Trehalose Metabolism: From Osmoprotection to Signaling
Abstract
:1. Introduction
2. The Multiple Roles of Trehalose
3. Biotechnological Applications of Trehalose
- Protector of enzyme activity: Trehalose can be used to store thermolabile enzymes such as DNA polymerase, restriction enzymes and DNA ligase at ambient temperature [15].
- Stabilizer and protector for complex molecules: Unstable molecules such as antibodies can be dehydrated at room temperature or 37 °C in the presence of trehalose, maintaining their activity after various months in storage [8].
- Foods additive: Trehalose can be used in dried or processed foods such as fruits and vegetables, in order to preserve aromas and their organoleptic properties; it should also be pointed out that this disaccharide is not toxic and it is already consumed as part of the human diet, as it is present in bread, honey, mushrooms, wine, beer, etc [16].
- Selection marker in the production of transgenic plants: The AtTPS1 gene of A. thaliana encodes the TPS1 enzyme, which confers glucose insensibility to seeds and tissues of plants overexpressing this gene when cultivated under tissue-culture conditions [21]. Germination and differentiation in wild type plants are inhibited by glucose, thus AtTPS1 can be used as a selectable marker gene during the process of plant transformation, using this monosaccharide as a selective agent [22].
- Cosmetics industry: Trehalose traps and reduces bad odors emitting from human skin by up to 70%, making it a useful additive for facial or body creams and for deodorants [7].
