Interactions of Laurylated and Myristoylated KR12 Fragment of the LL37 Peptide with Polyoxidovanadates
Abstract
1. Introduction
2. Results and Discussion
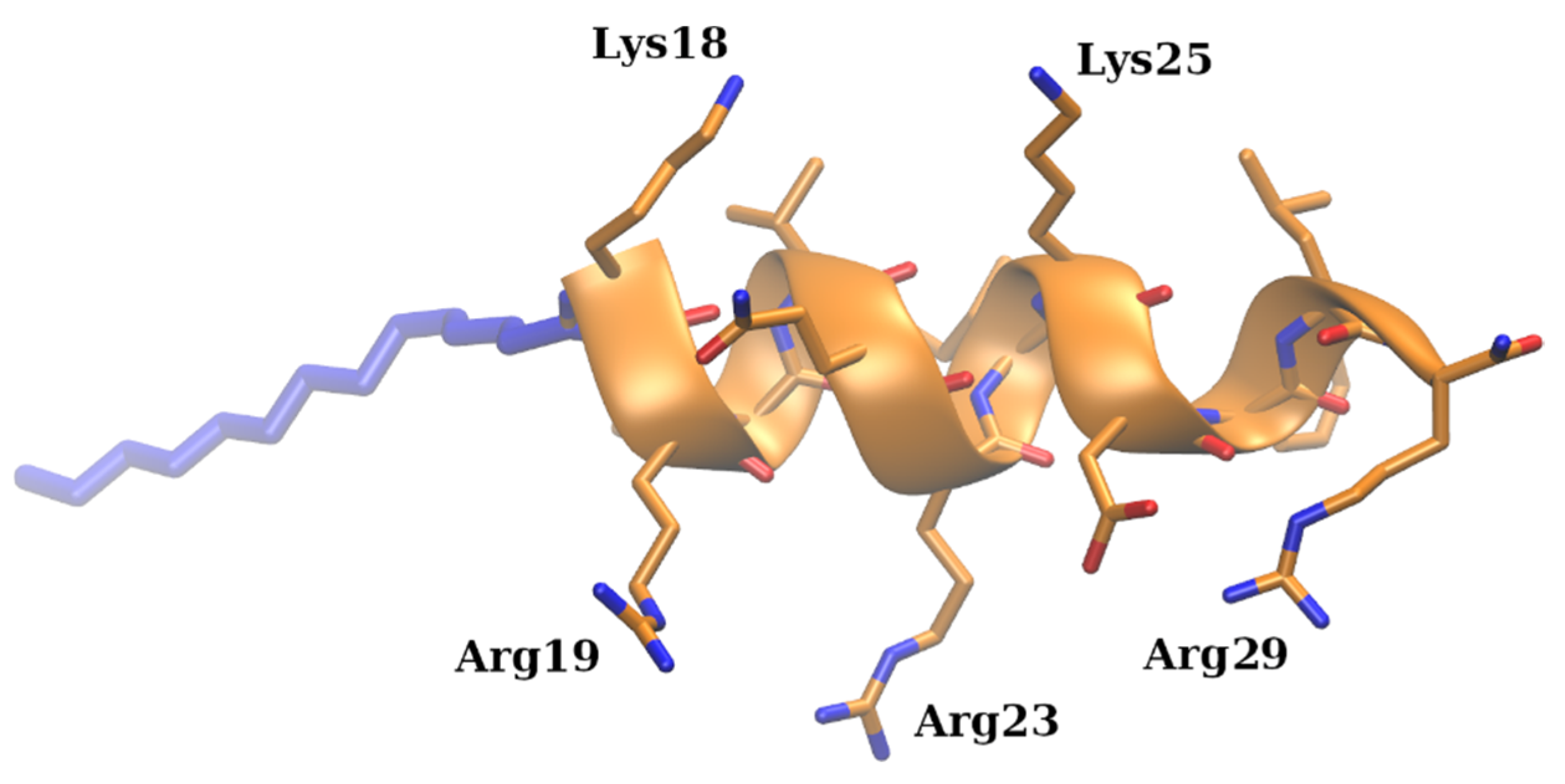
2.1. Experimental Conditions Affect the Structure and Charge of the Investigated Molecules
2.2. Thermodynamic Parameters of Lipopeptide–Polyoxidovanadate Interactions
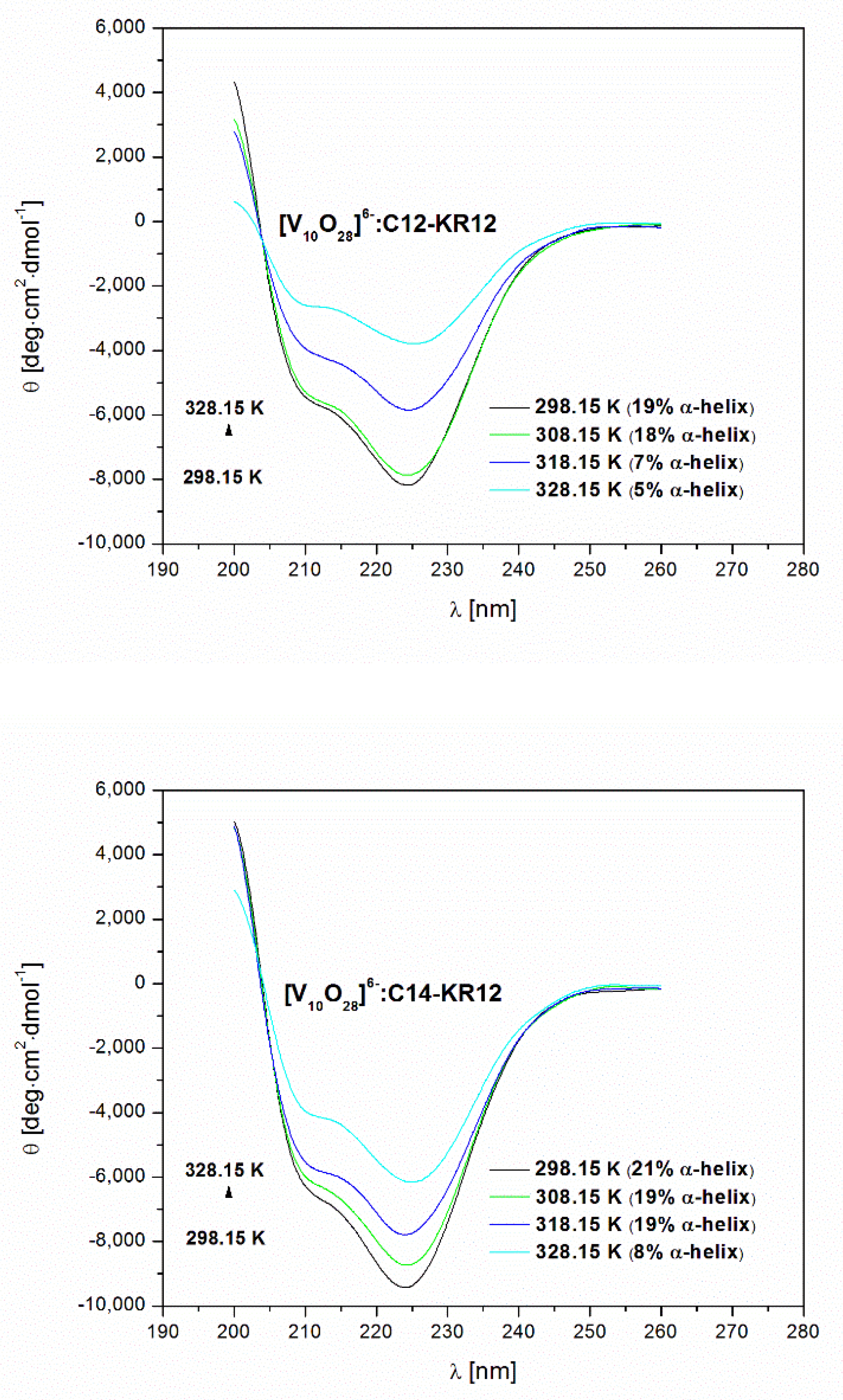
2.3. The Effect of Environmental Conditions on the Secondary Structure of Lipopeptides
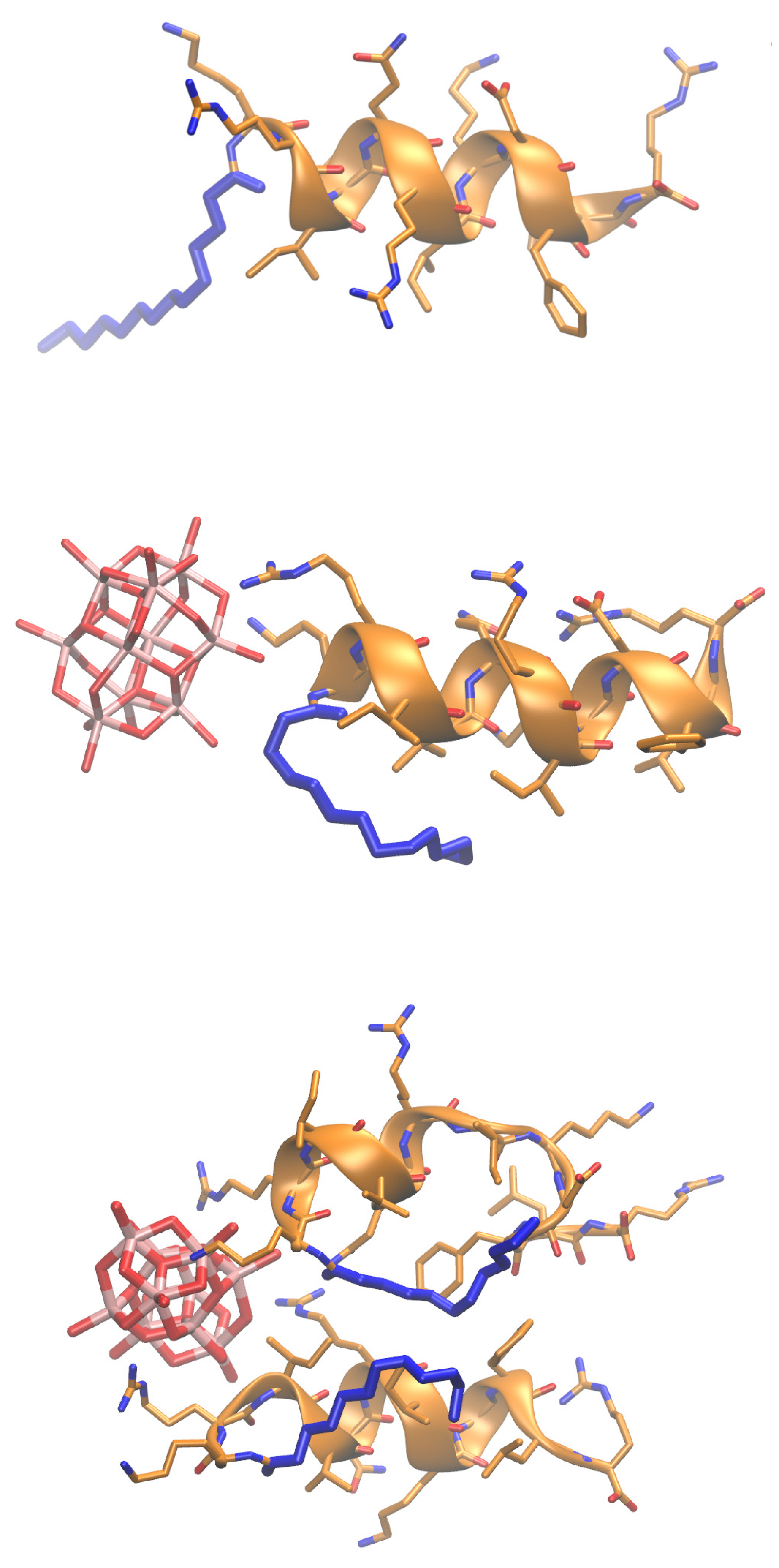
2.4. Molecular Insights into Decavanadate–Lipopeptide Interactions
3. Materials and Methods
3.1. Reagents
3.2. Synthesis of (NH4)6[V10O28](H2O)6
3.3. Synthesis of the C12-KR12 and C14-KR12 Peptides
3.4. Isothermal Titration Calorimetry (ITC)
3.5. Circular Dichroism Spectroscopy (CD)
3.6. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aureliano, M.; Gumerova, N.I.; Sciortino, G.; Garribba, E.; McLauchlan, C.C.; Rompel, A.; Crans, D.C. Polyoxidovanadates’ interactions with proteins: An overview. Coord. Chem. Rev. 2022, 454, 214344. [Google Scholar]
- Bijelic, A.; Aureliano, M.; Rompel, A. Polyoxometalates as Potential Next-Generation Metallodrugs in the Combat Against Cancer. Angew. Chem. Int. Ed. Engl. 2019, 58, 2980–2999. [Google Scholar] [PubMed]
