Iohexol as an Emerging Contaminant Potentially Influencing Structural and Functional Changes in the Activated Sludge Microbiome
Abstract
1. Introduction
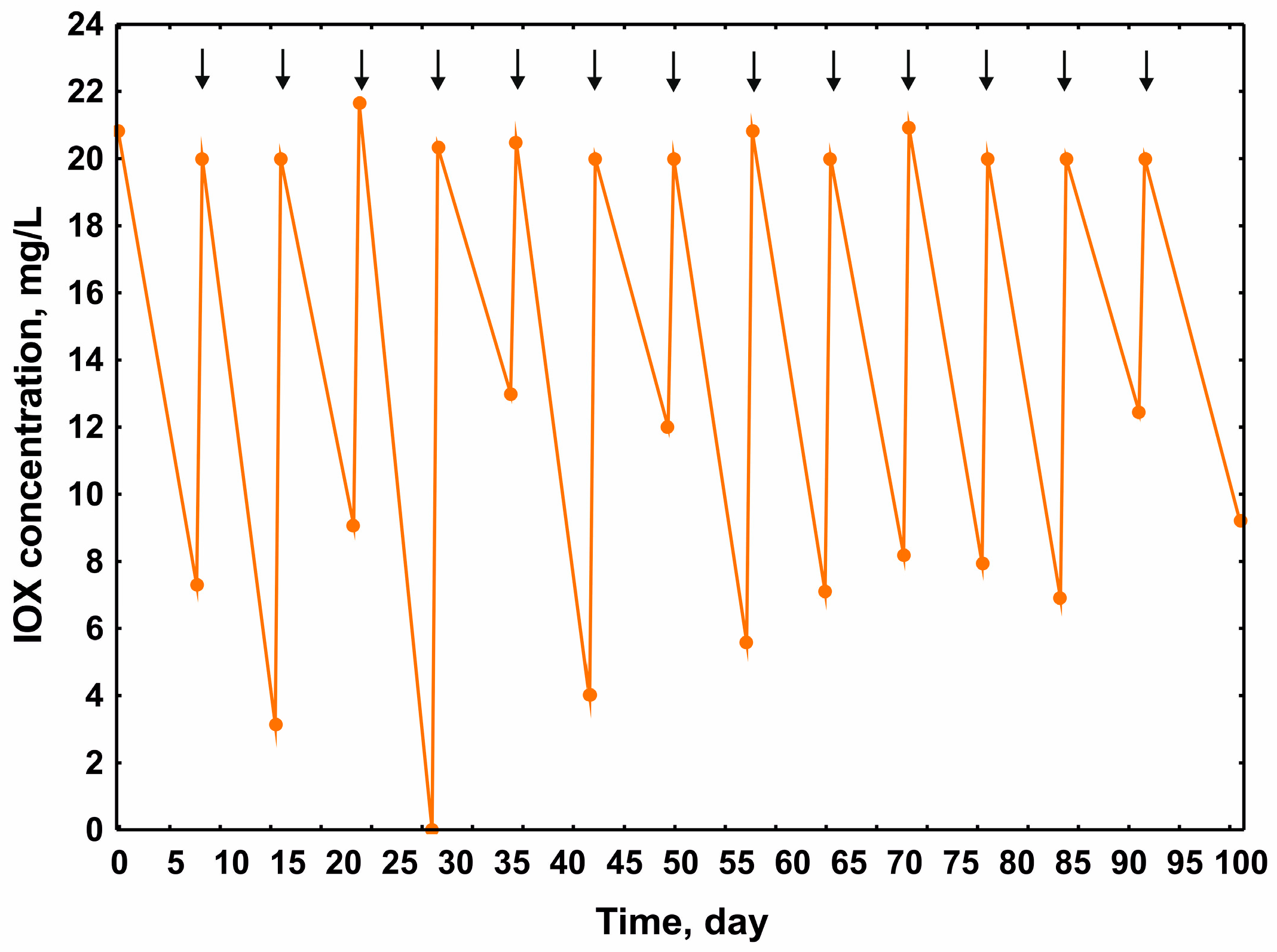
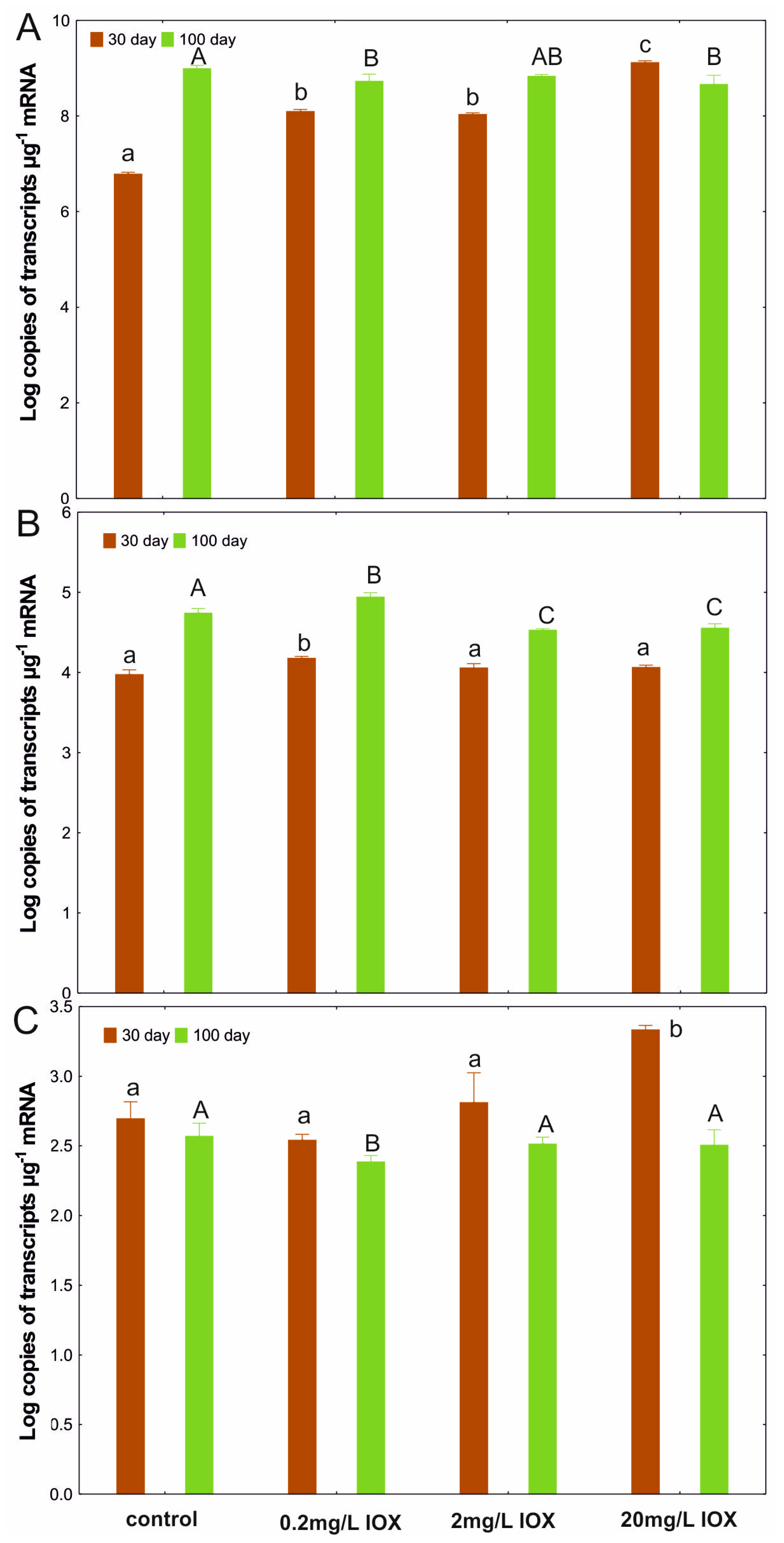
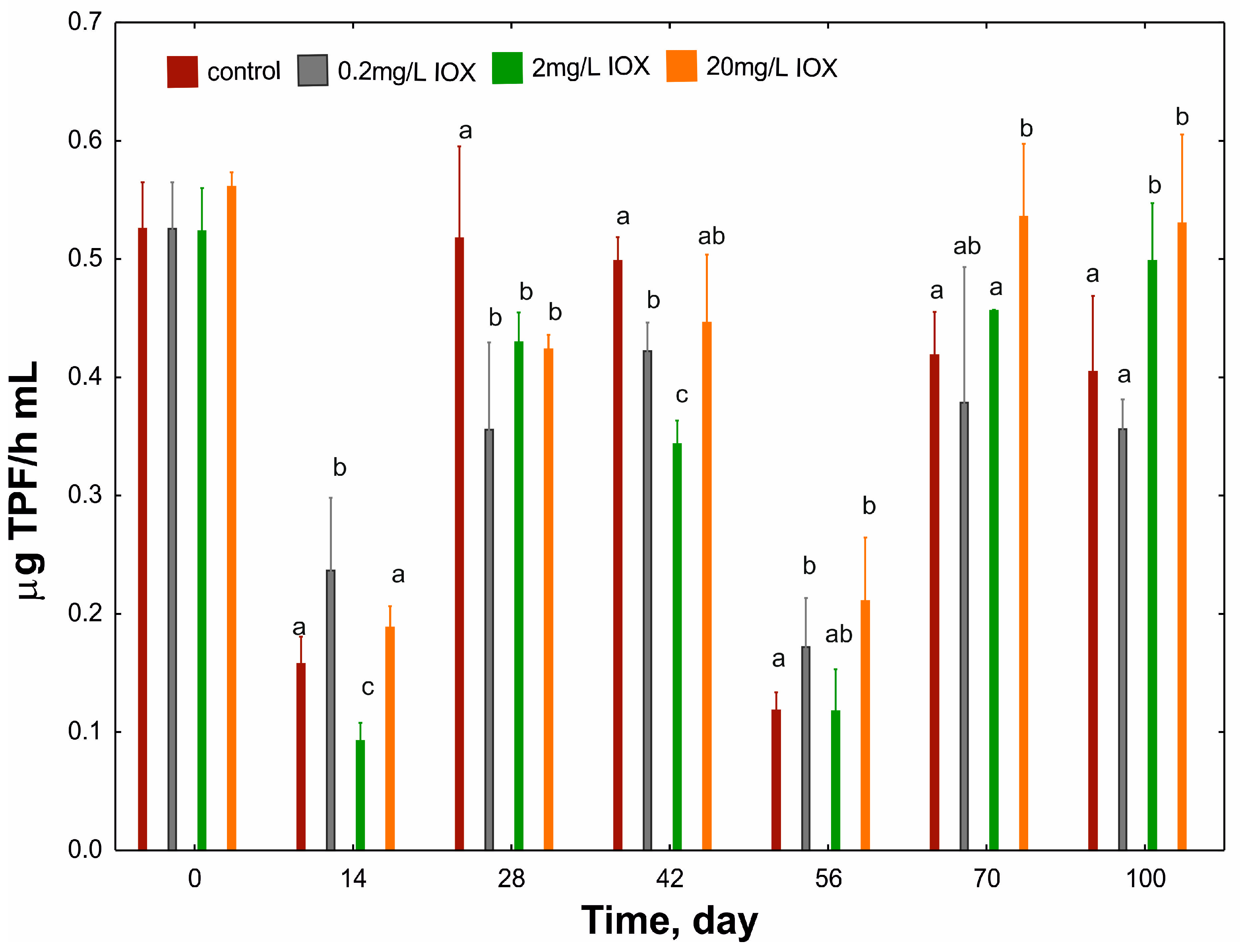
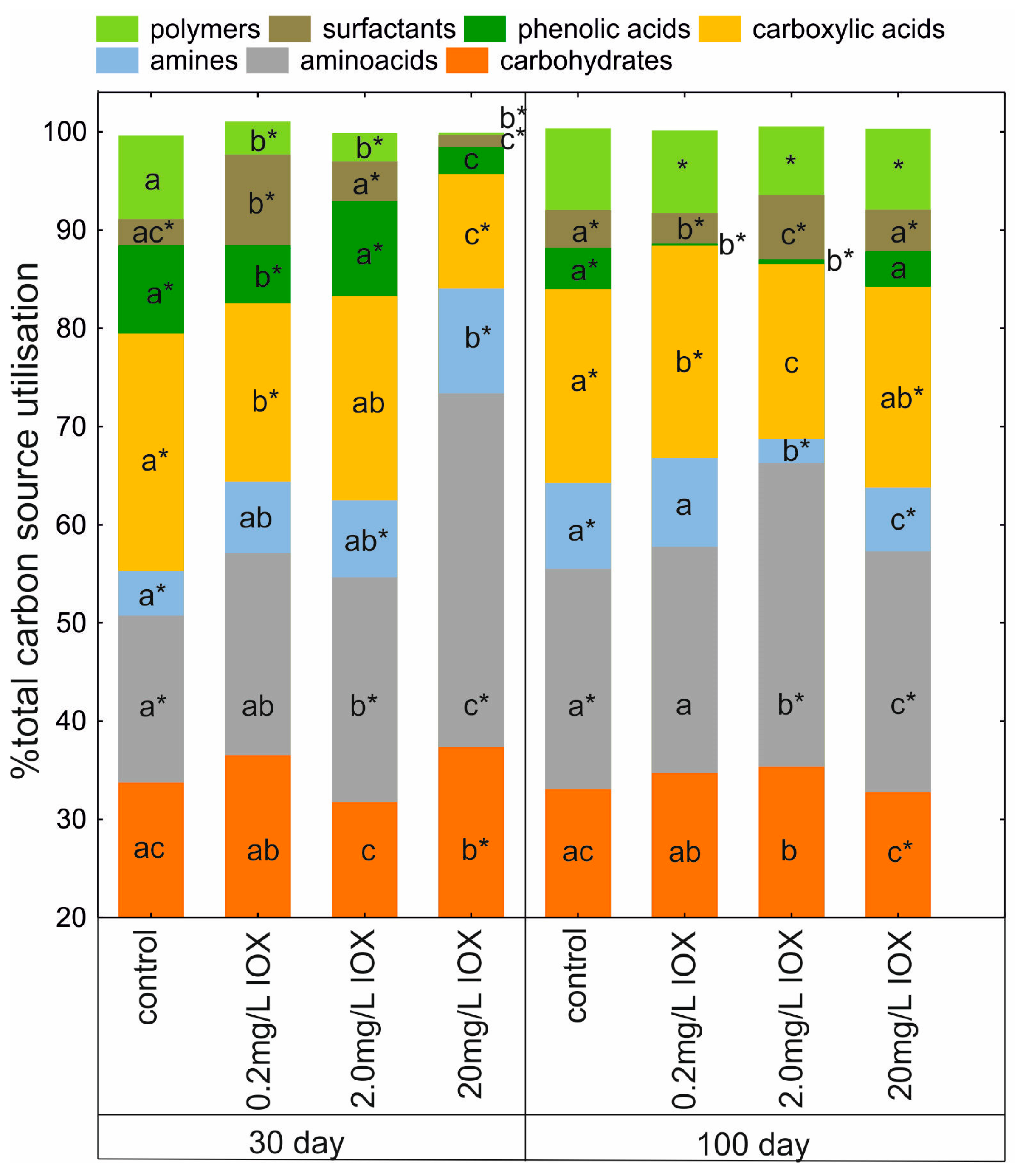
2. Results and Discussion
3. Materials and Methods
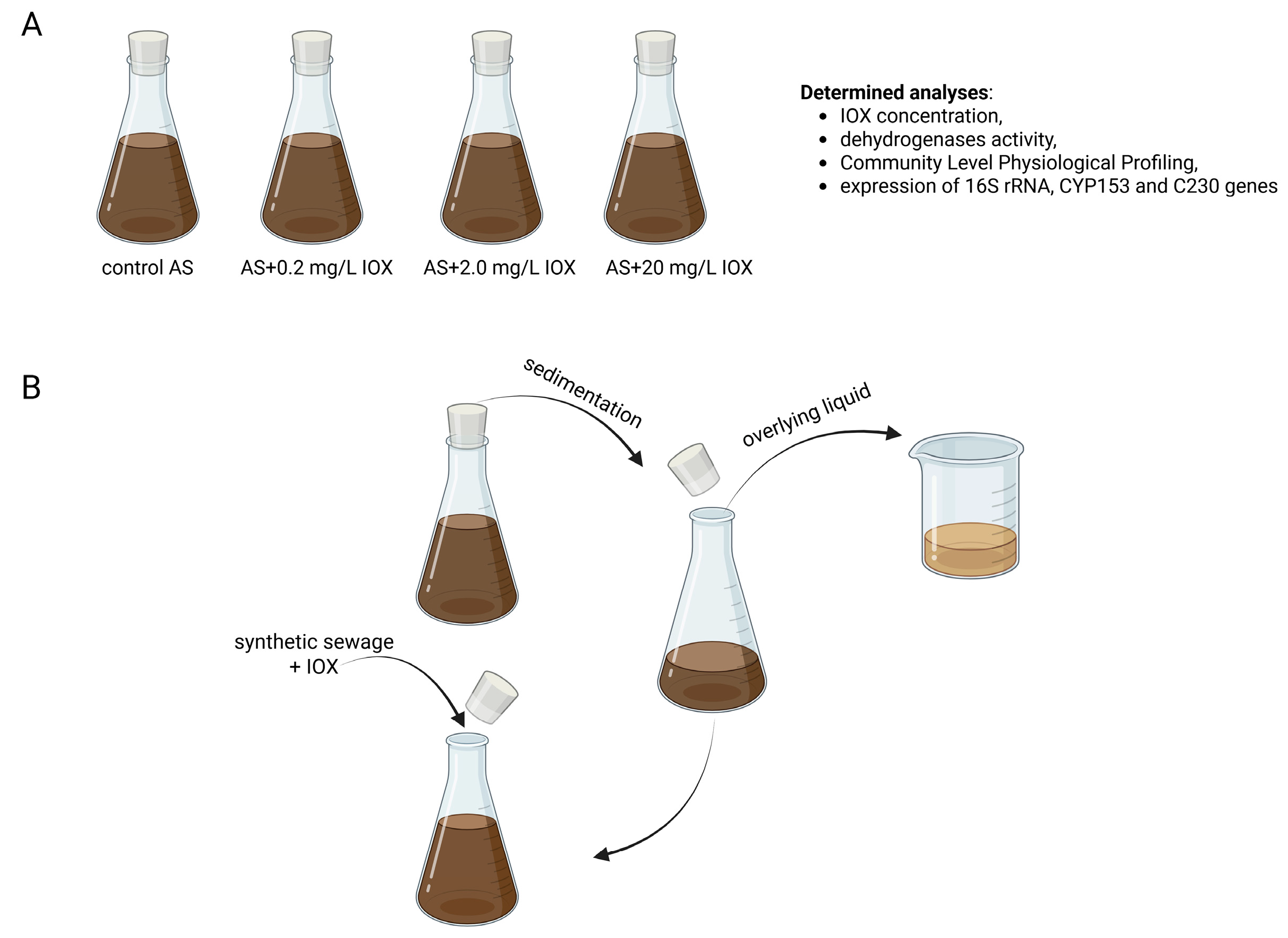
3.1. Sample Collection and Experimental Setup
3.2. Sewage Physico-Chemical Analysis
3.3. Measuring Dehydrogenase Activity
3.4. Determination of IOX Concentration
3.5. Community-Level Physiological Profiling
3.6. RNA Extraction and Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
3.7. Data Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rusandu, A.; Bustadmo, L.; Gravvold, H.; Anvik, M.S.; Skilleas Olsen, K.; Hanger, N. Iodinated contrast media waste management in hospitals in central Norway. Radiography 2024, 30, 1272–1276. [Google Scholar] [CrossRef]
- Nowak, A.; Pacek, G.; Mrozik, A. Transformation and ecotoxicological effects of iodinated X-ray contrast media. Rev. Environ. Sci. Biotechnol. 2020, 19, 337–354. [Google Scholar] [CrossRef]
- Xiao, Y.D.; Paudel, R.; Liu, J.; Ma, C.; Zhang, Z.S.; Zhou, S.K. MRI contrast agents: Classification and application (Review). Int. J. Mol. Med. 2016, 38, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Daftary, A. Iodinated contrast media and their adverse reactions. J. Nucl. Med. Technol. 2008, 36, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Kormos, J.L.; Schulz, M.; Kohler, H.P.E.; Ternes, T.A. Biotransformation of selected iodinated X-ray contrast media and characterization of microbial transformation pathways. Environ. Sci. Technol. 2010, 44, 4998–5007. [Google Scholar] [CrossRef]
