Design and Synthesis of Novel Candidate CK1δ Proteolysis Targeting Chimeras (PROTACs)
Abstract
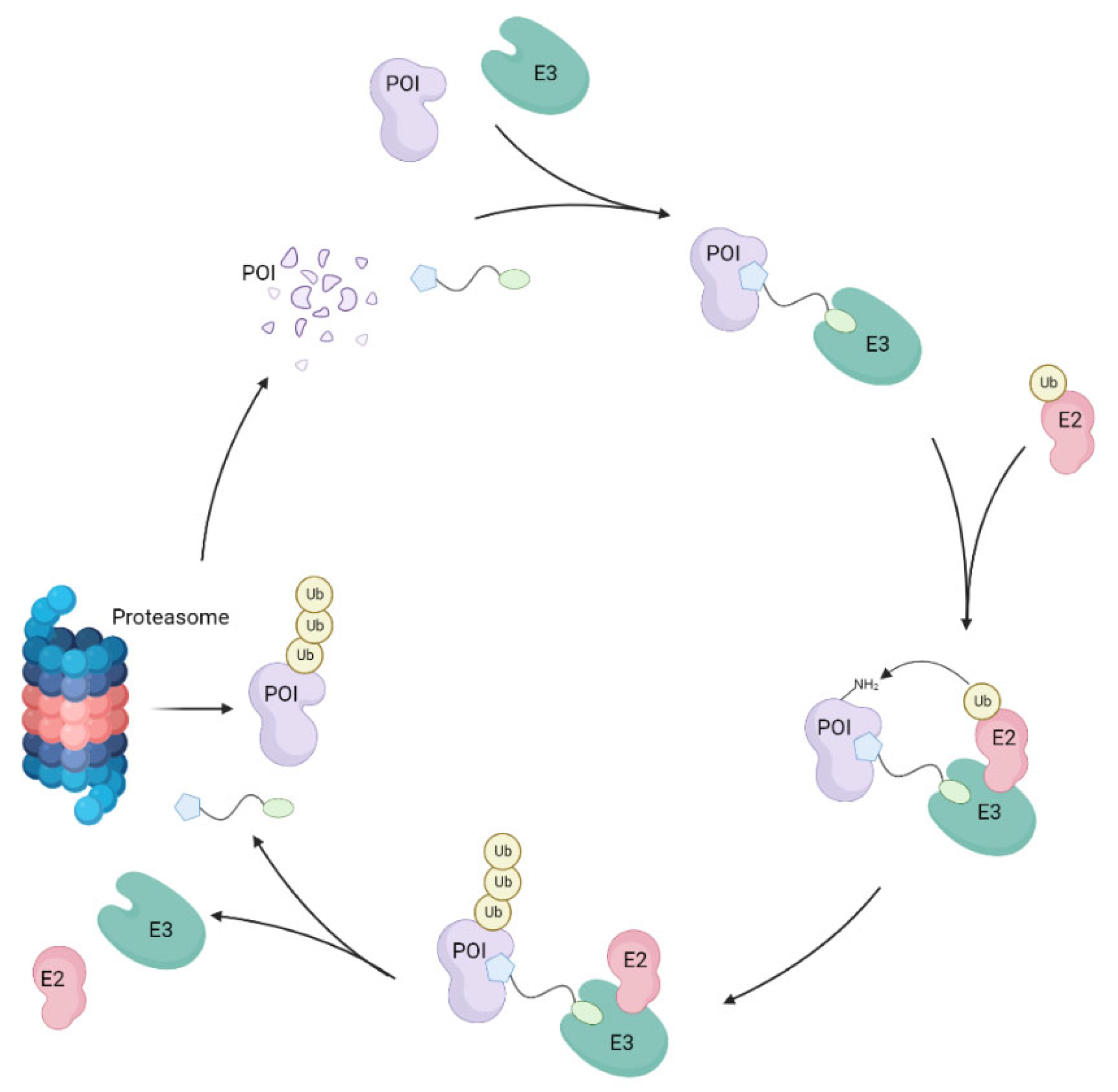