- Wang, X.; Wei, S.; Zhao, C.; Li, X.; Jin, J.; Shi, X.; Su, Z.; Li, J.; Wang, J. Promising application of polyoxometalates in the treatment of cancer, infectious diseases and Alzheimer’s disease. J. Biol. Inorg. Chem. 2021, 27, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, N.; Kamata, K. Catalytic oxidation of hydrocarbons with hydrogen peroxide by vanadium-based polyoxometalates. Coord. Chem. Rev. 2011, 19, 2358–2370. [Google Scholar]
- Li, J.; Yu, Z.; Zhang, J.; Liu, C.; Zhang, Q.; Shi, H.; Wu, D. Rapid, Massive, and Green Synthesis of Polyoxometalate-Based Metal–Organic Frameworks to Fabricate POMOF/PAN Nanofiber Membranes for Selective Filtration of Cationic Dyes. Molecules 2024, 29, 1493. [Google Scholar] [CrossRef]
- Díaz, J.; Pizzio, L.R.; Pecchi, G.; Campos, C.H.; Azócar, L.; Briones, R.; Romero, R.; Troncoso, E.; Méndez-Rivas, C.; Melín, V.; et al. Catalytic Selective Oxidation of β-O-4 Bond in Phenethoxybenzene as a Lignin Model Using (TBA)5[PMo10V2O40] Nanocatalyst: Optimization of Operational Conditions. Molecules 2023, 28, 6368. [Google Scholar] [CrossRef]
- Gao, Y.; Choudhari, M.; Such, G.K.; Ritchie, C. Polyoxometalates as chemically and structurally versatile components in self-assembled materials. Chem. Sci. 2022, 13, 2510–2527. [Google Scholar]
- Wang, H.; Li, B. Recent Advances on the Functionalities of Polyoxometalate-Based Ionic Liquids. Molecules 2024, 29, 3216. [Google Scholar] [CrossRef]
- Jones, C.F.; Hood, B.R.; de Coene, Y.; Lopez-Poves, I.; Champagne, B.; Clays, K.; Fielden, J. Bridge improvement work: Maximising non-linear optical performance in polyoxometalate derivatives. Chem. Commun. 2024, 60, 1731–1734. [Google Scholar]