- Akao, P.K.; Mamane, H.; Kaplan, A.; Gozlan, I.; Yehoshua, Y.; Kinel-Tahan, Y.; Avisar, D. Iohexol removal and degradation-product formation via biodegradation by microalga Chlorella vulgaris. Algal Res. 2020, 51, 102050. [Google Scholar] [CrossRef]
- Lopez-Prieto, I.; Wu, S.; Ji, W.; Daniels, K.D.; Snyder, S.A. A direct injection liquid chromatography tandem mass spectrometry method for the kinetic study on ionidated contrast media (ICMs) removal in natural water. Chemosphere 2020, 243, 125311. [Google Scholar] [CrossRef]
- Zhang, W.; Fourcade, F.; Amrane, A.; Geneste, F. Removal of iodine-containing X-ray contrast media from environment: The challenge of a total mineralization. Molecules 2023, 28, 341. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Z.; Tang, Y.; Xiao, D.; Zhang, D.; Huang, Y.; Guo, Y.; Liu, J. Oxidative degradation of iodinated X-ray contrast media (iomeprol and iohexol) with sulfate radical: An experimental and theoretical study. Chem. Eng. J. 2019, 368, 999–1012. [Google Scholar] [CrossRef]
- Dekker, H.M.; Stroomberg, G.J.; Prokop, M. Tackling the increasing contamination of the water supply by iodinated contrast media. Insights Imaging 2022, 13, 30. [Google Scholar] [CrossRef]
- Zinicovscaia, I.; Yushin, N.; Sifelani, D.; Humelnicu, D. Efficiency of biological versus mesoporous adsorbents for gadolinium removal from wastewater. Sci. Rep. 2025, 15, 30500. [Google Scholar] [CrossRef]
- Sousa, J.; Pinto, J.; Barbosa, H.; Tavares, D.S.; Freitas, R.; Trindade, T.; Rocha, J.; Pereira, E. Efficient recovery of gadolinium from contaminated waters using manganese ferrite nanoparticles. Recycling 2025, 10, 57. [Google Scholar] [CrossRef]
