Biomarkers in Alzheimer’s Disease Analysis by Mass Spectrometry-Based Proteomics
Abstract
:1. Introduction
2. Sources of Biomarkers
2.1. DNA-Based Biomarkers
2.2. Blood-Based Biomarkers
2.3. CSF-Based Biomarkers
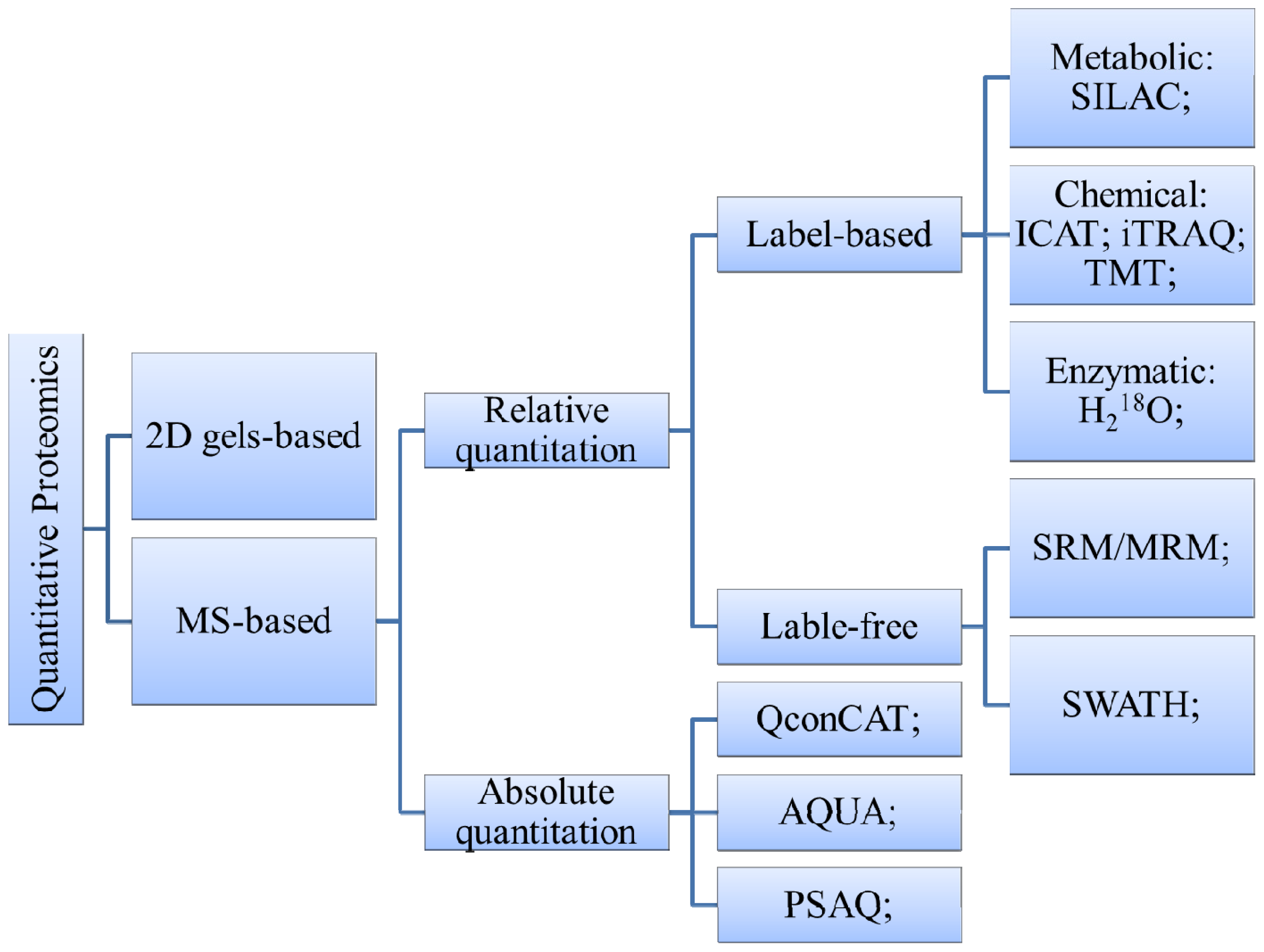
3. Technologies for Proteomic Analysis
3.1. Biomarkers Discovery
3.1.1. Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-Gel) Based Method
3.1.2. MS-Based Method
3.2. Biomarkers Verification and Validation
4. Conclusions and Future Perspectives
4.1. Peptidomics
4.2. Modification-Specific Proteomics
4.3. Metabonomics
4.4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Zhang, X.; Li, L.; Wei, D.; Yap, Y.; Chen, F. Moving cancer diagnostics from bench to bedside. Trends Biotechnol 2007, 25, 166–173. [Google Scholar]
- Rifai, N.; Gillette, M.A.; Carr, S.A. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat. Biotechnol 2006, 24, 971–983. [Google Scholar]
- Poste, G. Bring on the biomarkers. Nature 2011, 469, 156–157. [Google Scholar]
- Puntmann, V.O. How to guide on biomarkers: Biomarker definitions, Validation and applications with examples from cardiovascular disease. Postgrad. Med. J 2009, 85, 538–545. [Google Scholar]
- Anderson, D.C.; Kodukula, K. Biomarkers in pharmacology and drug discovery. Biochem. Pharmacol 2014, 87, 172–188. [Google Scholar]
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar]
- Ghidoni, R.; Paterlini, A.; Benussi, L. Translational proteomics in Alzheimer’s disease and related disorders. Clin. Biochem 2013, 46, 480–486. [Google Scholar]
- Podlesniy, P.; Figueiro-Silva, J.; Llado, A.; Antonell, A.; Sanchez-Valle, R.; Alcolea, D.; Lleo, A.; Molinuevo, J.L.; Serra, N.; Trullas, R. Low cerebrospinal fluid concentration of mitochondrial DNA in preclinical alzheimer disease. Ann. Neurol 2013, 74, 655–668. [Google Scholar]
- Blennow, K.; Hampel, H.; Weiner, M.; Zetterberg, H. Cerebrospinal fluid and plasma biomarkers in alzheimer disease. Nat. Rev. Neurol 2010, 6, 131–144. [Google Scholar]
- Hampel, H.; Blennow, K.; Shaw, L.M.; Hoessler, Y.C.; Zetterberg, H.; Trojanowski, J.Q. Total and phosphorylated tau protein as biological markers of Alzheimer’s disease. Exp. Gerontol 2010, 45, 30–40. [Google Scholar]
- Trojanowski, J.Q.; Vandeerstichele, H.; Korecka, M.; Clark, C.M.; Aisen, P.S.; Petersen, R.C.; Blennow, K.; Soares, H.; Simon, A.; Lewczuk, P.; et al. Update on the biomarker core of the Alzheimer’s disease neuroimaging initiative subjects. Alzheimer’s Dement 2010, 6, 230–238. [Google Scholar]
- Weiner, M.W.; Veitch, D.P.; Aisen, P.S.; Beckett, L.A.; Cairns, N.J.; Green, R.C.; Harvey, D.; Jack, C.R.; Jagust, W.; Liu, E.; et al. The Alzheimer’s disease neuroimaging initiative: A review of papers published since its inception. Alzheimer’s Dement 2012, 8, S1–S68. [Google Scholar]
- Whiteaker, J.R.; Lin, C.; Kennedy, J.; Hou, L.; Trute, M.; Sokal, I.; Yan, P.; Schoenherr, R.M.; Zhao, L.; Voytovich, U.J.; et al. A targeted proteomics–based pipeline for verification of biomarkers in plasma. Nat. Biotechnol 2011, 29, 625–634. [Google Scholar]
- Anderson, N.L. The clinical plasma proteome: A survey of clinical assays for proteins in plasma and serum. Clin. Chem 2010, 56, 177–185. [Google Scholar]
- Nature editorial office. Valid concerns. The reporting of candidate biomarkers for disease must be rigorous to drive translational research. Nature 2010, 463, 401–402.
- Prvulovic, D.; Hampel, H. Amyloid β (Aβ) and phospho-tau (p-tau) as diagnostic biomarkers in Alzheimer’s disease. Clin. Chem. Lab. Med 2011, 49, 367–374. [Google Scholar]
- Watt, A.D.; Perez, K.A.; Faux, N.G.; Pike, K.E.; Rowe, C.C.; Bourgeat, P.; Salvado, O.; Masters, C.L.; Villemagne, V.L.; Barnham, K.J. Increasing the predictive accuracy of amyloid-beta blood-borne biomarkers in Alzheimer’s disease. J. Alzheimer’s Dis 2011, 24, 47–59. [Google Scholar]
