Development of Proteomics-Based Fungicides: New Strategies for Environmentally Friendly Control of Fungal Plant Diseases
Abstract
:1. Introduction
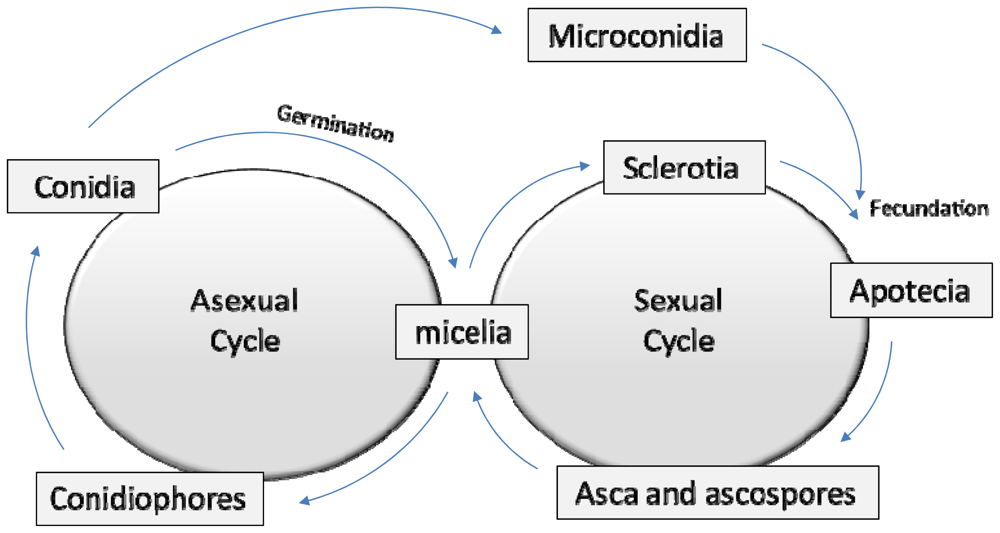
2. A Brief Fungal Biology Review
3. Pathogenicity and Virulence Factors
4. Classic Chemical Antifungal Biocides
4.1. Mode of Action of Antifungal Biocides
4.2. Mechanisms of Fungal Resistance to Biocides
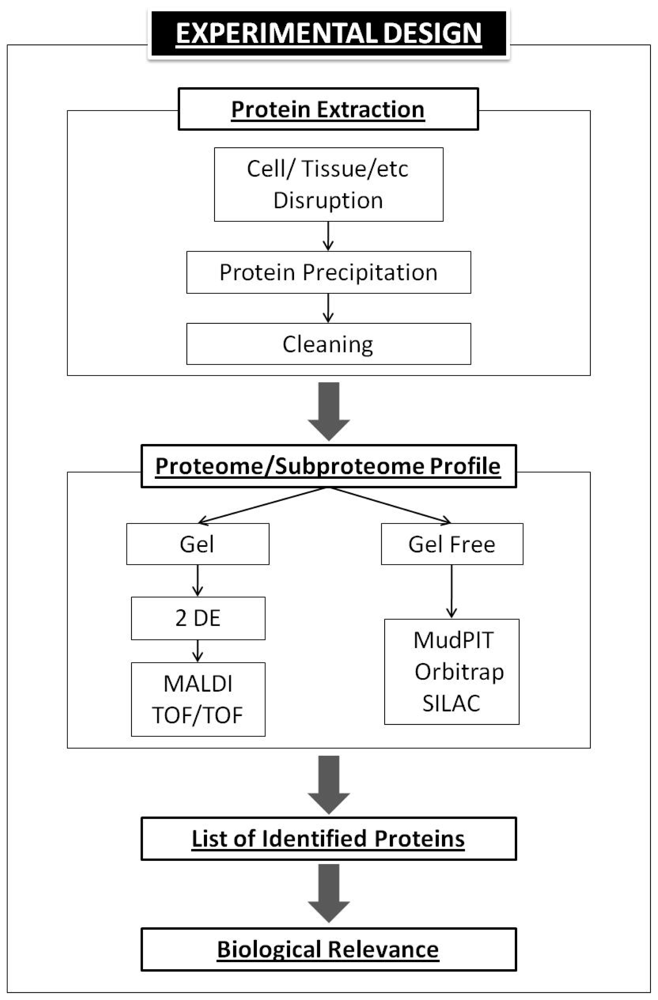
5. Basic Proteomics Workflow
6. Bioinformatic Approaches
7. New Protein-Based Strategies to Classical Chemical Fungicide Design
8. Peptide Aptamers
9. RNA Silencing
10. Peptide Probes
11. Conclusions
References
- Fernández-Acero, FJ; Carbú, M; Garrido, C; Vallejo, I; Cantoral, JM. Proteomic advances in phytopathogenic fungi. Curr Proteomics 2007, 79–88. [Google Scholar]
- Garrido, C; Cantoral, JM; Carbú, M; González-Rodríguez, VE; Fernández-Acero, FJ. New proteomic approaches to plant pathogenic fungi. Curr Proteomics 2011, 2011, 306–315. [Google Scholar]
- Santos, M; Diánez, F; de Cara, M; Tello, JC. Possibilities of the use of vinasses in the control of fungi phytopathogens. Bioresour. Technol 2008, 99, 9040–9043. [Google Scholar]
- Massart, S; J, HM. Use of molecular techniques to elucidate the mechanisms of action of fungal biocontrol agents: A review. J. Microbiol Methods 2007, 69, 229–241. [Google Scholar]
- Tournu, H; Serneels, J; Dijck, PV. Fungal Pathogens Research: Novel and Improved Molecular Approaches for the Discovery of Antifungal Drug Targets. Curr Drug Targets 2005, 909–922. [Google Scholar]
- Fernández-Acero, FJ; Colby, T; Harzen, A; Carbú, M; Wieneke, U; Cantoral, JM; Schmidt, J. 2-DE proteomic approach to the Botrytis cinerea secretome induced with different carbon sources and plant-based elicitors. Proteomics 2010, 10, 2270–2280. [Google Scholar]
- Yajima, W; Kav, NNV. The proteome of the phytopathogenic fungus Sclerotinia sclerotiorum. Proteomics 2006, 6, 5995–6007. [Google Scholar]
- Wang, Y; Chiu, J-F; He, Q-Y; Miles, H; William, M; Kenneth, B. Genomics and proteomics in drug design and discovery. In Pharmacology; Academic Press: San Diego, CA USA, 2009; pp. 561–573. [Google Scholar]
- Ferrer-Alcón, M; Arteta, D; Guerrero, MJ; Fernandez-Orth, D; Simón, L; Martinez, A. The use of gene array technology and proteomics in the search of new targets of diseases for therapeutics. Toxicol. Lett 2009, 186, 45–51. [Google Scholar]
- Fernández-Acero, FJ; Jorge, I; Calvo, E; Vallejo, I; Carbú, M; Camafeita, LE; Garrido, C; López, JA; Cantoral, JM; Jorrin, J. Proteomic analysis of phytopathogenic fungus Botrytis cinerea as a potential tool for identifying pathogenicity factors, therapeutic targets and for basic research. Arch. Microbiol 2007, 187, 207–215. [Google Scholar]
- Elad, Y; Williamson, B; Tudzynski, P; Delen, N. Botrytis: Biology, Pathology and Control; Dordrecht, The Netherland, 2004. [Google Scholar]
- Agrios, GN. Plant Pathology, 5th ed; Academic Press: San Diego, CA USA, 2005. [Google Scholar]
- Idnurm, A; Howlett, BJ. Pathogenicity genes of phytopathogenic fungi. Mol. Plant Pathol 2001, 2, 241–255. [Google Scholar]