- Puertas, E.C.; Peyot, M.L.; Pineda, M.; Volk, K.; Coulombe, S.; Yargeau, V. Degradation of diatrizoate in a pin-to-liquid plasma reactor, its transformation products and their residual toxicity. Sci. Total Environ. 2021, 782, 146895. [Google Scholar] [CrossRef]
- Singh, R.R.; Rajnarayanan, R.; Aga, D.S. Binding of ionidated contrast media (ICM) and their transformation products with hormone receptors: Are ICM the new EDCs? Sci. Total Environ. 2019, 692, 32–36. [Google Scholar] [CrossRef]
- Oppenheimer, J.A.; Prasse, C.; Newmeyer, M.; Schwab, K.J.; Jacangelo, J.G. Monitoring iohexol and its transformation products as evidence of reclaimed water irrigation input to contiguous waterbodies. Sci. Total Environ. 2024, 946, 174351. [Google Scholar] [CrossRef]
- Scheider, I.; Yotinov, I.; Dinova, N.; Geneva, B.; Daskalova, E.; Lincheva, S.; Topalova, Y. Assessment of denitrification and nitrification processes during landfill leachate treatment. Processes 2023, 11, 2960. [Google Scholar] [CrossRef]
- Dzionek, A.; Wojcieszyńska, D.; Menashe, O.; Szada, D.; Potocka, I.; Jesionowski, T.; Guzik, U. The influence of activated sludge augmentation on its ability to degrade paracetamol. Molecules 2024, 29, 4520. [Google Scholar] [CrossRef]
- Derre, M.G.; Steinbach, S.M.; Hendrix, G.K.; Stanton, M.; Adams, L.G. Iodinated non-ionic contrast medium does not inhibit Escherichia coli growth in ex vivo inoculated canine urine. Am. J. Vet. Res. 2023, 14, 84. [Google Scholar] [CrossRef]