- Rosenmann, H. CSF biomarkers for amyloid and tau pathology in Alzheimer’s disease. J. Mol. Neurosci 2012, 47, 1–14. [Google Scholar]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.C.; Yates, J.R., III. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev 2013, 113, 2343–2394. [Google Scholar]
- Ramaswamy, S.; Perou, C.M. DNA microarrays in breast cancer: The promise of personalised medicine. Lancet 2003, 361, 1576–1577. [Google Scholar]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteomics 2009, 6, 267–283. [Google Scholar]
- Kit, H.A.; Nielsen, H.M.; Tost, J. DNA methylation based biomarkers: Practical considerations and applications. Biochimie 2012, 94, 2314–2337. [Google Scholar]
- Thambisetty, M.; Lovestone, S. Blood-based biomarkers of Alzheimer’s disease: Challenging but feasible. Biomark. Med 2010, 4, 65–79. [Google Scholar]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.L.; Schmittgen, T.D.; et al. Detection of microrna expression in human peripheral blood microvesicles. PLoS One 2008, 3. [Google Scholar] [CrossRef]
- Muller, M.; Kuiperij, H.B.; Claassen, J.A.; Kusters, B.; Verbeek, M.M. MicroRNAs in Alzheimer’s disease: Differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol. Aging 2014, 35, 152–158. [Google Scholar]
- Lukiw, W.J. Micro–RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport 2007, 18, 297–300. [Google Scholar]
- Wang, X.; Liu, P.; Zhu, H.; Xu, Y.; Ma, C.; Dai, X.; Huang, L.; Liu, Y.; Zhang, L.; Qin, C. MiR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res. Bull 2009, 80, 268–273. [Google Scholar]
- Lukiw, W.J.; Alexandrov, P.N.; Zhao, Y.; Hill, J.M.; Bhattacharjee, S. Spreading of Alzheimer’s disease inflammatory signaling through soluble micro-RNA. Neuroreport 2012, 23, 621–626. [Google Scholar]
- Alexandrov, P.N.; Dua, P.; Hill, J.M.; Bhattacharjee, S.; Zhao, Y.; Lukiw, W.J. MicroRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int. J. Biochem. Mol. Biol 2012, 3, 365–373. [Google Scholar]
- Wang, W.X.; Rajeev, B.W.; Stromberg, A.J.; Ren, N.; Tang, G.; Huang, Q.; Rigoutsos, I.; Nelson, P.T. The expression of microRNA MIR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci 2008, 28, 1213–1223. [Google Scholar]
- Geekiyanage, H.; Jicha, G.A.; Nelson, P.T.; Chan, C. Blood serum miRNA: Non-invasive biomarkers for Alzheimer’s disease. Exp. Neurol 2012, 235, 491–496. [Google Scholar]
- Liu, W.; Liu, C.; Zhu, J.; Shu, P.; Yin, B.; Gong, Y.; Qiang, B.; Yuan, J.; Peng, X. MicroRNA-16 targets amyloid precursor protein to potentially modulate alzheimer’s-associated pathogenesis in samp8 mice. Neurobiol. Aging 2012, 33, 522–534. [Google Scholar]
- Cogswell, J.P.; Ward, J.; Taylor, I.A.; Waters, M.; Shi, Y.; Cannon, B.; Kelnar, K.; Kemppainen, J.; Brown, D.; Chen, C.; et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimer’s Dis 2008, 14, 27–41. [Google Scholar]
- Schonrock, N.; Matamales, M.; Ittner, L.M.; Gotz, J. MicroRNA networks surrounding APP and amyloid-β metabolism-implications for Alzheimer’s disease. Exp. Neurol 2012, 235, 447–454. [Google Scholar]
- Tan, L.; Yu, J.T.; Hu, N. Non-coding RNAs in Alzheimer’s disease. Mol. Neurobiol 2013, 47, 382–393. [Google Scholar]
- Sheinerman, K.S.; Tsivinsky, V.G.; Abdullah, L.; Crawford, F.; Umansky, S.R. Plasma microRNA biomarkers for detection of mild cognitive impairment: Biomarker validation study. Aging 2013, 5, 925–938. [Google Scholar]
- Sheinerman, K.S.; Tsivinsky, V.G.; Crawford, F.; Mullan, M.J.; Abdullah, L.; Umansky, S.R. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging 2012, 4, 590–605. [Google Scholar]