- Possible medical uses: The role of trehalose in reducing the symptoms in illnesses such as Huntington’s chorea and in osteoporosis has been explored. In the former, trehalose prevented the formation of polyglutamine protein in the brain [23] and in the second study, the consumption of trehalose was found to reduce the degeneration of bones in female rats whose ovaries had been removed. However, the mechanisms involved in these uses are not fully understood [7].
4. Transgenic Manipulation of Trehalose Metabolism
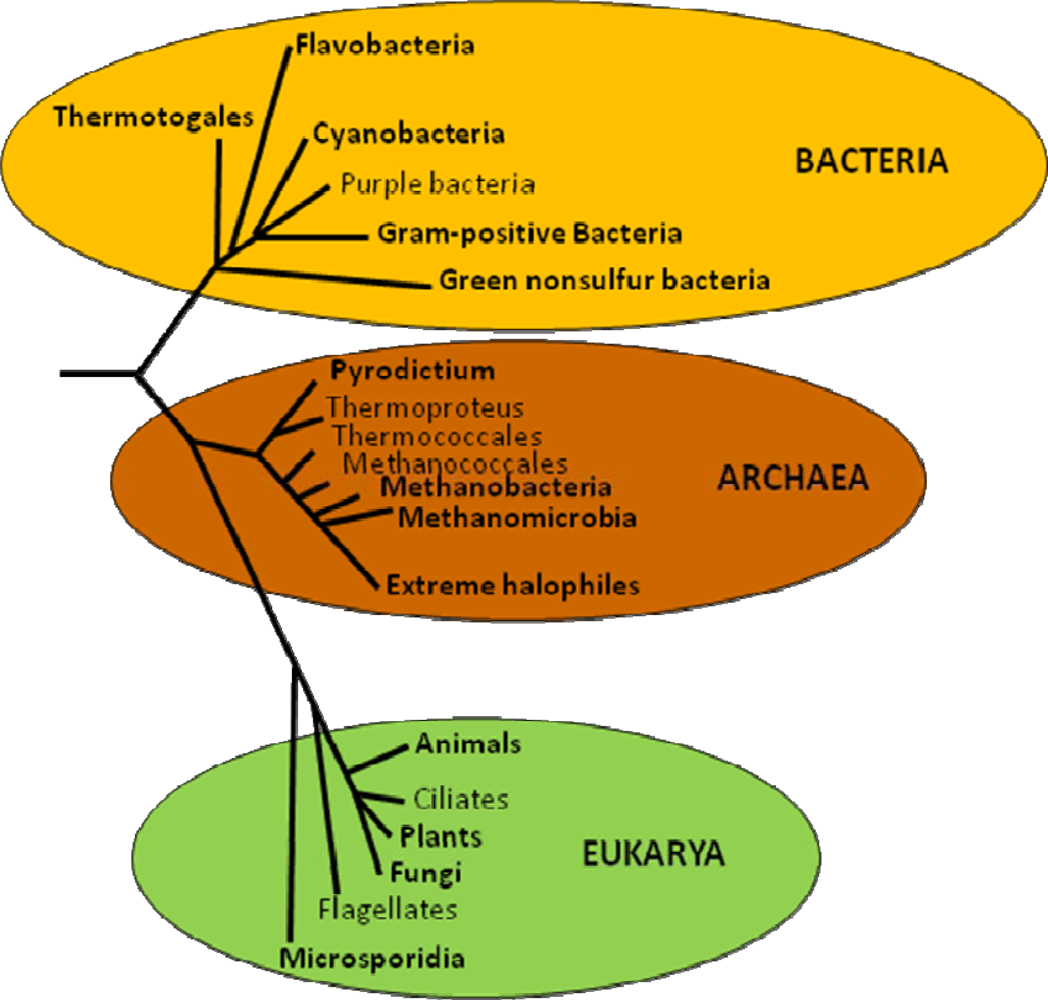
5. Evolution of Trehalose Biosynthesis
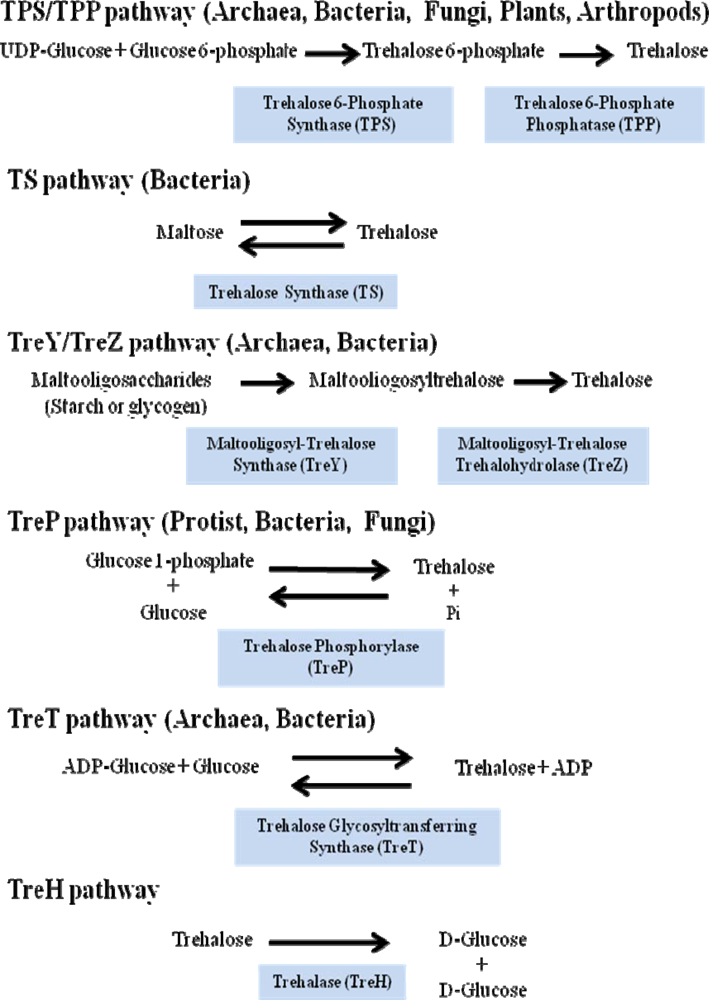
- The most common and the best studied route among different species involves the enzyme trehalose-6-phosphate synthase (TPS), which catalyses the transfer of glucose by means of UDP-glucose to glucose-6-phosphate, leading to trehalose-6-phosphate (T6P). In a second stage trehalose-6-phosphate phosphatase (TPP) catalyzes the hydrolysis of the phosphate group from the intermediate disaccharide to generate trehalose [49]. This TPS-TPP route is found in a variety of organisms; for example insects [50], plants such as the “resurrection” plant Selaginella lepidophylla and Arabidopsis thaliana [38,51], Escherichia coli [40] and Saccharomyces cerevisiae [35].
- Trehalose synthase (TS) catalyses an intramolecular arrange of maltose, in order to convert the glycosidic bond α-(1–4) of this disaccharide to the α-(1-1) trehalose bond [52]. This enzyme is found in several organisms such as Pimelobacter sp, Pseudomonas syringae and Thermus caldophilus.