- Nowak, A.; Pacwa-Płociniczak, M.; Marchlewicz, A. The effect of long-term exposure to diatrizoate on microbial communities in activated sludge. Sci. Rep. 2025, 15, 17441. [Google Scholar] [CrossRef]
- Johnston, J.; Behrens, S. Seasonal dynamics of the activated sludge microbiome in sequencing batch reactors, assessed using 16S rRNA transcript amplicon sequencing. Appl. Environ. Microbiol. 2020, 86, e00597-20. [Google Scholar] [CrossRef]
- Johnston, J.; Vilardi, K.; Cotto, I.; Sudarshan, A.; Bian, K.; Klaus, S.; Bachmann, M.; Parsons, M.; Wilson, C.; Bott, C.; et al. Metatranscriptomic analysis reveals synergistic activities of comammox and anammox bacteria in full-scale attached growth nitrogen removal system. Environ. Sci. Technol. 2024, 58, 13023–13034. [Google Scholar] [CrossRef] [PubMed]
- Hupert-Kocurek, K.; Wojcieszyńska, D.; Guzik, U.; Borowski, T. A single amino acid substitution within catalytically non-active N-terminal domain of catechol 2,3-dioxygenase (C23O) increases enzyme activity towards 4-chlorocatechol. J. Mol. Catal. B Enzym. 2015, 122, 64–71. [Google Scholar] [CrossRef]
- Patil, V.V.; Park, K.H.; Lee, S.G.; Woo, E. Structural analysis of the phenol-responsive sensory domain of the transcription activator PoxR. Structure 2016, 24, 624–630. [Google Scholar] [CrossRef][Green Version]
- Liwrska-Bizukojic, E. Influence of imidazolium ionic liquids on dehydrogenase activity of activated sludge microorganisms. Water Air Soil. Pollut. 2011, 221, 327–335. [Google Scholar] [CrossRef][Green Version]
- Tomska, A. Influence of selected pharmaceuticals on activated sludge dehydrogenase activity. Ecol. Eng. Environ. Technol. 2016, 28, 214–218. (In Polish) [Google Scholar] [CrossRef]