- Kiko, T.; Nakagawa, K.; Tsuduki, T.; Furukawa, K.; Arai, H.; Miyazawa, T. MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J. Alzheimer’s Dis 2014, 39, 253–259. [Google Scholar]
- Thadikkaran, L.; Siegenthaler, M.A.; Crettaz, D.; Queloz, P.A.; Schneider, P.; Tissot, J.D. Recent advances in blood-related proteomics. Proteomics 2005, 5, 3019–3034. [Google Scholar]
- Zürbig, P.; Jahn, H. Use of proteomic methods in the analysis of human body fluids in Alzheimer research. Electrophoresis 2012, 33, 3617–3630. [Google Scholar]
- Mayeux, R.; Schupf, N. Blood-based biomarkers for Alzheimer’s disease: Plasma Aβ40 and Aβ42, and genetic variants. Neurobiol. Aging 2011, 32, S10–S19. [Google Scholar]
- Issaq, H.J.; Xiao, Z.; Veenstra, T.D. Serum and plasma proteomics. Chem. Rev 2007, 107, 3601–3620. [Google Scholar]
- Koyama, A.; Okereke, O.I.; Yang, T.; Blacker, D.; Selkoe, D.J.; Grodstein, F. Plasma amyloid-β as a predictor of dementia and cognitive decline: A systematic review and meta-analysis. Arch. Neurol 2012, 69, 824–831. [Google Scholar]
- Gupta, V.B.; Laws, S.M.; Villemagne, V.L.; Ames, D.; Bush, A.I.; Ellis, K.A.; Lui, J.K.; Masters, C.; Rowe, C.C.; Szoeke, C.; et al. Plasma apolipoprotein E and alzheimer disease risk. Neurology 2011, 76, 1091–1098. [Google Scholar]
- Yang, M.H.; Yang, Y.H.; Lu, C.Y.; Jong, S.B.; Chen, L.J.; Lin, Y.F.; Wu, S.J.; Chu, P.Y.; Chung, T.W.; Tyan, Y.C. Activity-dependent neuroprotector homeobox protein: A candidate protein identified in serum as diagnostic biomarker for Alzheimer’s disease. J. Proteomics 2012, 75, 3617–3629. [Google Scholar]
- Du, Y.; Dodel, R.C.; Eastwood, B.J.; Bales, K.R.; Gao, F.; Lohmuller, F.; Muller, U.; Kurz, A.; Zimmer, R.; Evans, R.M.; et al. Association of an interleukin 1α polymorphism with Alzheimer’s disease. Neurology 2000, 55, 480–483. [Google Scholar]
- Adams, S.; Harold, A.; Bremner, W.; Bhatti, A. Immediate post-parathyroidectomy stridor resolved with intravenous calcium. BMJ Case Rep 2009, 2009. [Google Scholar] [CrossRef]
- Lambert, J.C.; Heath, S.; Even, G.; Campion, D.; Sleegers, K.; Hiltunen, M.; Combarros, O.; Zelenika, D.; Bullido, M.J.; Tavernier, B.; et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet 2009, 41, 1094–1099. [Google Scholar]
- Oishi, M.; Mochizuki, Y.; Yoshihashi, H.; Takasu, T.; Nakano, E. Laboratory examinations correlated with severity of dementia. Ann. Clin. Lab. Sci 1996, 26, 340–345. [Google Scholar]
- Karsidag, T.; Tuzun, S.; Kemik, A.S.; Purisa, S.; Unlu, A. Alpha-1 protease inhibitor and antichymotrypsin levels in acute pancreatitis. Turkish J. Trauma Emerg. Surg 2012, 18, 195–199. [Google Scholar]
- Guan, F.; Gu, J.; Hu, F.; Zhu, Y.; Wang, W. Association between α1-antichymotrypsin signal peptide-15A/T polymorphism and the risk of Alzheimer’s disease: A meta-analysis. Mol. Biol. Rep 2012, 39, 6661–6669. [Google Scholar]
- Zamostiano, R.; Pinhasov, A.; Gelber, E.; Steingart, R.A.; Seroussi, E.; Giladi, E.; Bassan, M.; Wollman, Y.; Eyre, H.J.; Mulley, J.C.; et al. Cloning and characterization of the human activity-dependent neuroprotective protein. J. Biol. Chem 2001, 276, 708–714. [Google Scholar]
- Fernandez-Montesinos, R.; Torres, M.; Baglietto-Vargas, D.; Gutierrez, A.; Gozes, I.; Vitorica, J.; Pozo, D. Activity-dependent neuroprotective protein (ADNP) expression in the amyloid precursor protein/presenilin 1 mouse model of Alzheimer’s disease. J. Mol. Neurosci 2010, 41, 114–120. [Google Scholar]
- Shaw, L.M.; Korecka, M.; Clark, C.M.; Lee, V.M.; Trojanowski, J.Q. Biomarkers of neurodegeneration for diagnosis and monitoring therapeutics. Nat. Rev. Drug Discov 2007, 6, 295–303. [Google Scholar]
- Sonnen, J.A.; Keene, C.D.; Montine, K.S.; Li, G.; Peskind, E.R.; Zhang, J.; Montine, T.J. Biomarkers for Alzheimer’s disease. Expert Rev. Neurother 2007, 7, 1021–1028. [Google Scholar]