- Rehder, D. Import and Implications of Vanadium in Live Aspects. Inorganics 2023, 11, 256. [Google Scholar] [CrossRef]
- Pessoa, J.C.; Correia, I. Misinterpretations in Evaluating Interactions of Vanadium Complexes with Proteins and Other Biological Targets. Inorganics 2021, 9, 17. [Google Scholar] [CrossRef]
- Aureliano, M.; Crans, D.C. Decavanadate (V10O286−) and oxovanadates: Oxometalates with many biological activities. J. Inorg. Biochem. 2009, 103, 536–546. [Google Scholar] [PubMed]
- Greijer, B.H.; Nestor, G.; Eriksson, J.E.; Seisenbaeva, G.A.; Kessler, V.G. Factors influencing stoichiometry and stability of polyoxometalate—Peptide complexes. Dalton Trans. 2022, 51, 9511–9521. [Google Scholar]
- Turner, T.L.; Nguyen, V.H.; McLauchlan, C.C.; Dymon, Z.; Dorsey, B.M.; Hooker, J.D.; Jones, M.A. Inhibitory effects of decavanadate on several enzymes and Leishmania tarentolae In Vitro. J. Inorg. Biochem. 2012, 108, 96–104. [Google Scholar]
- Aureliano, M.; Ohlin, C.A. Decavanadate in vitro and in vivo effects: Facts and opinions. J. Inorg. Biochem. 2014, 137, 123–130. [Google Scholar]
- Lakshmaiah Narayana, J.; Mechesso, A.F.; Rather, I.I.G.; Zarena, D.; Luo, J.; Xie, J.; Wang, G. Origami of KR-12 Designed Antimicrobial Peptides and Their Potential Applications. Antibiotics 2024, 13, 816. [Google Scholar] [CrossRef]
- Kamysz, E.; Sikorska, E.; Jaśkiewicz, M.; Bauer, M.; Neubauer, D.; Bartoszewska, S.; Barańska-Rybak, W.; Kamysz, W. Lipidated Analogs of the LL-37-Derived Peptide Fragment KR12—Structural Analysis, Surface-Active Properties and Antimicrobial Activity. Int. J. Mol. Sci. 2020, 21, 887. [Google Scholar] [CrossRef]