- Feng, Q.; Xiao, Y.; Li, X.; Xue, Z.; Fang, F.; Cao, J.; Oleyiblo, J.O.; Hu, Z. Using the dehydrogenase activity for alert of activated sludge system under different copper concentrations. Desal Water Treat. 2016, 57, 17836–17843. [Google Scholar] [CrossRef]
- Hu, C.-Y.; Xu, L.; Lin, Y.-L.; Li, C. A comparative study on the degradation of iohexol and diatrizoate during UV/persulfate process: Kinetics, degradation pathways and iodinated disinfection by-products. Environ. Sci. Water Res. Technol. 2024, 10, 718. [Google Scholar] [CrossRef]
- Hapeshi, E.; Lambrianides, A.; Koutsoftas, P.; Kastanos, E.; Michael, C.; Fatta-Kassinos, D. Investigating the fate of iodinated X-ray contrast media iohexol and diatrizoate during microbial degradation in an MBBR system treating urban wastewater. Environ. Sci. Pollut. Res. 2013, 20, 3592–3606. [Google Scholar] [CrossRef]
- Yang, X.; Yin, M.; Zhu, Y.; Zhao, S.; Xi, H. Key genomes, transcriptomes, proteins, and metabolic factors involved in the detoxification/tolerance of TNT and its intermediates by bacteria in anaerobic/aerobic environments. J. Hazard. Mater. 2024, 478, 135489. [Google Scholar] [CrossRef]
- Phale, P.S.; Malhotra, H.; Shah, B.A. Chapter One—Degradation strategies and associated regulatory mechanisms/features for aromatic compound metabolism in bacteria. Adv. Appl. Microbiol. 2020, 112, 1–65. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Dai, Y.; Wu, L.; Qi, C.; Hou, L.; Zhang, D.; Li, Y.; Xu, C.; He, H.; Yang, S. Degradation of iohexol in the Co(II)/peracetic acid system under neutral conditions: Influencing factors, degradation pathways and toxicity. Sep. Purif. Technol. 2023, 319, 124083. [Google Scholar] [CrossRef]