- Clark, C.M.; Davatzikos, C.; Borthakur, A.; Newberg, A.; Leight, S.; Lee, V.M.; Trojanowski, J.Q. Biomarkers for early detection of Alzheimer pathology. Neurosignals 2008, 16, 11–18. [Google Scholar]
- De Almeida, S.M.; Shumaker, S.D.; LeBlanc, S.K.; Delaney, P.; Marquie-Beck, J.; Ueland, S.; Alexander, T.; Ellis, R.J. Incidence of post-dural puncture headache in research volunteers. Headache 2011, 51, 1503–1510. [Google Scholar]
- Oreskovic, D.; Klarica, M. The formation of cerebrospinal fluid: Nearly a hundred years of interpretations and misinterpretations. Brain Res. Rev 2010, 64, 241–262. [Google Scholar] [Green Version]
- Kroksveen, A.C.; Opsahl, J.A.; Aye, T.T.; Ulvik, R.J.; Berven, F.S. Proteomics of human cerebrospinal fluid: Discovery and verification of biomarker candidates in neurodegenerative diseases using quantitative proteomics. J. Proteomics 2011, 74, 371–388. [Google Scholar]
- Miller, L.L.; Bale, W.F. Synthesis of all plasma protein fractions except gamma globulins by the liver; The use of zone electrophoresis and lysine-epsilon-C14 to define the plasma proteins synthesized by the isolated perfused liver. J. Exp. Med 1954, 99, 125–132. [Google Scholar]
- Blennow, K.; Wallin, A.; Fredman, P.; Karlsson, I.; Gottfries, C.G.; Svennerholm, L. Blood-brain barrier disturbance in patients with Alzheimer’s disease is related to vascular factors. Acta Neurol. Scand 1990, 81, 323–326. [Google Scholar]
- Chalbot, S.; Zetterberg, H.; Blennow, K.; Fladby, T.; Grundke-Iqbal, I.; Iqbal, K. Cerebrospinal fluid secretory Ca2+-Dependent phospholipase A2 activity is increased in alzheimer disease. Clin. Chem 2009, 55, 2171–2179. [Google Scholar]
- Chalbot, S.; Zetterberg, H.; Blennow, K.; Fladby, T.; Grundke-Iqbal, I.; Iqbal, K. Cerebrospinal fluid secretory Ca2+-dependent phospholipase A2 activity: A biomarker of blood-cerebrospinal fluid barrier permeability. Neurosci. Lett 2010, 478, 179–183. [Google Scholar]
- Laterza, O.F.; Modur, V.R.; Crimmins, D.L.; Olander, J.V.; Landt, Y.; Lee, J.M.; Ladenson, J.H. Identification of novel brain biomarkers. Clin. Chem 2006, 52, 1713–1721. [Google Scholar]
- Lee, J.M.; Blennow, K.; Andreasen, N.; Laterza, O.; Modur, V.; Olander, J.; Gao, F.; Ohlendorf, M.; Ladenson, J.H. The brain injury biomarker VLP-1 is increased in the cerebrospinal fluid of Alzheimer disease patients. Clin. Chem 2008, 54, 1617–1623. [Google Scholar]
- Weingarten, M.D.; Lockwood, A.H.; Hwo, S.Y.; Kirschner, M.W. A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862. [Google Scholar]
- Blennow, K.; Zetterberg, H. Cerebrospinal fluid biomarkers for Alzheimer’s disease. J. Alzheimer’s Dis 2009, 18, 413–417. [Google Scholar]
- Blom, E.S.; Giedraitis, V.; Zetterberg, H.; Fukumoto, H.; Blennow, K.; Hyman, B.T.; Irizarry, M.C.; Wahlund, L.O.; Lannfelt, L.; Ingelsson, M. Rapid progression from mild cognitive impairment to Alzheimer’s disease in subjects with elevated levels of tau in cerebrospinal fluid and the APOE epsilon4/epsilon4 genotype. Dement. Geriatr. Cogn. Disord 2009, 27, 458–464. [Google Scholar]
- Samgard, K.; Zetterberg, H.; Blennow, K.; Hansson, O.; Minthon, L.; Londos, E. Cerebrospinal fluid total tau as a marker of Alzheimer’s disease intensity. Int. J. Geriatr. Psychiatry 2010, 25, 403–410. [Google Scholar]
- Wallin, A.K.; Hansson, O.; Blennow, K.; Londos, E.; Minthon, L. Can CSF biomarkers or pre-treatment progression rate predict response to cholinesterase inhibitor treatment in Alzheimer’s disease? Int. J. Geriatr. Psychiatry 2009, 24, 638–647. [Google Scholar]
- Liu, Q.; Xie, F.; Siedlak, S.L.; Nunomura, A.; Honda, K.; Moreira, P.I.; Zhua, X.; Smith, M.A.; Perry, G. Neurofilament proteins in neurodegenerative diseases. Cell Mol. Life Sci 2004, 61, 3057–3075. [Google Scholar]