- The TreY-TreZ pathway is present in some bacteria where the conversion of maltooligosaccharides present in starch, are broken down to trehalose. The thermophilic archaebacteria belonging to the Sulfolobus genus, as well as Arthrobacter sp Q36 and Rhizobium sp M-11, display amylolytic activity which leads to trehalose. This is a two-step pathway involving maltooligosyl-trehalose synthase (TreY) catalyzing the conversion of maltodextrines to maltooligosyl-trehalose and subsequently the maltooligosyl-trehalose trehalohydrolase (TreZ) breaks this intermediate to generate trehalose [53].
- Trehalose phosphorylase (TreP) has been reported in Agaricus bisporus, Catellatospora ferruginea, Euglena gracilis and Flammulina velutipes. TreP catalyzes a reversible reaction in vitro, which hydrolyzes trehalose and transfers a glucose molecule to the inorganic phosphate, to form glucose-1-phosphate and release free glucose [54]. This reaction can go in one direction or another, depending on the species [55].
- TreT is the trehalose glycosyltransferring synthase, which results in trehalose formation from ADP-glucose and glucose. This reaction is very similar to that described for UDP-glucose and glucose-6-phosphate, but varies because the synthesis occurs in a single reaction and the products are trehalose and ADP. This reversible reaction has been detected in various organisms, such as Thermococcus litoralis and Sulfolobus solfataricus KM1 [56].
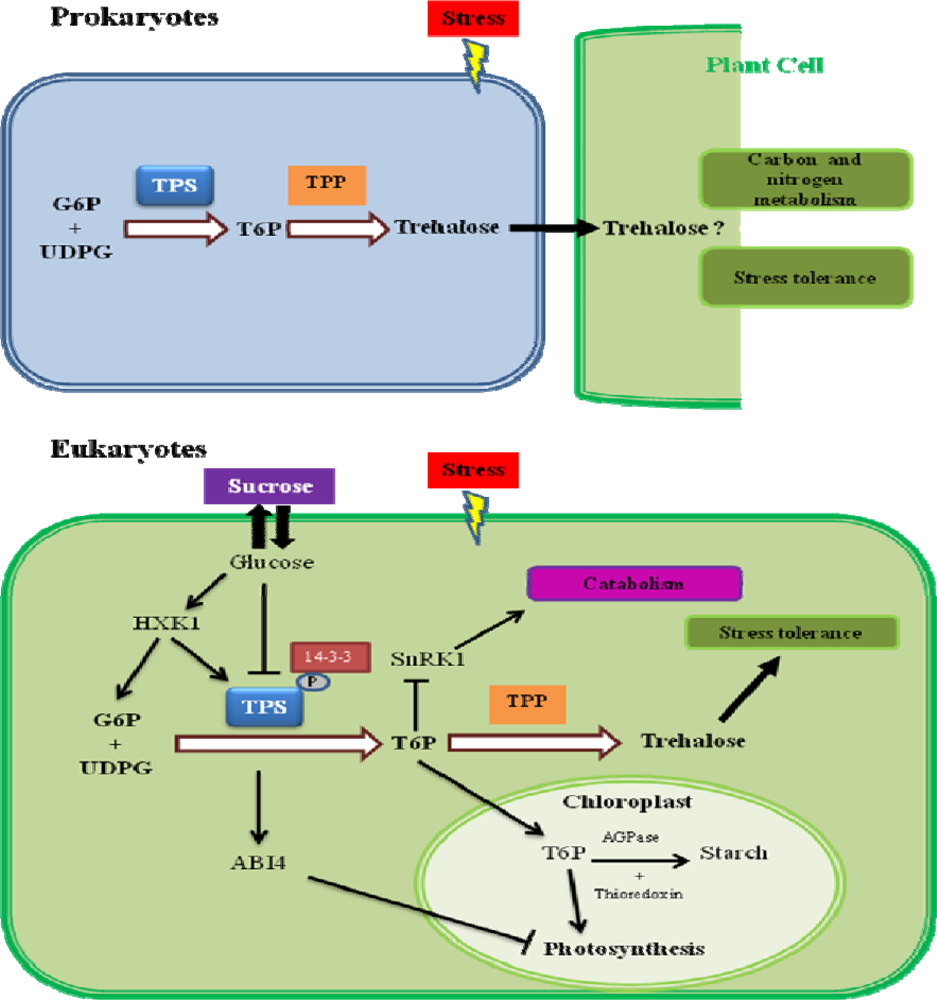
6. Trehalose-6-Phosphate as a Signaling Molecule
7. Involvement of Trehalose in Pathogenesis and Symbiosis
8. Conclusions
Acknowledgments
References
- Crowe, JH; Carpenter, JF; Crowe, LM. The role of vitrification in anhydrobiosis. Annu. Rev. Physiol 1998, 60, 73–103. [Google Scholar]
- Wharton, DA. Life at the Limits Organisms in Extreme Environments; Cambridge University Press: Cambridge, UK, 2002; pp. 93–128. [Google Scholar]
- Singer, MA; Lindquist, S. Thermotolerance in Saccharomyces cerevesiae: The yin and yang of trehalose. Trends Biotechnol 1998, 16, 460–468. [Google Scholar]
- Elbein, AD; Pan, YT; Pastuszak, I; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar]
- Crowe, JH; Crowe, LM; Chapman, D. Preservation of membranes in anhydrobiotics organism. The role of trehalose. Science 1984, 223, 209–217. [Google Scholar]
- Crowe, JH; Hoekstra, FA; Crowe, LM. Anhydrobiosis. Annu. Rev. Physiol 1992, 54, 579–599. [Google Scholar]
- Higashiyama, T. Novel functions and applications of trehalose. Pure Appl. Chem 2002, 74, 1263–1269. [Google Scholar]
- Roser, B; Colaço, C. A sweeter way to fresh food. New Sci 1993, 138, 25–28. [Google Scholar]
- Clegg, JS. The physical properties and metabolic status of Artemia cysts at low water contents: “The water replacement hypothesis”. In Membranes, Metabolism and Dry Organisms; Leopold, AC, Ed.; Cornell University Press: Ithaca, NY, USA, 1985; pp. 169–187. [Google Scholar]
- Donnamaria, MC; Howard, EI; Grigera, JR. Interaction of water with α-α trehalose in solution: Molecular dynamics simulation approach. J. Chem. Soc., Faraday Trans 1994, 90, 2731–2735. [Google Scholar]
- Green, JL; Angell, CA. Phase relations and vitrification in saccharide-water solution and the trehalose anomally. J. Phys. Chem 1989, 93, 2880–2882. [Google Scholar]