- Michalska, J.; Greń, I.; Żur, J.; Wasilkowski, D.; Mrozik, A. Impact of the biological cotreatment of the Kalina Pond leachate on laboratory sequencing batch reactor operation and activated sludge quality. Water 2019, 11, 1539. [Google Scholar] [CrossRef]
- Miksch, K. Application of dehydrogenase activity determinations in biodegradation of refinery sewage. Gas Water Civ. Technol. 1977, 51, 234–235. (In Polish) [Google Scholar]
- Nowak, A.; Mrozik, A. Degradation of 4-chlorophenol and microbial diversity in soil inoculated with single Pseudomonas sp. CF600 and Stenotrophomonas maltophilia KB2. J. Environ. Manag. 2018, 215, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Al-Mutairi, N.Z. Variable distributional characteristics of substrate utilisation patterns in activated sludge plants in Kuwait. Biores Technol. 2008, 100, 1524–1532. [Google Scholar] [CrossRef]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef] [PubMed]
- Sei, K.; Asano, K.I.; Tateishi, N.; Mori, K.; Ike, M.; Fujita, M. Design of PCR primers and gene probes for the general detection of bacterial populations capable of degrading aromatic compounds via catechol cleavage pathways. J. Biosci. Bioeng. 1999, 88, 542–550. [Google Scholar] [CrossRef]
- Yousaf, S.; Ripka, K.; Reichenauer, T.G.; Andria, V.; Afzal, M.; Sessitsch, A. Hydrocarbon degradation and plant colonization by selected bacterial strains isolated from Italian ryegrass and birdsfoot trefoil. J. Appl. Microbiol. 2010, 109, 1389–1401. [Google Scholar] [CrossRef]
- Gupta, V.B.; Shrivastava, A. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron. Young Sci. 2011, 2, 21–25. [Google Scholar] [CrossRef]