- Brettschneider, J.; Petzold, A.; Schottle, D.; Claus, A.; Riepe, M.; Tumani, H. The neurofilament heavy chain (NFH) in the cerebrospinal fluid diagnosis of Alzheimer’s disease. Dement. Geriatr. Cogn. Disord 2006, 21, 291–295. [Google Scholar]
- Cheon, M.S.; Kim, S.H.; Fountoulakis, M.; Lubec, G. Heart type fatty acid binding protein (H-FABP) is decreased in brains of patients with Down syndrome and Alzheimer’s disease. J. Neural. Transm. Suppl 2003, 225–234. [Google Scholar]
- Perrin, R.J.; Craig-Schapiro, R.; Malone, J.P.; Shah, A.R.; Gilmore, P.; Davis, A.E.; Roe, C.M.; Peskind, E.R.; Li, G.; Galasko, D.R.; et al. Identification and validation of novel cerebrospinal fluid biomarkers for staging early Alzheimer’s disease. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Mattsson, N.; Savman, K.; Osterlundh, G.; Blennow, K.; Zetterberg, H. Converging molecular pathways in human neural development and degeneration. Neurosci. Res 2010, 66, 330–332. [Google Scholar]
- Davidsson, P.; Blennow, K. Neurochemical dissection of synaptic pathology in Alzheimer’s disease. Int. Psychogeriatr 1998, 10, 11–23. [Google Scholar]
- Thorsell, A.; Bjerke, M.; Gobom, J.; Brunhage, E.; Vanmechelen, E.; Andreasen, N.; Hansson, O.; Minthon, L.; Zetterberg, H.; Blennow, K. Neurogranin in cerebrospinal fluid as a marker of synaptic degeneration in Alzheimer’s disease. Brain Res 2010, 1362, 13–22. [Google Scholar]
- Herrmann, M.; Ebert, A.D.; Galazky, I.; Wunderlich, M.T.; Kunz, W.S.; Huth, C. Neurobehavioral outcome prediction after cardiac surgery: Role of neurobiochemical markers of damage to neuronal and glial brain tissue. Stroke 2000, 31, 645–650. [Google Scholar]
- Peskind, E.R.; Griffin, W.S.; Akama, K.T.; Raskind, M.A.; van Eldik, L.J. Cerebrospinal fluid S-100B is elevated in the earlier stages of Alzheimer’s disease. Neurochem. Int 2001, 39, 409–413. [Google Scholar]
- Jesse, S.; Steinacker, P.; Cepek, L.; von Arnim, C.A.; Tumani, H.; Lehnert, S.; Kretzschmar, H.A.; Baier, M.; Otto, M. Glial fibrillary acidic protein and protein S-100B: Different concentration pattern of glial proteins in cerebrospinal fluid of patients with Alzheimer’s disease and Creutzfeldt-Jakob disease. J. Alzheimer’s Dis 2009, 17, 541–551. [Google Scholar]
- Wang, H.; Li, R.; Shen, Y. Beta-secretase: Its biology as a therapeutic target in diseases. Trends Pharmacol. Sci 2013, 34, 215–225. [Google Scholar]
- Snyder, H.M.; Carrillo, M.C.; Grodstein, F.; Henriksen, K.; Jeromin, A.; Lovestone, S.; Mielke, M.M.; O’Bryant, S.; Sarasa, M.; Sjogren, M.; et al. Developing novel blood-based biomarkers for Alzheimer’s disease. Alzheimer’s Dement 2014, 10, 109–114. [Google Scholar]
- Hilpert, H.; Guba, W.; Woltering, T.J.; Wostl, W.; Pinard, E.; Mauser, H.; Mayweg, A.V.; Rogers-Evans, M.; Humm, R.; Krummenacher, D.; et al. βsecretase (BACE1) inhibitors with high in vivo efficacy suitable for clinical evaluation in Alzheimer’s disease. J. Med. Chem 2013, 56, 3980–3995. [Google Scholar]
- Altelaar, A.F.; Munoz, J.; Heck, A.J. Next-generation proteomics: Towards an integrative view of proteome dynamics. Nat. Rev. Genet 2013, 14, 35–48. [Google Scholar]
- Chevallet, M.; Luche, S.; Diemer, H.; Strub, J.M.; van Dorsselaer, A.; Rabilloud, T. Sweet silver: A formaldehyde-free silver staining using aldoses as developing agents, with enhanced compatibility with mass spectrometry. Proteomics 2008, 8, 4853–4861. [Google Scholar] [Green Version]
- Fey, S.J.; Larsen, P.M. 2D or not 2D. Two-dimensional gel electrophoresis. Curr. Opin. Chem. Biol 2001, 5, 26–33. [Google Scholar]
- Song, F.; Poljak, A.; Smythe, G.A.; Sachdev, P. Plasma biomarkers for mild cognitive impairment and Alzheimer’s disease. Brain Res. Rev 2009, 61, 69–80. [Google Scholar]