- Brzeski, J.; Wyrzykowski, D.; Makowska, J. Application of a modern theoretical approach to the study of the interaction of KR-12 peptides derived from human cathelicidins with Cu(II) ions. Dalton Trans. 2024, 23, 9942–9951. [Google Scholar]
- Paredes-Pérez, L.F.; Mendoza, A.; García-García, A.; Serrano-De la Rosa, L.E.; Méndez-Rojas, M.A.; Melendez, F.J. Guanidinium and spermidinium decavanadates: As small biomimetic models to understand non-covalent interactions between decavanadate and arginine and lysine side chains in proteins. Front. Chem. Biol. 2024, 3, 1451167. [Google Scholar]
- Crans, C.D.; Mahroof-Tahir, M.; Anderson, P.O.; Miller, M.M. X-ray Structure of (NH4)6(Gly-Gly)2V10O28∙4H2O: Model Studies for Polyoxometalate-Protein Interactions. Inorg. Chem. 1994, 33, 5586–5590. [Google Scholar]
- Zhao, L.; Cao, Z.; Bian, Y.; Hu, G.; Wang, J.; Zhou, Y. Molecular Dynamics Simulations of Human Antimicrobial Peptide LL-37 in Model POPC and POPG Lipid Bilayers. Int. J. Mol. Sci. 2018, 19, 1186. [Google Scholar] [CrossRef] [PubMed]
- Salazar Marcano, D.E.; Savić, N.D.; Abdelhameed, S.A.M.; de Azambuja, F.; Parac-Vogt, T.N. Exploring the Reactivity of Polyoxometalates toward Proteins: From Interactions to Mechanistic Insights. JACS Au 2023, 3, 978–990. [Google Scholar] [PubMed]