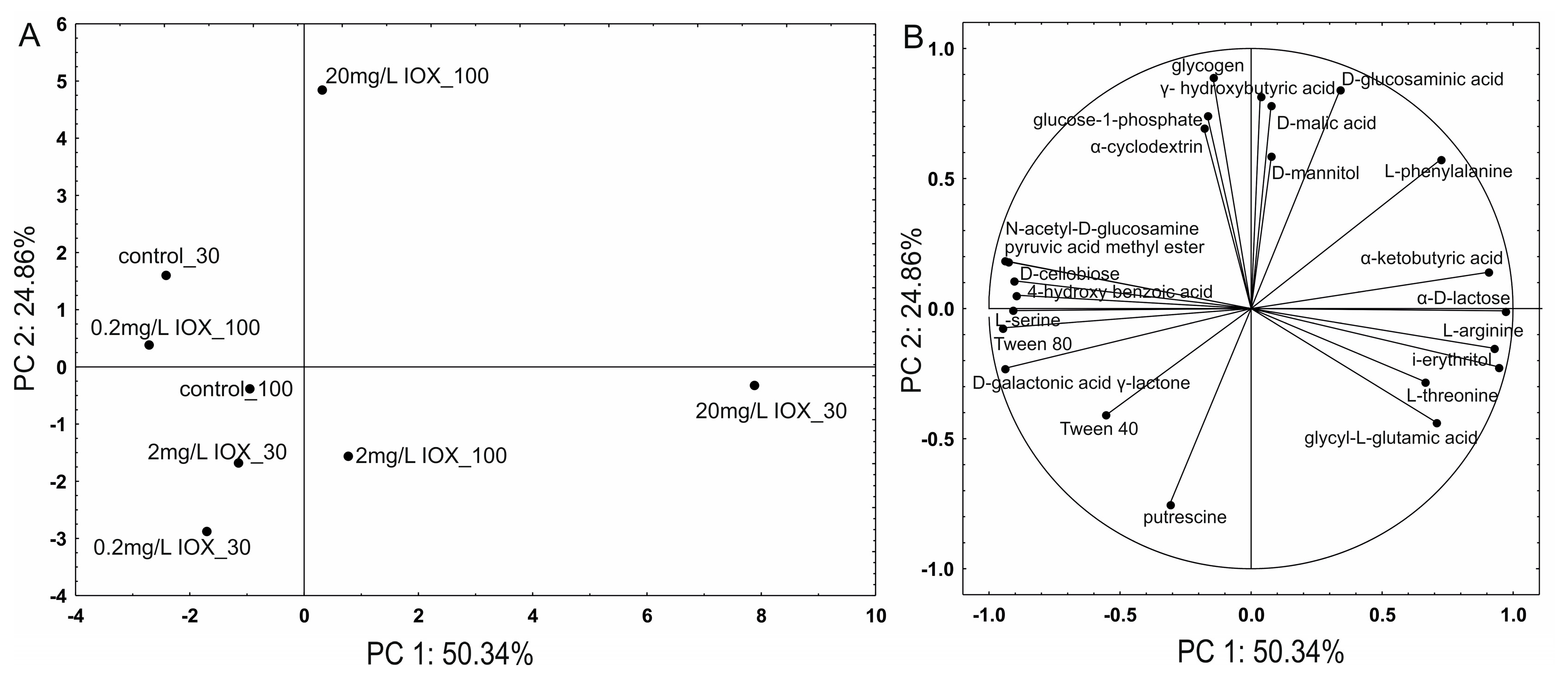
| G | H′ | R | AWCD | N-Use, % | P-Use, % | |
|---|---|---|---|---|---|---|
| start | 0.54 ± 0.02 | 1.27 ± 0.02 | 27.5 ± 1.6 | 1.11 ± 0.14 | 35.8 ± 4.2 | 3.76 ± 0.42 |
| control_30 | 0.60 ± 0.07 a | 1.25 ± 0.02 a | 24.0 ± 2.2 a | 0.93 ± 0.07 a | 28.8 ± 0.5 a | 7.86 ± 0.00 a |
| 0.2 IOX_30 | 0.71 ± 0.11 b | 1.27 ± 0.01 ab | 21.5 ± 4.9 ab | 0.84 ± 0.21 a | 33.2 ± 4.7 c | 0.09 ± 0.05 b |
| 2.0 IOX_30 | 0.59 ± 0.09 a | 1.29 ± 0.03 b | 23.5 ± 4.9 ab | 0.84 ± 0.22 a | 35.9 ± 3.5 bc | 0.98 ± 0.09 c |
| 20 IOX_30 | 0.87 ± 0.09 c | 1.05 ± 0.01 d | 20.0 ± 4.4 b | 0.24 ± 0.08 b | 38.2 ± 5.6 b | 1.89 ± 0.00 d |
| control_100 | 0.54 ± 0.09 a | 1.34 ± 0.01 a | 25.5 ± 2.7 ac | 0.80 ± 0.19 a | 34.7 ± 4.1 a | 7.01 ± 2.60 a |
| 0.2 IOX_100 | 0.64 ± 0.02 b | 1.24 ± 0.01 b | 19.5 ± 1.6 b | 0.75 ± 0.02 a | 33.8 ± 2.7 a | 0.00 ± 0.00 b |
| 2.0 IOX_100 | 0.45 ± 0.01 c | 1.33 ± 0.01 a | 23.5 ± 1.6 a | 1.34 ± 0.05 b | 40.3 ± 3.7 b | 0.00 ± 0.00 b |
| 20 IOX_100 | 0.46 ± 0.10 ac | 1.37 ± 0.00 c | 26.0 ± 0.0 c | 1.04 ± 0.21 c | 33.5 ± 2.4 a | 10.73 ± 0.50 c |
| Parameter | Start | Control | 0.2 mg/L IOX | 2 mg/L IOX | 20 mg/L IOX |
|---|---|---|---|---|---|
| pH | 6.7 ± 0.2 | 7.4 ± 0.2 | 7.0 ± 0.2 | 7.4 ± 0.2 | 7.2 ± 0.2 |
| COD | 2880 ± 432 | 988 ± 148 | 1570 ± 236 | 1300 ± 195 | 1500 ± 225 |
| BOD | 570 ± 114 | 150 ± 30 | 43 ± 9 | 120 ± 24 | 230 ± 46 |
| SVI | 34.3 ± 6.8 | 76.9 ± 15.2 | 68.3 ± 13.6 | 89.7 ± 17.4 | 90.2 ± 18.0 |
| TSS | >4000 ± 40 | 1200 ± 117 | 1600 ± 161 | 1600 ± 156 | 1300 ± 133 |
| TOC | 825 ± 165 | 431 ± 86 | 499 ± 100 | 307 ± 61 | 457 ± 91 |
| DR | 5030 ± 503 | 1930 ± 193 | 2430 ± 243 | 3150 ± 315 | 2000 ± 200 |