- Apweiler, R.; Aslanidis, C.; Deufel, T.; Gerstner, A.; Hansen, J.; Hochstrasser, D.; Kellner, R.; Kubicek, M.; Lottspeich, F.; Maser, E.; et al. Approaching clinical proteomics: Current state and future fields of application in fluid proteomics. Clin. Chem. Lab. Med 2009, 47, 724–744. [Google Scholar]
- Hye, A.; Lynham, S.; Thambisetty, M.; Causevic, M.; Campbell, J.; Byers, H.L.; Hooper, C.; Rijsdijk, F.; Tabrizi, S.J.; Banner, S.; et al. Proteome-based plasma biomarkers for Alzheimer’s disease. Brain 2006, 129, 3042–3050. [Google Scholar]
- Zhang, R.; Barker, L.; Pinchev, D.; Marshall, J.; Rasamoelisolo, M.; Smith, C.; Kupchak, P.; Kireeva, I.; Ingratta, L.; Jackowski, G. Mining biomarkers in human sera using proteomic tools. Proteomics 2004, 4, 244–256. [Google Scholar]
- Thambisetty, M.; Hye, A.; Foy, C.; Daly, E.; Glover, A.; Cooper, A.; Simmons, A.; Murphy, D.; Lovestone, S. Proteome-based identification of plasma proteins associated with hippocampal metabolism in early Alzheimer’s disease. J. Neurol 2008, 255, 1712–1720. [Google Scholar]
- Thambisetty, M.; Simmons, A.; Hye, A.; Campbell, J.; Westman, E.; Zhang, Y.; Wahlund, L.O.; Kinsey, A.; Causevic, M.; Killick, R.; et al. Plasma biomarkers of brain atrophy in Alzheimer’s disease. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Thambisetty, M.; An, Y.; Kinsey, A.; Koka, D.; Saleem, M.; Guntert, A.; Kraut, M.; Ferrucci, L.; Davatzikos, C.; Lovestone, S.; et al. Plasma clusterin concentration is associated with longitudinal brain atrophy in mild cognitive impairment. Neuroimage 2012, 59, 212–217. [Google Scholar]
- Henkel, A.W.; Muller, K.; Lewczuk, P.; Muller, T.; Marcus, K.; Kornhuber, J.; Wiltfang, J. Multidimensional plasma protein separation technique for identification of potential Alzheimer’s disease plasma biomarkers: A pilot study. J. Neural Transm 2012, 119, 779–788. [Google Scholar]
- Buts, K.; Michielssens, S.; Hertog, M.L.; Hayakawa, E.; Cordewener, J.; America, A.H.; Nicolai, B.M.; Carpentier, S.C. Improving the identification rate of data independent label-free quantitative proteomics experiments on non-model crops: A case study on apple fruit. J. Proteomics 2014. [Google Scholar] [CrossRef]
- Cole, L.M.; Bluff, J.E.; Carolan, V.A.; Paley, M.N.; Tozer, G.M.; Clench, M.R. MALDI-MSI and label free LC-ESI-MS/MS shotgun proteomics to investigate protein induction in a murine fibrosarcoma model following treatment with a vascular disrupting agent. Proteomics 2014, 14, 890–903. [Google Scholar]
- Mohayeji, M.; Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; Samperi, R.; Stampachiacchiere, S.; Toorchi, M.; Lagana, A. Heterosis profile of sunflower leaves: A label free proteomics approach. J. Proteomics 2014, 99, 101–110. [Google Scholar]
- Hakimi, A.; Auluck, J.; Jones, G.D.; Ng, L.L.; Jones, D.J. Assessment of reproducibility in depletion and enrichment workflows for plasma proteomics using label-free quantitative data-independent LC–MS. Proteomics 2014, 14, 4–13. [Google Scholar]
- Liu, X.; Hu, Y.; Pai, P.J.; Chen, D.; Lam, H. Label-free quantitative proteomics analysis of antibiotic response in staphylococcus aureus to oxacillin. J. Proteome Res 2014, 13, 1223–1233. [Google Scholar]
- Zhang, J.; Goodlett, D.R.; Quinn, J.F.; Peskind, E.; Kaye, J.A.; Zhou, Y.; Pan, C.; Yi, E.; Eng, J.; Wang, Q.; et al. Quantitative proteomics of cerebrospinal fluid from patients with alzheimer disease. J. Alzheimer’s Dis 2005, 7, 125–133. [Google Scholar]
- Fu, Y.J.; Xiong, S.; Lovell, M.A.; Lynn, B.C. Quantitative proteomic analysis of mitochondria in aging PS-1 transgenic mice. Cell Mol. Neurobiol 2009, 29, 649–664. [Google Scholar]
- Choe, L.; D’Ascenzo, M.; Relkin, N.R.; Pappin, D.; Ross, P.; Williamson, B.; Guertin, S.; Pribil, P.; Lee, K.H. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer’s disease. Proteomics 2007, 7, 3651–3660. [Google Scholar]