- Odds, FC; Gow, NA; Brown, AJ. Fungal virulence studies come of age. Genome Biol 2001, 2. [Google Scholar]
- Fernández-Acero, FJ; Colby, T; Harzen, A; Wieneke, U; Garrido, C; Carbú, M; Vallejo, I; Schmidt, J; Cantoral, JM. Analysis of Botrytis cinerea secretome. J. Plant Pathol 2008, 90, 197. [Google Scholar]
- Durán-Patrón, R; Cantoral, JM; Hernández-Galán, R; Hanson, JR; Collado, IG. The biodegradation of the phytotoxic metabolite botrydial by its parent organism. Botrytis cinerea J Chem Res 2004, 441–443. [Google Scholar]
- Tudzynski, B; Gronover, CS. Signalling in Botrytis cinerea . In Botrytis: Biology, Pathology and Control; Elad, Y, Williamson, B, Tudzynski, P, Delen, N, Eds.; Kluwer Academic Publishers: Dordrecht, The Netherland, 2004. [Google Scholar]
- Tudzynski, P; Kokkelink, L. Botrytis cinerea: Molecular aspects of a necrotrophic life style. In The Mycota; Deising, HB, Ed.; Springer: Berlin Heidelberg, Germany, 2009; pp. 29–50. [Google Scholar]
- Del Sorbo, G; Schoonbeek, H-J; De Waard, MA. Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Genet. Biol 2000, 30, 1–15. [Google Scholar]
- Rui, O; Hahn, M. The Botrytis cinerea hexokinase, Hxk1, but not the glucokinase, Glk1, is required for normal growth and sugar metabolism, and for pathogenicity on fruits. Microbiology 2007, 153, 2791–2802. [Google Scholar]
- Santos, M; Rebordinos, L; Gutiérrez, S; Cardoza, R-E; Martín, J-F; Cantoral, JM. Characterization of the gdhA Gene from the Phytopathogen Botrytis cinerea. Fungal Genet. Biol 2001, 34, 193–206. [Google Scholar]
- Schumacher, J; Viaud, M; Simon, A; Tudzynski, B. The G alpha subunit BCG1, the phospholipase C (BcPLC1) and the calcineurin phosphatase co-ordinately regulate gene expression in the grey mould fungus Botrytis cinerea. Mol. Microbiol 2008, 67, 1027–1050. [Google Scholar]
- Shih, YJ; Li, J; Goodwin, PH. Two pectin lyase genes, pnI-1 and pnI2, from Colletotrichum gloeosporioides f. sp. malvae differ in a cellulose-binding domain and in their expression during infection of Malva pusilla. Microbiology 2002, 148, 2149–2157. [Google Scholar]
- El-Bebany, AF; Rampitsch, C; Daayf, F. Proteomic analysis of the phytopathogenic soilborne fungus Verticillium dahliae reveals differential protein expression in isolates that differ in aggressiveness. Proteomics 2010, 10, 289–303. [Google Scholar]
- Fernández-Acero, FJ; Colby, T; Harzen, A; Cantoral, JM; Schmidt, J. Proteomic analysis of the phytopathogenic fungus Botrytis cinerea during cellulose degradation. Proteomics 2009, 9, 2892–2902. [Google Scholar]
- Maddi, A; Bowman, SM; Free, SJ. Trifluoromethanesulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genet. Biol 2009, 46, 768–781. [Google Scholar]
- Shah, P; Atwood, JA; Orlando, R; El Mubarek, H; Podila, GK; Davis, MR. Comparative proteomic analysis of botrytis cinerea secretome. J. Proteome Res 2009, 8, 1123–1130. [Google Scholar]
- Fahien, L; Kmiotek, E; MacDonald, M; Fibich, B; Mandic, M. Regulation of malate dehydrogenase activity by glutamate, citrate, alpha-ketoglutarate, and multienzyme interaction. J. Biol. Chem 1988, 263, 10687–10697. [Google Scholar]