- Paiva, CLA; Panek, AD. Biotechnological applications of the disaccharide trehalose. Biotechnol. Annu. Rev 1996, 2, 293–314. [Google Scholar]
- Richards, AB; Krakowka, S; Dexter, LB; Schimdt, H; Wolterbeek, APM; Wallakenns-Berendsen, DH; Shigoyuki, A; Kurimoto, M. Trehalose: A review of properties, history of use and humans tolerance, and results of multiple safety studies. Food Chem. Toxicol 2002, 40, 871–898. [Google Scholar]
- Schiraldi, C; Di Lernia, I; de Rosa, M. Trehalose production: Exploiting novel approaches. Trends Biotechnol 2002, 20, 420–425. [Google Scholar]
- Colaço, C; Sen, S; Thangavelu, M; Pinder, S; Roser, B. Extraordinary stability of enzymes dried in trehalose: Simplified molecular biology. Biotechnology 1992, 10, 1007–1111. [Google Scholar]
- Kidd, G; Devorak, J. Trehalose is a sweet target for agbiotech. Biotechnology 1994, 12, 1328–1329. [Google Scholar]
- Eroglu, A; Russo, MJ; Biegansky, R; Fowler, A; Cheley, S; Bayley, H; Toner, M. Intracellular trehalose improves the survival of cryopreserved mammalian cells. Nat. Biotechnol 2000, 18, 163–167. [Google Scholar]
- Guo, N; Puhlev, I; Brown, DR; Mansbridge, J; Levine, F. Trehalose expression confers desiccation tolerance on human cells. Nat. Biotech 2000, 18, 168–171. [Google Scholar]
- Iwaya-Inoue, M; Tataka, M. Trehalose plus chloramphenicol prolong the base life of tulip flowers. Hort. Sci 2001, 36, 946–950. [Google Scholar]
- Otsubo, M; Iwaya-Inoue, M. Trehalose delays senescence in cut gladiolus spikes. Hort. Sci 2000, 35, 1107–1110. [Google Scholar]
- Avonce, N; Leyman, B; Mascorro Gallardo, JO; van Dijck, P; Thevelein, J; Iturriaga, G. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 2004, 136, 3649–3659. [Google Scholar]
- Leyman, B; Avonce, N; Ramon, M; van Dijck, P; Iturriaga, G; Thevelein, JM. Trehalose-6-phosphate synthase as an intrinsic selection marker for plant transformation. J. Biotechnol 2006, 121, 309–317. [Google Scholar]
- Katsuno, M; Adachi, H; Sobue, G. Sweet relief for Huntington disease. Nat. Med 2004, 10, 123–124. [Google Scholar]
- Holmström, KO; Welin, B; Mandal, A; Kristiansdottir, I; Teeri, T; Lamark, T; Strom, AR; Palva, ET. Production of the Escherichia coli betaine-aldehyde dehydrogenase, an enzyme required for synthesis of the osmoprotectant glycine betaine, in transgenic plants. Plant J 1996, 5, 749–758. [Google Scholar]
- Pilon-Smits, EAH; Terry, N; Sears, T; Kim, H; Zayed, A; Hwang, S; van Dun, K; Voogd, E; Verwoerd, TC; Krutwagen, RWHH; Goddijn, OJM. Trehalose-producing transgenic tobacco plants show improved growth performance under drought stress. J. Plant Physiol 1998, 152, 525–532. [Google Scholar]
- Yeo, E-T; Kwon, H-B; Han, S-E; Lee, J-T; Ryu, J-C; Byun, M-O. Genetic engineering of drought resistant potato plants by introduction of the trehalose-6-phosphate synthase (TPS1) gene from Saccharomyces cerevisiae. Mol. Cells 2000, 10, 263–268. [Google Scholar]
- Cortina, C; Culiáñez-Macià, FA. Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci 2005, 169, 75–82. [Google Scholar]
- Garg, AK; Kim, JK; Owens, TG; Ranwala, A; Choi, Y; Kochian, L; Wu, RJ. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2002, 99, 15898–15903. [Google Scholar]
- Miranda, JA; Avonce, N; Suárez, R; Thevelein, J; van Dijck, P; Iturriaga, G. A bifunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis. Planta 2007, 226, 1411–1421. [Google Scholar]