- Beynon, R.J.; Doherty, M.K.; Pratt, J.M.; Gaskell, S.J. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2005, 2, 587–589. [Google Scholar]
- Ding, C.; Li, Y.; Kim, B.J.; Malovannaya, A.; Jung, S.Y.; Wang, Y.; Qin, J. Quantitative analysis of cohesin complex stoichiometry and SMC3 modification-dependent protein interactions. J. Proteome Res 2011, 10, 3652–3659. [Google Scholar]
- Chen, J.; Wang, M.; Turko, I.V. Mass spectrometry quantification of clusterin in the human brain. Mol. Neurodegener 2012, 7, 41. [Google Scholar] [CrossRef]
- Chen, J.; Wang, M.; Turko, I.V. Quantification of amyloid precursor protein isoforms using quantification concatamer internal standard. Anal. Chem 2013, 85, 303–307. [Google Scholar]
- Kingsmore, S.F. Multiplexed protein measurement: Technologies and applications of protein and antibody arrays. Nat. Rev. Drug Discov 2006, 5, 310–320. [Google Scholar]
- Vitzthum, F.; Behrens, F.; Anderson, N.L.; Shaw, J.H. Proteomics: From basic research to diagnostic application. A review of requirements & needs. J. Proteome Res 2005, 4, 1086–1097. [Google Scholar]
- Lista, S.; Faltraco, F.; Prvulovic, D.; Hampel, H. Blood and plasma-based proteomic biomarker research in Alzheimer’s disease. Prog. Neurobiol 2013, 101, 1–17. [Google Scholar]
- Wei, X.; Li, L. Mass spectrometry-based proteomics and peptidomics for biomarker discovery in neurodegenerative diseases. Int. J. Clin. Exp. Pathol 2009, 2, 132–148. [Google Scholar]
- Khorvash, F.; Abdi, F.; Dialami, K.; Kooshki, A.M. Can serum procalcitonin and C-reactive protein as nosocomial infection markers in hospitalized patients without localizing signs? J. Res. Med. Sci 2011, 16, 1280–1285. [Google Scholar]
- Abdi, F.; Quinn, J.F.; Jankovic, J.; McIntosh, M.; Leverenz, J.B.; Peskind, E.; Nixon, R.; Nutt, J.; Chung, K.; Zabetian, C.; et al. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J. Alzheimer’s Dis 2006, 9, 293–348. [Google Scholar]
- Aebersold, R.; Burlingame, A.L.; Bradshaw, R.A. Western blots versus selected reaction monitoring assays: Time to turn the tables? Mol. Cell Proteomics 2013, 12, 2381–2382. [Google Scholar]
- Shi, M.; Caudle, W.M.; Zhang, J. Biomarker discovery in neurodegenerative diseases: A proteomic approach. Neurobiol. Dis 2009, 35, 157–164. [Google Scholar]
- Zhang, K.; Schrag, M.; Crofton, A.; Trivedi, R.; Vinters, H.; Kirsch, W. Targeted proteomics for quantification of histone acetylation in Alzheimer’s disease. Proteomics 2012, 12, 1261–1268. [Google Scholar]
- Pannee, J.; Portelius, E.; Oppermann, M.; Atkins, A.; Hornshaw, M.; Zegers, I.; Hojrup, P.; Minthon, L.; Hansson, O.; Zetterberg, H.; et al. A selected reaction monitoring (SRM)-based method for absolute quantification of Aβ38, Aβ40, and Aβ42 in cerebrospinal fluid of Alzheimer’s disease patients and healthy controls. J. Alzheimer’s Dis 2013, 33, 1021–1032. [Google Scholar]
- Menschaert, G.; Vandekerckhove, T.T.; Baggerman, G.; Schoofs, L.; Luyten, W.; van Criekinge, W. Peptidomics coming of age: A review of contributions from a bioinformatics angle. J. Proteome Res 2010, 9, 2051–2061. [Google Scholar]
- Tinoco, A.D.; Saghatelian, A. Investigating endogenous peptides and peptidases using peptidomics. Biochemistry 2011, 50, 7447–7461. [Google Scholar]
- Monacelli, F.; Borghi, R.; Pacini, D.; Serrati, C.; Traverso, N.; Odetti, P. Pentosidine determination in CSF: A potential biomarker of alzheimer’disease? Clin. Chem. Lab. Med 2014, 52, 1–4. [Google Scholar]
- Kingwell, K. Alzheimer disease: CSF levels of mitochondrial DNA—A new biomarker for preclinical alzheimer disease? Nat. Rev. Neurol 2013, 9, 420. [Google Scholar]