- Kamysz, E.; Sikorska, E.; Bauer, M.; Sikora, K.; Neubauer, D. Influence of Lipidation Pattern of the KR12 Fragment of Peptide LL-37 on Its Antibacterial and Hemolytic Activities. Int. J. Mol. Sci. 2023, 24, 5505. [Google Scholar] [CrossRef]
- Kamysz, W.; Okrój, M.; Łempicka, E.; Ossowski, T.; Łukasiak, J. Fast and efficient purification of synthetic peptides by solid-phase extraction. Acta Chromatogr. 2004, 14, 180–186. [Google Scholar]
- Makowska, M.; Kosikowska-Adamus, P.; Zdrowowicz, M.; Wyrzykowski, D.; Prahl, A.; Sikorska, E. Lipidation of Naturally Occurring α-Helical Antimicrobial Peptides as a Promising Strategy for Drug Design. Int. J. Mol. Sci. 2023, 24, 3951. [Google Scholar] [CrossRef]
- Case, D.A.; Cerutti, D.S.; III Cheatham, T.E.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Greene, D.; Homeyer, N.; et al. AMBER 2017; University of California: San Francisco, CA, USA, 2017. [Google Scholar]
- Bayly, I.C.; Cieplak, P.; Cornell, W.; Kollman, A.P. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Wang, J.; Wolf, M.R.; Caldwell, W.J.; Kollman, A.P.; Case, A.D. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C.J. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar]

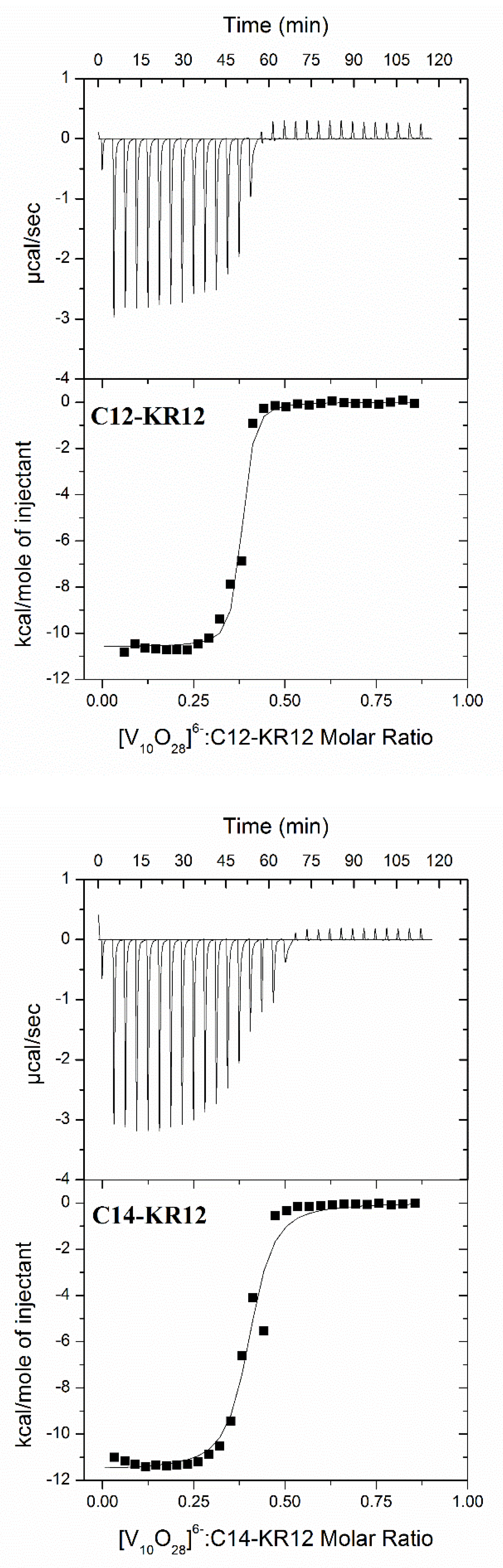

| Parameter | [V10O286−] + C12-KR12 | [V10O286−] + C14-KR12 |
|---|---|---|
| N (stoichiometry) | 0.37 (±0.01) | 0.39 (±0.01) |
| logK(ITC) | 6.84 (±0.15) | 6.16 (±0.15) |
| ΔG(ITC) [kcal mol−1] | −9.33 (±0.20) | −8.41 (±0.18) |
| ΔH(ITC) [kcal mol−1] | −10.58 (±0.15) | −11.54 (±0.25) |
| TΔS(ITC) [kcal mol−1] | −1.25 | −3.13 |
| Temperature [K] | The Percentage of α-Helical Content [%] | |||