- Fernández-Acero, FJ; Carbú, M; Garrido, C; Collado, IG; Cantoral, JM; Vallejo, I. Screening study of potential lead compounds for natural product-based fungicides against Phytophthora species. J. Phytopathol 2006, 154, 616–621. [Google Scholar]
- Bhadauria, V; Zhao, W-S; Wang, L-X; Zhang, Y; Liu, J-H; Yang, J; Kong, L-A; Peng, Y-L. Advances in fungal proteomics. Microbiol. Res 2007, 162, 193–200. [Google Scholar]
- Fernández-Acero, FJ; Jorge, I; Calvo, E; Vallejo, I; Carbú, M; Camafeita, LE; López, JA; Cantoral, JM; Jorrin, J. Two-dimensional electrophoresis protein profile of the phytopathogenic fungus Botrytis cinerea. Proteomics 2006, 6, S88–S96. [Google Scholar]
- Choquer, M; Fournier, E; Kunz, C; Levis, C; Pradier, J-M; Simon, A; Viaud, M. Botrytis cinerea virulence factors:newinsights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett 2007, 277, 1–10. [Google Scholar]
- Gadd, GM; White, C. Heavy metal and radionuclide accumulation and toxicity in fungi and yeasts. In Metal-Microbe Interaction; Poole, RK, Gadd, GM, Eds.; IRL Press: Oxford, UK, 1989; pp. 19–38. [Google Scholar]
- Russell, A. Biocide use and antibiotic resistance: The relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis 2003, 3, 794–803. [Google Scholar]
- Russell, AD; Furr, JR. Biocides: mechanisms of antifungal action and fungal resistance. Sci. Prog 1996, 79, 27–48. [Google Scholar]
- Russell, AD; Furr, RJ; Maillard, JY. Microbial susceptibility and resistance to biocides. ASM News 1997, 63, 481–487. [Google Scholar]
- McDonnell, G; Russell, AD. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev 1999, 12, 147–179. [Google Scholar]
- Russell, AD. Antifungal activity of biocides. In Principles and Practice of Disinfection, Preservation, and Sterilization; Russell, AD, Hugo, WB, Ayliffe, GAJ, Eds.; Blackwll Scientific Publications: Oxford UK, 1999; pp. 149–167. [Google Scholar]
- Russell, AD. Mechanisms of bacterial resistance to non-antibiotics: Food additives and food and pharmaceutical preservatives. J. Appl. Bacteriol 1990, 71, 191–201. [Google Scholar]
- Maillard, JY. Antibacterial mechanisms of action of biocides. J. Appl. Microbiol 2000, 92, 16–27. [Google Scholar]
- Simons, C; Walsh, SE; Maillard, JY; Russell, AD. A note: Ortho-Phthalaldehyde: Proposed mechanism of action of a new antimicrobial agent. Lett. Appl. Microbiol 2000, 31, 299–302. [Google Scholar]
- Navarro, JM; Monsan, P. Étude du mécanisme d’interaction du glutaraldéhyde avec les microorganismes. Ann. Microbiol 1976, 127B, 295–307. [Google Scholar]
- Hiom, SJ; Furr, JR; Russell, AD. Uptake of 14C-chlorhexidine gluconate by Sacharomyces cerevisiae, Candida albicans, and Candida glabrata. Lett. Appl. Microbiol 1995, 21, 20–22. [Google Scholar]
- Bobichon, H; Bouchet, P. Action of chlorhexidine on budding candida albicans: Scanning and transmission electron microscopie study. Mycopathologia 1978, 100, 27–35. [Google Scholar]
- Sautour, M; Mathieu, G; Delcourt, A; Divies, C; Bensoussan, M. Action de la chlorhexidine sur l’expression de la virulence de Candida albicans. Cryptogam. Mycol 1999, 20, 179–188. [Google Scholar]