- Suárez, R; Calderón, C; Iturriaga, G. Improved tolerance to multiple abiotic stresses in transgenic alfalfa accumulating trehalose. Crop Sci 2009. [Google Scholar]
- Avonce, N; Leyman, B; Thevelein, J; Iturriaga, G. Trehalose metabolism and glucose sensing in plants. Biochem. Soc. Trans 2005, 33, 276–279. [Google Scholar]
- Nwaka, S; Holzer, H. Molecular biology of trehalose and trehalases in the yeast, Saccharomyces cereviciae. Prog. Nucleic Acid Res. Mol. Biol 1998, 58, 197–237. [Google Scholar]
- Gancedo, C; Flores, CL. The importance of a functional biosynthetic pathway for the life of yeast and fungi. FEMS Yeast Res 2004, 4, 351–359. [Google Scholar]
- Thevelein, JM. Regulation of trehalose mobilization in fungi. Microbiol. Rev 1984, 48, 42–59. [Google Scholar]
- de Virgilio, C; Hottiger, T; Dominguez, J; Boller, T; Wiemken, A. The role of trehalose synthesis for the adquisition of thermotolerance in yeast. I. Genetic evidence that trehalose is a thermoprotectant. Eur. J. Biochem 1994, 219, 179–186. [Google Scholar]
- Hottiger, T; de Virgilio, C; Hall, N; Boller, T; Wiemken, A. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur. J. Biochem 1994, 219, 187–193. [Google Scholar]
- Adams, RP; Kendall, E; Kartha, KK. Comparison of free sugars in growing and desiccated plants of Selaginella lepidophylla. Biochem. Syst. Ecol 1990, 18, 107–110. [Google Scholar]
- Blázquez, MA; Santos, E; Lisset-Flores, C; Martínez-Zapater, J; Salinas, J; Gancedo, C. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J 1998, 13, 685–689. [Google Scholar]
- Iturriaga, G; Gaff, DF; Zentella, R. New desiccation-tolerant plants, including a grass, in the central highlands of Mexico, accumulate trehalose. Aust. J. Bot 2000, 48, 153–158. [Google Scholar]
- Ström, AR; Kaasen, I. Trehalose metabolism in Escherichia coli: Stress protection and stress regulation of gene expression. Mol. Microbiol 1993, 8, 205–210. [Google Scholar]
- Purvis, JE; Yomano, LP; Ingram, LO. Enhanced trehalose production improves growth of Escherichia coli under osmotic stress. Appl. Environ. Microbiol 2005, 71, 3761–3769. [Google Scholar]
- Wyatt, GR; Kalf, GF. The chemistry of insect hemolymph. Trehalose and other carbohydrates. J. Gen. Physiol 1957, 40, 833–846. [Google Scholar]
- Fairbairn, D; Passey, RF. Occurrence and distribution of trehalose and glycogen in the eggs and tissues of Ascaris lumbricoides. Exp. Parasitol 1957, 6, 566–574. [Google Scholar]
- Clegg, JS; Evans, DR. Blood trehalose and flight metabolism in the blowfly. Science 1961, 134, 54–55. [Google Scholar]
- Cavalier-Smith, T. Obcells as proto-organism: Membranes heredity, lithophosphorylation, and the origins of the genetic code, the first cells, and photosynthesis. J. Mol. Evol 2001, 53, 555–595. [Google Scholar]
- McMullan, G; Christie, JJ; Rahman, TJ; Banta, I; Terman, N; Marchant, R. Habitat, applications and genomics of the aerobic, thermophilic genus Geobacillus. Biochem. Soc. Trans 2004, 32, 214–217. [Google Scholar]
- Madigan, MT; Oren, A. Thermophilic and halophilic extremophiles. Curr. Opin. Microbiol 1999, 2, 265–269. [Google Scholar]
- Yancey, PH. Organic osmolytes as compatible, metabolic and counteracting cryoprotectants in high osmolarity and other stresses. J. Exp. Biol 2005, 208, 2819–2830. [Google Scholar]
- Cabib, E; Leloir, L. The biosynthesis of trehalose phosphate. J. Biochem 1958, 231, 259–275. [Google Scholar]