1. Introduction
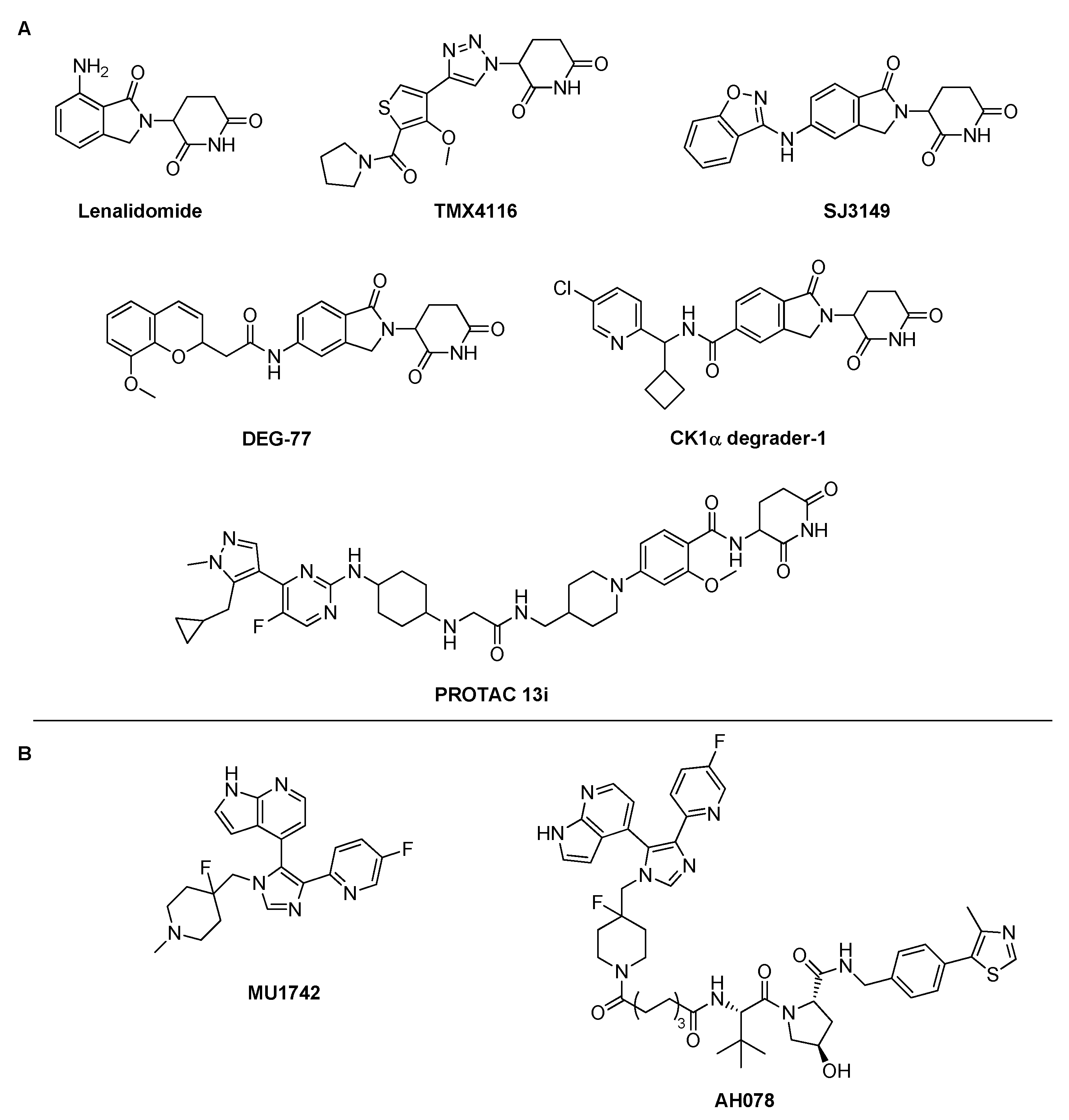
CK1 Isoforms as Proteins of Interest for PROTAC Design