|---|---|---|---|---|
| C12-KR12 | [V10O28]6−:C12-KR12 | C14-KR12 | [V10O28]6−:C14-KR12 | |
| 298.15 | 36 | 19 | 49 | 21 |
| 308.15 | 32 | 18 | 46 | 19 |
| 318.15 | 26 | 7 | 42 | 19 |
| 328.15 | 7 | 5 | 38 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapica, M.; Kamysz, E.; Grabowska, O.; Tesmar, A.; Pająk, M.; Chmur, K.; Brzeski, J.; Samsonov, S.A.; Wyrzykowski, D. Interactions of Laurylated and Myristoylated KR12 Fragment of the LL37 Peptide with Polyoxidovanadates. Molecules 2025, 30, 1589. https://doi.org/10.3390/molecules30071589
Kapica M, Kamysz E, Grabowska O, Tesmar A, Pająk M, Chmur K, Brzeski J, Samsonov SA, Wyrzykowski D. Interactions of Laurylated and Myristoylated KR12 Fragment of the LL37 Peptide with Polyoxidovanadates. Molecules. 2025; 30(7):1589. https://doi.org/10.3390/molecules30071589
Chicago/Turabian StyleKapica, Martyna, Elżbieta Kamysz, Ola Grabowska, Aleksandra Tesmar, Marek Pająk, Katarzyna Chmur, Jakub Brzeski, Sergey A. Samsonov, and Dariusz Wyrzykowski. 2025. "Interactions of Laurylated and Myristoylated KR12 Fragment of the LL37 Peptide with Polyoxidovanadates" Molecules 30, no. 7: 1589. https://doi.org/10.3390/molecules30071589
APA StyleKapica, M., Kamysz, E., Grabowska, O., Tesmar, A., Pająk, M., Chmur, K., Brzeski, J., Samsonov, S. A., & Wyrzykowski, D. (2025). Interactions of Laurylated and Myristoylated KR12 Fragment of the LL37 Peptide with Polyoxidovanadates. Molecules, 30(7), 1589. https://doi.org/10.3390/molecules30071589