- Elferink, JGR; Booij, HL. Interaction of yeast cells with chloroxidine. Biochem. Pharmacol 1974, 23, 1413–1419. [Google Scholar]
- Cartwright, CP; Juorszek, JR; Beavan, MJ; Ruby, FMS; De Morais, SMF; Rose, AH. Ethanol dissipates the proton motive force across the plasma membrane of Saccharomyces cerevisiae. J. Gene. Microbiol 1986, 132, 369–377. [Google Scholar]
- Salueiro, SP; S-Correia, I; Novias, JM. Etanol induced leakage in Saccharomyces cerevisiae: kinetics and relationship to yeast ethanol tolerance and alcohol fermentation productivity. Appl. Environ. Microbiol 1988, 54, 903–909. [Google Scholar]
- Brul, S; Klis, FM. Mechanistic and mathematical inactivation studies of food spoilage fungi. Fungal Genet. Biol 1999, 27, 199–208. [Google Scholar]
- Eklund, T. Organic acids and esters. In Food Preservation Procedures; Gould, GW, Ed.; Elsevier Applied Science: London, UK, 1991; pp. 161–200. [Google Scholar]
- Fernández-Ortuño, D; Torés, JA; de Vicente, A; Pérez-García, A. Mechanisms of resistance to Qol fungicides in phytopathogenic fungi. Int. Microbiol 2008, 11, 1–9. [Google Scholar]
- Gorman, SP; Scott, EM; Russell, AD. Antimicrobial activity, uses, and mechanisms of action of glutaraldehyde. J. Appl. Bacteriol 1980, 48, 161–190. [Google Scholar]
- Hiom, SJ; Furr, JR; Russell, AD. Effects of chlorhexidine diacetate and cetylpyridinium chloride on whole cells and protoplasts of Saccharomyces cerevisiae. Microbios 1993, 74, 111–120. [Google Scholar]
- Holyoak, CD; Stratford, M; McMullin, Z. Activity of the plasma membrane H+-ATPase and optimal glycolytic flux are required for rapid adaptation and growth in the presence of the weak acid preservative sorbic acid. Appl. Environ. Microbiol 1996, 62, 3158–3164. [Google Scholar]
- Piper, P; Mahe, Y; Thompson, S. The Pdr12 ATP-binding cassette ABC is required for the development of weak acid resistance to Saccharomyces cerevisiae. EMBO J 1998, 17, 4257–4265. [Google Scholar]
- Steffens, JJ; Pell, EJ; Tien, M. Mechanisms of fungicide resistance in phytopathogenic fungi. Curr. Opin. Biotechnol 1996, 7, 348–355. [Google Scholar]
- Kataoka, S; Takagaki, M; Kaku, K; Shimizo, T. Mechanisms of action and selectivity of a novel fungicide, pyribencarb. J. Pestic. Sci 2010, 35, 99–106. [Google Scholar]
- Bennett, JE. Flucytosine. Ann. Intern. Med 1977, 86, 319–322. [Google Scholar]
- Vermes, A; Guchelaar, HJ; Dankert, J. Flucytosine: A review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother 2000, 46, 171–179. [Google Scholar]
- Lukens, RJ. Antimicrobial agents in crop production. In Disinfection, Sterilization and Preservation; Block, SS, Ed.; Lea & Febiger: Philadelphia, PA, USA, 1983; pp. 695–713. [Google Scholar]
- Ishii, H. Studies on fungicide resistance in phytopathogenic fungi. J. Gen. Plant Pathol 2004, 70, 379–381. [Google Scholar]
- Russell, AD; Hugo, WB. Antimicrobial activity and action of silver. Prog. Med. Chem 1994, 31, 351–371. [Google Scholar]