2. Results and Discussion
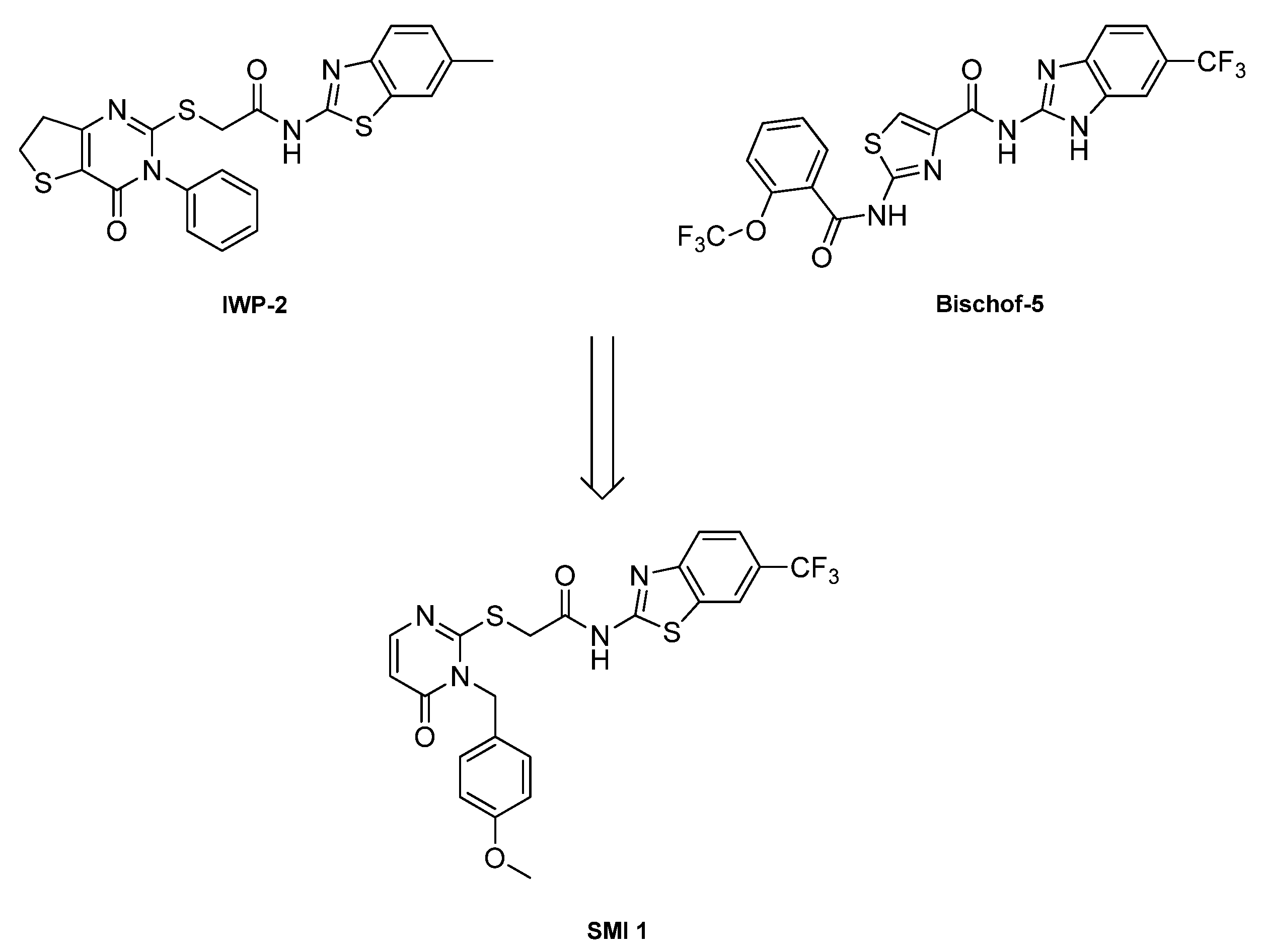
2.1. CK1δ PROTAC Design Based on SMI 1
2.2. Synthesis of Key Aniline CK1δ POI Ligand I9