- Candy, DJ; Kilby, BA. Site and mode of trehalose biosynthesis in the locust. Nature 1958, 183, 1584–1595. [Google Scholar]
- Zentella, R; Mascorro-Gallardo, JO; van Dijck, P; Folch-Mallol, J; Bonini, B; van Vaeck, C; Gaxiola, R; Covarrubias, AA; Nieto-Sotelo, J; Thevelein, JM; Iturriaga, G. Selaginella lepidophylla trehalose-6-phosphate synthase complements growth and stress-tolerance defects in a yeast tps1 mutant. Plant Physiol 1999, 119, 1473–1482. [Google Scholar]
- Nishimoto, T; Nakada, T; Chaen, H; Fukuda, S; Kurimoto, M; Tsujisaka, Y. Purification and properties of a novel enzyme, trehalose synthase, from Pimelobacter sp R48. Biosci. Biotechnol., Biochem 1996, 60, 640–644. [Google Scholar]
- Maruta, K; Mitsuzumi, H; Kubota, M; Chaen, H; Fukuda, S; Sugimoto, T; Kurimoto, M. Cloning and sequencing of a cluster of genes encoding novel enzymes of trehalose biosynthesis from termophilic archaebacterium Sulfolobus acidocaldarius. Biochem. Biophys. Acta 1996, 1291, 177–181. [Google Scholar]
- Wannet, WJB; Op den Camp, HJM; Wisselink, HW; van der Drift, C; van Griensven, LJLD; Vogels, GD. Purification and characterization of trehalose phosphorylase from the commercial mushrooms. Agaricus bisporus. Biochem. Biophys. Acta 1998, 1425, 177–188. [Google Scholar]
- Ren, Y; Dai, X; Zhou, J; Liu, J; Pei, H; Xiang, H. Gene expression and molecular characterization of a thermostable trehalose phosphorylase from Termoanaerobacter tengcongensis. Sci. China, Ser. C Life Sci 2005, 48, 221–227. [Google Scholar]
- Qu, Q; Lee, SJ; Boss, W. TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archeon Thermococcus litoralis. J. Biol. Chem 2004, 279, 47890–47897. [Google Scholar]
- Goddijn, OJM; Verwoerd, TC; Voogd, E; Krutwagen, RWHH; de Graaf, PTHM; van Dun, K; Poels, J; Ponstein, AS; Damm, B; Pen, J. Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol 1997, 113, 181–190. [Google Scholar]
- Avonce, N; Mendoza-Vargas, A; Morett, E; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol 2006, 6, 109–124. [Google Scholar]
- Leyman, B; van Dijck, P; Thevelein, JM. An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci 2001, 6, 510–513. [Google Scholar]
- Vogel, G; Aeschbacher, RA; Muller, J; Boller, T; Wiemken, A. Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: Identification by functional complementation of the yeast tps2 mutant. Plant J 1998, 13, 673–683. [Google Scholar]
- Lunn, JE. Gene families and evolution of trehalose metabolism in plants. Funct. Plant Biol 2007, 34, 550–563. [Google Scholar]
- Ramon, M; de Smet, I; Vandesteene, L; Naudts, M; Leyman, B; van Dijck, P; Rolland, F; Beeckman, T; Thevelein, JM. Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from. Arabidopsis thaliana Plant Cell Environ 2009. [Google Scholar]
- Thevelein, JM; Hohmann, S. Trehalose synthase: Guard to the gate of glycolysis in yeast? Trends Biochem. Sci 1995, 20, 3–10. [Google Scholar]
- Blázquez, MA; Lagunas, R; Gancedo, C; Gancedo, JM. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett 1993, 329, 51–54. [Google Scholar]
- Eastmond, PJ; van Dijken, AJH; Spielman, M; Kerr, A; Dickinson, HG; Jones, JDG; Smeekens, S; Graham, IA. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 2002, 29, 225–235. [Google Scholar]