- Gisi, U; Chin, KM; Knapova, G; Farber, RK; Mohr, U; Parisi, S; Sierotzki, H; Steinfeld, H. Recent developments in elucidating modes of resistance of phenylamide, DMI and strobilurin fungicides. Crop Prot 2000, 19, 863–867. [Google Scholar]
- Fluit, AC; Visser, MR; Schmitz, FJ. Molecular detection of antimicrobial resistance. Clin. Microbiol. Rev 2001, 14, 836–871. [Google Scholar]
- Gullino, ML; Leroux, P; Smith, CM. Uses and challenges of novel compounds for plant disease control. Crop Prot 2000, 19, 1–11. [Google Scholar]
- MgGrath, MT. Fungicide resistance in cucurbit powdery mildew, experiences and challenges. Plant Disease 2001, 85, 236–245. [Google Scholar]
- Ghannoum, MA; Rice, LB. Antifungal Agents: Mode of Action, Mechanisms of Resistance, and Correlation of These Mechanisms with Bacterial Resistance. Clin. Microbiol. Rev 1999, 12, 501–517. [Google Scholar]
- Brent, KJ. Fungicide Resistance in Crop Pathogens, How Can It Be Managed; Global Crop Protection Federation: Brussels, Belgium, 1995. [Google Scholar]
- Ma, Z; Michailides, T. Advances in understanding molecular mechanisms of fungicide resistance and molecular detection of resistant genotypes in phytopathogenic fungi. Crop Protection 2005, 24, 853–863. [Google Scholar]
- Marichal, P; Van den Bossche, V; Odds, FC; Nobels, G; Warnock, DW; Timmerman, V; Broeckhoven, C; Fay, S; Mose-Larsen, P. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother 1997, 41, 2229–2237. [Google Scholar]
- Schnabel, G; Jones, AL. The 14α-demethylase (CYP51A1) gene is overexpressed in Venturia inaequalis strains resistance to myclobutanil. Phytopathology 2001, 91, 102–110. [Google Scholar]
- Zwiers, LH; Stergiopoulos, I; Gielkens, MMC; Goodall, SD; Waard, MAD. ABC transporters of the wheat pathogen Mycosphaerella graminicola function as protectants against biotic and xenobiotic toxic compounds. Mol. Genet Genomics 2003, 269, 499–507. [Google Scholar]
- White, TC. Mechanisms of resistance to antifungal agents. In Manual of Clinical Microbiology; Murray, PR, Baron, EJ, Jorgensen, JH, Pfaller, MA, Yolken, RH, Eds.; ASM press: Washington, DC, USA, 2003; pp. 1869–1879. [Google Scholar]
- De Waard, MA; Andrade, AC; Hayashi, K; Schoonbeek, H; Stergiopoulos, I; Zwiers, L. Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest. Manag. Sci 2006, 62, 195–207. [Google Scholar]
- Andrade, AC; Del Sorbo, G; Van Nistelrooy, JGM; De Waard, MA. The ABC transporter AtrB from Aspergillus nidulans mediates resistance to all major classes of fungicides and some natural toxic compounds. Microbiology 2000, 146, 1987–1997. [Google Scholar]
- Andrade, AC; van Nistelrooy, JGM; Peery, R-B; Skatrud, PL; De Waard, MA. The role of ABC transporters from Aspergillus nidulans in protection against cytotoxic agents and antibiotic production. Mol. Gen. Genet 2000, 263, 966–977. [Google Scholar]
- Kato, N; Miyawaki, N; Sakasawa, C. Formaldehyde dehydrogenase from formaldehyde-resistant debaryomyces vanriji FT-1 and Pseudomonas putida F61. Agric. Biol. Chem 1983, 47, 415–416. [Google Scholar]
- Hundt, K; Martin, D; Hammer, E; Jonas, U; Kindermann, MK; Schauer, E. Transformation of triclosan by Trametes versicolor and Pycnoporus cinnabarinus. Appl. Environ. Microbiol 2000, 66, 4157–4160. [Google Scholar]