| PO4− | 129 ± 19 | 47.5 ± 7.1 | 50.2 ± 7.5 | 44.2 ± 6.6 | 42.6 ± 6.4 |
| Total N | 139 ± 1 | 78 ± 12 | 100 ± 15 | 100 ± 15 | 74 ± 11 |
| N-NH4+ | 23 ± 2 | <0.1 ± 0.02 | 0.22 ± 0.06 | 0.17 ± 0.04 | 0.21 ± 0.01 |
| N-NO2− | 0.23 ± 0.02 | 0.04 ± 0.01 | 0.07 ± 0.03 | 0.02 ± 0.01 | 0.06 ± 0.02 |
| N-NO3− | 4.0 ± 0.6 | 29 ± 4 | 41 ± 6 | 34 ± 5 | 27 ± 4 |
| Chlorides | 140 ± 14 | 150 ± 15 | 180 ± 18 | 170 ± 17 | 150 ± 15 |
| Sulfates | 96 ± 10 | 60 ± 6 | 61 ± 6 | 54 ± 5 | 54 ± 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowak, A.; Pacwa-Płociniczak, M.; Guzik, U.; Wojcieszyńska, D. Iohexol as an Emerging Contaminant Potentially Influencing Structural and Functional Changes in the Activated Sludge Microbiome. Molecules 2025, 30, 4641. https://doi.org/10.3390/molecules30234641
Nowak A, Pacwa-Płociniczak M, Guzik U, Wojcieszyńska D. Iohexol as an Emerging Contaminant Potentially Influencing Structural and Functional Changes in the Activated Sludge Microbiome. Molecules. 2025; 30(23):4641. https://doi.org/10.3390/molecules30234641
Chicago/Turabian StyleNowak, Agnieszka, Magdalena Pacwa-Płociniczak, Urszula Guzik, and Danuta Wojcieszyńska. 2025. "Iohexol as an Emerging Contaminant Potentially Influencing Structural and Functional Changes in the Activated Sludge Microbiome" Molecules 30, no. 23: 4641. https://doi.org/10.3390/molecules30234641
APA StyleNowak, A., Pacwa-Płociniczak, M., Guzik, U., & Wojcieszyńska, D. (2025). Iohexol as an Emerging Contaminant Potentially Influencing Structural and Functional Changes in the Activated Sludge Microbiome. Molecules, 30(23), 4641. https://doi.org/10.3390/molecules30234641