- Gómez, LD; Baud, S; Gilday, A; Li, Y; Graham, IA. Delayed embryo development in the Arabidopsis TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J 2006, 46, 69–84. [Google Scholar]
- Satoh-Nagasawa, N; Nagasawa, N; Malcomber, S; Sakai, H; Jackson, D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 2006, 441, 227–230. [Google Scholar]
- Kormish, JD; McGhee, JD. C. elegans lethal gut-obstructed gob-1 gene is trehalose-6-phosphate phosphatase. Dev. Biol 2005, 287, 35–47. [Google Scholar]
- van Dijken, AJH; Schluepmann, H; Smeekens, SC. Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol 2004, 135, 969–977. [Google Scholar]
- Rolland, F; Baena-González, E; Sheen, J. Sugar sensing and signalling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol 2006, 57, 675–709. [Google Scholar]
- Schluepmann, H; Pellny, T; van Dijken, A; Smeekens, S; Paul, M. Trehalose-6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6849–6854. [Google Scholar]
- van Dijck, P; Mascorro-Gallardo, JO; de Bus, M; Royackers, K; Iturriaga, G; Thevelein, JM. Truncation of Arabidopsis thaliana and Selaginella lepidophylla trehalose-6-phosphatae synthase unlocks high catalytic activity and supports high trehalose levels on expression in yeast. Biochem. J 2002, 366, 63–71. [Google Scholar]
- Harthill, JE; Meek, SE; Morrice, N; Peggie, MW; Borch, J; Wong, B; Mackintosh, C. Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J 2006, 47, 211–223. [Google Scholar]
- Acevedo-Hernández, GJ; León, P; Herrera-Estrella, LR. Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J 2005, 43, 506–519. [Google Scholar]
- Ramon, M; Rolland, F; Thevelein, JM; van Dijck, P; Leyman, B. ABI4 mediates the effects of exogenous trehalose on Arabidopsis growth and starch breakdown. Plant Mol. Biol 2007, 63, 195–206. [Google Scholar]
- Wingler, A; Fritzius, T; Wiemken, A; Boller, T; Aeschbacher, A. Trehalose induces the ADP-Glc pyrophosphorylase gene, Apl3, and starch synthesis in Arabidopsis. Plant Physiol 2000, 124, 105–114. [Google Scholar]
- Paul, M; Pellny, T; Goddijn, O. Enhancing photosynthesis with sugar signals. Trends Plant Sci 2001, 6, 197–200. [Google Scholar]
- Lunn, JE; Feil, R; Hendriks, JHM; Gibon, Y; Morcuende, R; Osuna, D; Scheible, WR; Carillo, P; Hajirezaei, MR; Stitt, M. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J 2006, 397, 139–148. [Google Scholar]
- Kolbe, A; Tiessen, A; Schluepmann, H; Paul, M; Ulrich, S; Geigenberger, P. Trehalose-6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc. Natl. Acad. Sci. USA 2005, 102, 11118–11123. [Google Scholar]
- Baena-González, E; Rolland, F; Thevelein, JM; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar]
- Zhang, Y; Primavesi, LF; Jhurreea, D; Andralojc, PJ; Mitchell, R; Powers, S; Schluepmann, H; Delatte, T; Wingler, A; Paul, MJ. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 2009, 149, 1860–1871. [Google Scholar]
- Zaragoza, O; Blázquez, MA; Gancedo, C. Disruption of the Candida albicans TPS1 gene encoding trehalose-6-phophate synthase impairs formation of hyphae and decreases infectivity. J. Bacteriol 1998, 180, 3809–3815. [Google Scholar]
- van Dijck, P; de Rop, L; Szlufcick, K; van Ael, E; Thevelein, JM. Disruption of the Candida albicans TPS2 gene encoding trehalose-6-phosphatae phosphatase decreases infectivity without affecting hypha formation. Int. Immunol 2002, 70, 1772–1782. [Google Scholar]