- Valix, M; Tang, J-Y; Malik, R. Heavy metal tolerance of fungi. Miner. Eng 2001, 14, 499–505. [Google Scholar]
- Gadd, GM. Metal tolerance. In Microbiology of Extreme Environments; Edwards, C, Ed.; Open University Press: Milton Keynes, London, UK, 1990; pp. 178–210. [Google Scholar]
- Murphy, RJ; Levy, JF. Production of copper oxalate by some copper tolerant fungi. Trans. Br. Mycol. Soc 1983, 81, 165–168. [Google Scholar]
- Warnock, DW; Arthington-Skaggs, BA; Li, RK. Antifungal drug susceptibility testing and resistance in Aspergillus. Drug Resist. Updat 1999, 2, 326–334. [Google Scholar]
- Kontoyiannis, DP; Lewis, RE. Antifungal drug resistance of pathogenic fungi. Lancet Infect. Dis 2002, 359, 1135–1144. [Google Scholar]
- Leroux, P; Gredt, M; Leroch, M; Walker, AS. Exploring mechanisms of resistance to respiratory inhibitors in field strains of Botrytis cinerea, the causal agent of gray mold. Appl. Environ. Microbiol 2010, 76, 6615–6630. [Google Scholar]
- Wilkins, MR; Sanchez, JC; Gooley, AA; Appel, RD; Humphery-Smith, I; Hochstrasser, DF; Williams, KL. Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev 1995, 13, 19–50. [Google Scholar]
- Xu, H; Xu, H; Lin, M; Wang, W; Li, Z; Huang, J; Chen, Y; Chen, X. Learning the drug target-likeness of a protein. Proteomics 2007, 7, 4255–4263. [Google Scholar]
- Chen, X; Fang, Y; Yao, L; Chen, Y; Xu, H. Does drug-target have a likeness? Methods Inf. Med 2007, 46, 360–363. [Google Scholar]
- Collado, IG; Sanchez, AJMA; Hanson, JR. Fungal terpene metabolites: Biosynthetic relationships and the control of the phytopathogenic fungus Botrytis cinerea. Natur. Product Rep 2007, 24, 674–686. [Google Scholar]
- Pinedo, C; Wang, C-M; Pradier, J-M; Dalmais, BRR; Choquer, M; Le PeÌ‚cheur, P; Morgant, G; Collado, IG; Cane, DE; Viaud, M. Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen botrytis cinerea. ACS Chem. Biol 2008, 3, 791–801. [Google Scholar]
- Beroza, P; Villar, HO; Wick, MM; Martin, GR. Chemoproteomics as a basis for post-genomic drug discovery. Drug Discov Today 2002, 7, 807–814. [Google Scholar]
- Rudolph, C; Schreier, PH; Uhrig, JF. Peptide-mediated broad-spectrum plant resistance to tospoviruses. Proc. Natl. Acad. Sci USA 2003, 100, 4429–4434. [Google Scholar]
- Crawford, M; Woodman, R; Ferrigno, PK. Peptide aptamers: Tools for biology and drug discovery. Brief. Funct Genomics Proteomics 2003, 2, 72–79. [Google Scholar]
- Nakayashiki, H; Kadotani, N; Shigeyuki, M. Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol 2006, 63, 127–135. [Google Scholar]
- Nakayashiki, H. RNA silencing in fungi: Mechanisms and applications. FEBS Lett 2005, 579, 5950–5957. [Google Scholar]
- Fattal, E; Bochot, A. Antisense oligonucleotides, aptamers and SiRNA: Promises for the treatment of ocular diseases. Arch. Soc. Esp. Oftalmol 2006, 81, 3–6. [Google Scholar]
- Richau, KH; van Der Hoorn, RAL. Studies on plant-pathogen interactions using activity-based proteomics. Curr Proteomics 2011, 7, 328–336. [Google Scholar]