- Ertekin-Taner, N. Alzheimer disease: The quest for Alzheimer disease genes—Focus on CSF tau. Nat. Rev. Neurol 2013, 9, 368–370. [Google Scholar]
- Schmidt, C.; Artjomova, S.; Hoeschel, M.; Zerr, I. CSF prion protein concentration and cognition in patients with Alzheimer disease. Prion 2013, 7, 229–234. [Google Scholar]
- Wijte, D.; McDonnell, L.A.; Balog, C.I.; Bossers, K.; Deelder, A.M.; Swaab, D.F.; Verhaagen, J.; Mayboroda, O.A. A novel peptidomics approach to detect markers of Alzheimer’s disease in cerebrospinal fluid. Methods 2012, 56, 500–507. [Google Scholar]
- Ballatore, C.; Lee, V.M.-Y.; Trojanowski, J.Q. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci 2007, 8, 663–672. [Google Scholar]
- Ando, K.; Dourlen, P.; Sambo, A.V.; Bretteville, A.; Belarbi, K.; Vingtdeux, V.; Eddarkaoui, S.; Drobecq, H.; Ghestem, A.; Begard, S.; et al. Tau pathology modulates PIN1 post-translational modifications and may be relevant as biomarker. Neurobiol. Aging 2013, 34, 757–769. [Google Scholar]
- Opii, W.O.; Joshi, G.; Head, E.; Milgram, N.W.; Muggenburg, B.A.; Klein, J.B.; Pierce, W.M.; Cotman, C.W.; Butterfield, D.A. Proteomic identification of brain proteins in the canine model of human aging following a long-term treatment with antioxidants and a program of behavioral enrichment: Relevance to Alzheimer’s disease. Neurobiol. Aging 2008, 29, 51–70. [Google Scholar]
- Zhang, J. Proteomics of human cerebrospinal fluid—The good, the bad, and the ugly. PROTEOMICS-Clin. Appl 2007, 1, 805–819. [Google Scholar]
- Deja, S.; Barg, E.; Mlynarz, P.; Basiak, A.; Willak-Janc, E. 1H NMR-based metabolomics studies of urine reveal differences between type 1 diabetic patients with high and low HbAc1 values. J Pharm. Biomed. Anal 2013, 83, 43–48. [Google Scholar]
- Huang, Y.; Tian, Y.; Li, G.; Li, Y.; Yin, X.; Peng, C.; Xu, F.; Zhang, Z. Discovery of safety biomarkers for realgar in rat urine using UFLC-IT-TOF/MS and 1H NMR based metabolomics. Anal. Bioanal. Chem 2013, 405, 4811–4822. [Google Scholar]
- Fukuhara, K.; Ohno, A.; Ota, Y.; Senoo, Y.; Maekawa, K.; Okuda, H.; Kurihara, M.; Okuno, A.; Niida, S.; Saito, Y.; et al. NMR-based metabolomics of urine in a mouse model of Alzheimer’s disease: Identification of oxidative stress biomarkers. J. Clin. Biochem. Nutr 2013, 52, 133–138. [Google Scholar]
- Li, N.J.; Liu, W.T.; Li, W.; Li, S.Q.; Chen, X.H.; Bi, K.S.; He, P. Plasma metabolic profiling of Alzheimer’s disease by liquid chromatography/mass spectrometry. Clin. Biochem 2010, 43, 992–997. [Google Scholar]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. Hmdb: A knowledgebase for the human metabolome. Nucleic Acids Res 2009, 37, 603–610. [Google Scholar]
- Gika, H.G.; Theodoridis, G.A.; Plumb, R.S.; Wilson, I.D. Current practice of liquid chromatography-mass spectrometry in metabolomics and metabonomics. J. Pharm. Biomed. Anal 2014, 87, 12–25. [Google Scholar]

© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, Y.; Qing, H.; Deng, Y. Biomarkers in Alzheimer’s Disease Analysis by Mass Spectrometry-Based Proteomics. Int. J. Mol. Sci. 2014, 15, 7865-7882. https://doi.org/10.3390/ijms15057865
Liu Y, Qing H, Deng Y. Biomarkers in Alzheimer’s Disease Analysis by Mass Spectrometry-Based Proteomics. International Journal of Molecular Sciences. 2014; 15(5):7865-7882. https://doi.org/10.3390/ijms15057865
Chicago/Turabian StyleLiu, Yahui, Hong Qing, and Yulin Deng. 2014. "Biomarkers in Alzheimer’s Disease Analysis by Mass Spectrometry-Based Proteomics" International Journal of Molecular Sciences 15, no. 5: 7865-7882. https://doi.org/10.3390/ijms15057865
APA StyleLiu, Y., Qing, H., & Deng, Y. (2014). Biomarkers in Alzheimer’s Disease Analysis by Mass Spectrometry-Based Proteomics. International Journal of Molecular Sciences, 15(5), 7865-7882. https://doi.org/10.3390/ijms15057865