- Murphy, H; Stewart, G; Mischenko, V; Apt, A; Harris, R; McAlister, M; Driscoll, D; Young, D; Robertson, B. The OtsAB is essential for trehalose biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem 2005, 280, 14524–14529. [Google Scholar]
- Foster, AJ; Jenkinson, JM; Talbot, NJ. Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J 2003, 22, 225–235. [Google Scholar]
- Brodmann, A; Schuller, A; Ludwig-Müller, J; Aeschbacher, R; Wiemken, A; Boller, T; Wingler, A. Induction of trehalase in Arabidopsis plants infected with the trehalose-producing pathogen Plasmodiophora brassicae. Mol. Plant-Microbe Interact 2002, 15, 693–700. [Google Scholar]
- Müller, J; Boller, T; Wiekman, A. Trehalose and trehalase in plants: Recent developments. Plant Sci 1995, 112, 1–9. [Google Scholar]
- Streeter, JG. Accumulation of α,α-trehalose by rhizobium bacteria and bacteroids. J. Bacteriol 1985, 164, 78–84. [Google Scholar]
- Müller, J; Boller, T; Wiemken, A. Trehalose becomes the most abundant non-structural carbohydrate during senescence of soybean nodules. J. Exp. Bot 2001, 52, 943–947. [Google Scholar]
- Hoelzle, I; Streeter, J. Increased accumulation of trehalose in rhizobia cultured under 1% oxygen. Appl. Environ. Microbiol 1990, 56, 3213–3215. [Google Scholar]
- Suárez, R; Wong, A; Ramírez, M; Barraza, A; Orozco, M; Cevallos, M; Lara, M; Hernández, G; Iturriaga, G. Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in Rhizobia. Mol. Plant-Microbe Interact 2008, 21, 958–966. [Google Scholar]
- Rodríguez-Salazar, SJ; Suárez, R; Caballero-Mellado, J; Iturriaga, G. Trehalose accumulation in Azospirillum improves drought tolerance and biomass in maize plants. FEMS Microbiol. Lett 2009, 296, 52–59. [Google Scholar]
| Used gene | Origin | Promoter | Transformed plant | Morphological alterations | Tolerance | Reference |
|---|---|---|---|---|---|---|
| TPS1 | Yeast | 35S | Tobacco | Yes | Drought | 24 |
| OtsAa | E. coli | 35S | Tobacco | Yes | Drought | 25, 28 |
| TPS1b | Yeast | 35S | Potato | Yes | Drought | 26 |
| TPS1 | Yeast | 35S | Tomato | Yes | Drought, salinity | 27 |
| TPS1 | Arabido psis | 35S | Arabidopsis | Flowering delay | Drought | 21 |
| OtsA-OtsBc | E. coli | ABRC1-actin1d | Rice | No | Drought, salinity, cold | 28 |
| TPS1-TPS2e | Yeast | 35S, RD29Af | Arabidopsis | No | Drought, salinity, heat, freezing | 29 |
| No | ||||||
| TPS1-TPS2 | Yeast | 35S, RD29A | Alfalfa | Stunted | Drought, salinity, heat, freezing | 30 |
| Larger |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Iturriaga, G.; Suárez, R.; Nova-Franco, B. Trehalose Metabolism: From Osmoprotection to Signaling. Int. J. Mol. Sci. 2009, 10, 3793-3810. https://doi.org/10.3390/ijms10093793
Iturriaga G, Suárez R, Nova-Franco B. Trehalose Metabolism: From Osmoprotection to Signaling. International Journal of Molecular Sciences. 2009; 10(9):3793-3810. https://doi.org/10.3390/ijms10093793
Chicago/Turabian StyleIturriaga, Gabriel, Ramón Suárez, and Barbara Nova-Franco. 2009. "Trehalose Metabolism: From Osmoprotection to Signaling" International Journal of Molecular Sciences 10, no. 9: 3793-3810. https://doi.org/10.3390/ijms10093793
APA StyleIturriaga, G., Suárez, R., & Nova-Franco, B. (2009). Trehalose Metabolism: From Osmoprotection to Signaling. International Journal of Molecular Sciences, 10(9), 3793-3810. https://doi.org/10.3390/ijms10093793