- Gu, C; Kolodziejek, I; Misas-Villamil, J; Shindo, T; Colby, T; Verdoes, M; Richau, KH; Schmidt, J; Overkleeft, HS; Van Der Hoorn, RAL. Proteasome activity profiling: A simple, robust and versatile method revealing subunit-selective inhibitors and cytoplasmic, defense-induced proteasome activities. Plant J 2010, 62, 160–170. [Google Scholar]

| Target site | Mechanisms of action | Example biocides | References |
|---|---|---|---|
| Cell wall | 1. Cross-linking of cell proteins and chitin | Gluteraldehyde | [35,41,52] |
| 2. Cell agglutination | [42] | ||
| Plasma membrane | 1. Induction leakage of intercellular materials and protoplast lysis; loss of structural organization and integrity; disruption to physiological function | Chlorhexidine, QAC’s Ethanol | [53] [46] [47] [48] |
| 2. Alteration of membrane properties and switching on an efflux pump system; membrane perturbation | Organic acids | [54] [55] [49] | |
| 3. Inhibition of the proton-motive force (Δcomponent); induction leakage of intercellular materials | Esters | [50] | |
| 4. Inhibition of respiration and energy transfer; inhibition of ATP synthesis | QoI’s | [51] | |
| 5. Interaction with ergosterol and destabilization of cell membrane functions; inhibition of cytochrome P450 in ergosterol biosynthetic pathway | DMI groups | [56] | |
| 6. Inhibition of the electron transport system | Benzylcarbamate | [57] | |
| DNA/RNA | Interferes with DNA and RNA synthesis | Pyrimidine analog: Flucytosine | [58,59] |
| Protein | 1. Interaction with alkylating and oxidizing agents; binding to key functional groups of fungal enzymes | Heavy metals (-SH groups) | [60] |
| 2. Inhibition of cell division; bind to proteins of tubulin; cytoskeleton formation | Benzimidazole | [61] |
© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Fernández Acero, F.J.; Carbú, M.; El-Akhal, M.R.; Garrido, C.; González-Rodríguez, V.E.; Cantoral, J.M. Development of Proteomics-Based Fungicides: New Strategies for Environmentally Friendly Control of Fungal Plant Diseases. Int. J. Mol. Sci. 2011, 12, 795-816. https://doi.org/10.3390/ijms12010795
Fernández Acero FJ, Carbú M, El-Akhal MR, Garrido C, González-Rodríguez VE, Cantoral JM. Development of Proteomics-Based Fungicides: New Strategies for Environmentally Friendly Control of Fungal Plant Diseases. International Journal of Molecular Sciences. 2011; 12(1):795-816. https://doi.org/10.3390/ijms12010795
Chicago/Turabian StyleFernández Acero, Francisco Javier, María Carbú, Mohamed Rabie El-Akhal, Carlos Garrido, Victoria E. González-Rodríguez, and Jesús M. Cantoral. 2011. "Development of Proteomics-Based Fungicides: New Strategies for Environmentally Friendly Control of Fungal Plant Diseases" International Journal of Molecular Sciences 12, no. 1: 795-816. https://doi.org/10.3390/ijms12010795
APA StyleFernández Acero, F. J., Carbú, M., El-Akhal, M. R., Garrido, C., González-Rodríguez, V. E., & Cantoral, J. M. (2011). Development of Proteomics-Based Fungicides: New Strategies for Environmentally Friendly Control of Fungal Plant Diseases. International Journal of Molecular Sciences, 12(1), 795-816. https://doi.org/10.3390/ijms12010795







