cells: A cDNA Microarray Analysis. International Journal of Molecular Sciences. 2014; 15(9):16284-16301. https://doi.org/10.3390/ijms150916284
Chicago/Turabian StyleZhou, Qian-Mei, Qi-Long Chen, Jia Du, Xiu-Feng Wang, Yi-Yu Lu, Hui Zhang, and Shi-Bing Su. 2014. "Synergistic Effect of Combinatorial Treatment with Curcumin and Mitomycin C on the Induction of Apoptosis of Breast Cancer Cells: A cDNA Microarray Analysis" International Journal of Molecular Sciences 15, no. 9: 16284-16301. https://doi.org/10.3390/ijms150916284
APA StyleZhou, Q.-M., Chen, Q.-L., Du, J., Wang, X.-F., Lu, Y.-Y., Zhang, H., & Su, S.-B. (2014). Synergistic Effect of Combinatorial Treatment with Curcumin and Mitomycin C on the Induction of Apoptosis of Breast Cancer Cells: A cDNA Microarray Analysis. International Journal of Molecular Sciences, 15(9), 16284-16301. https://doi.org/10.3390/ijms150916284