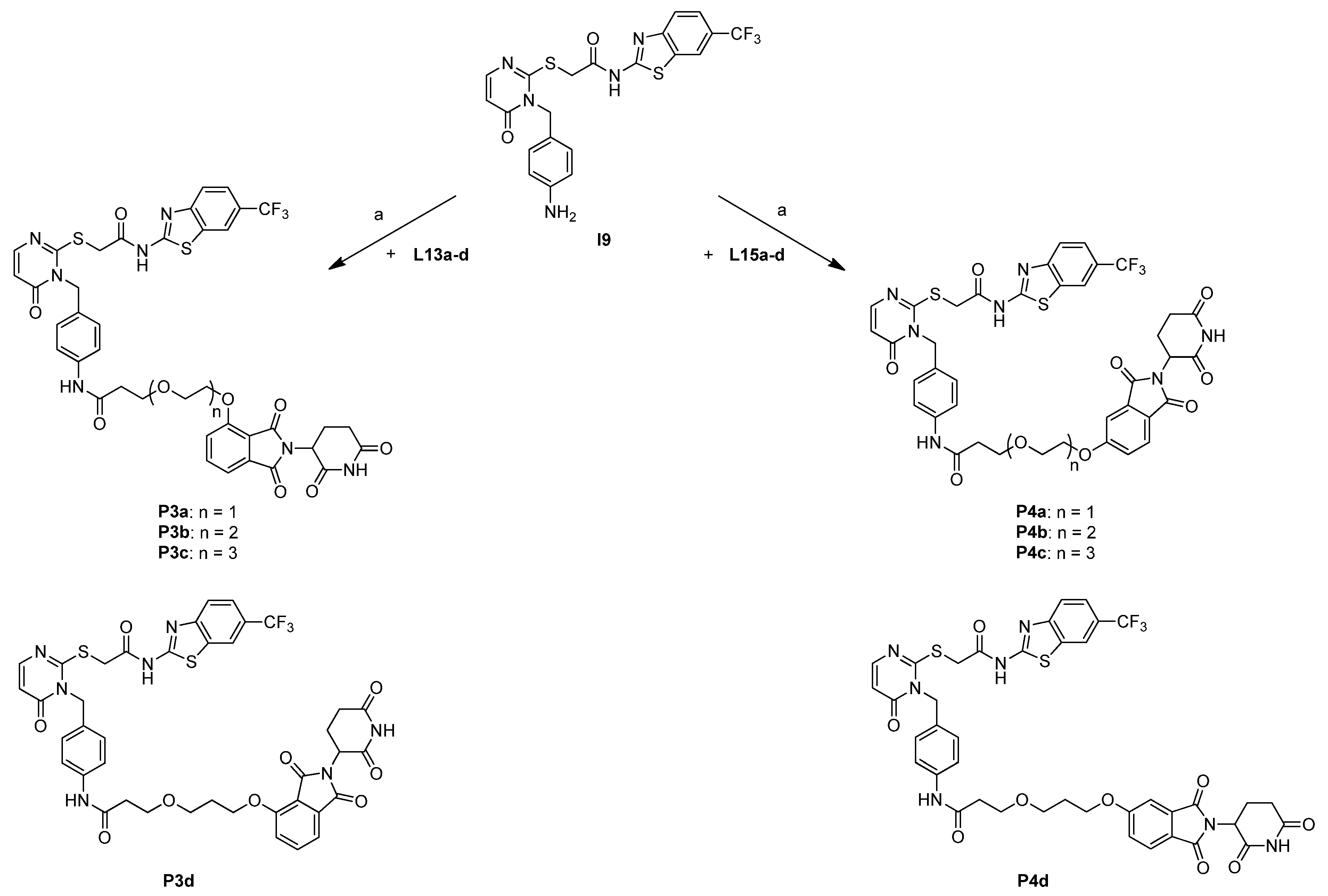
2.3. Synthesis of Candidate CK1δ PROTACs Containing Alkyl Linker Motifs
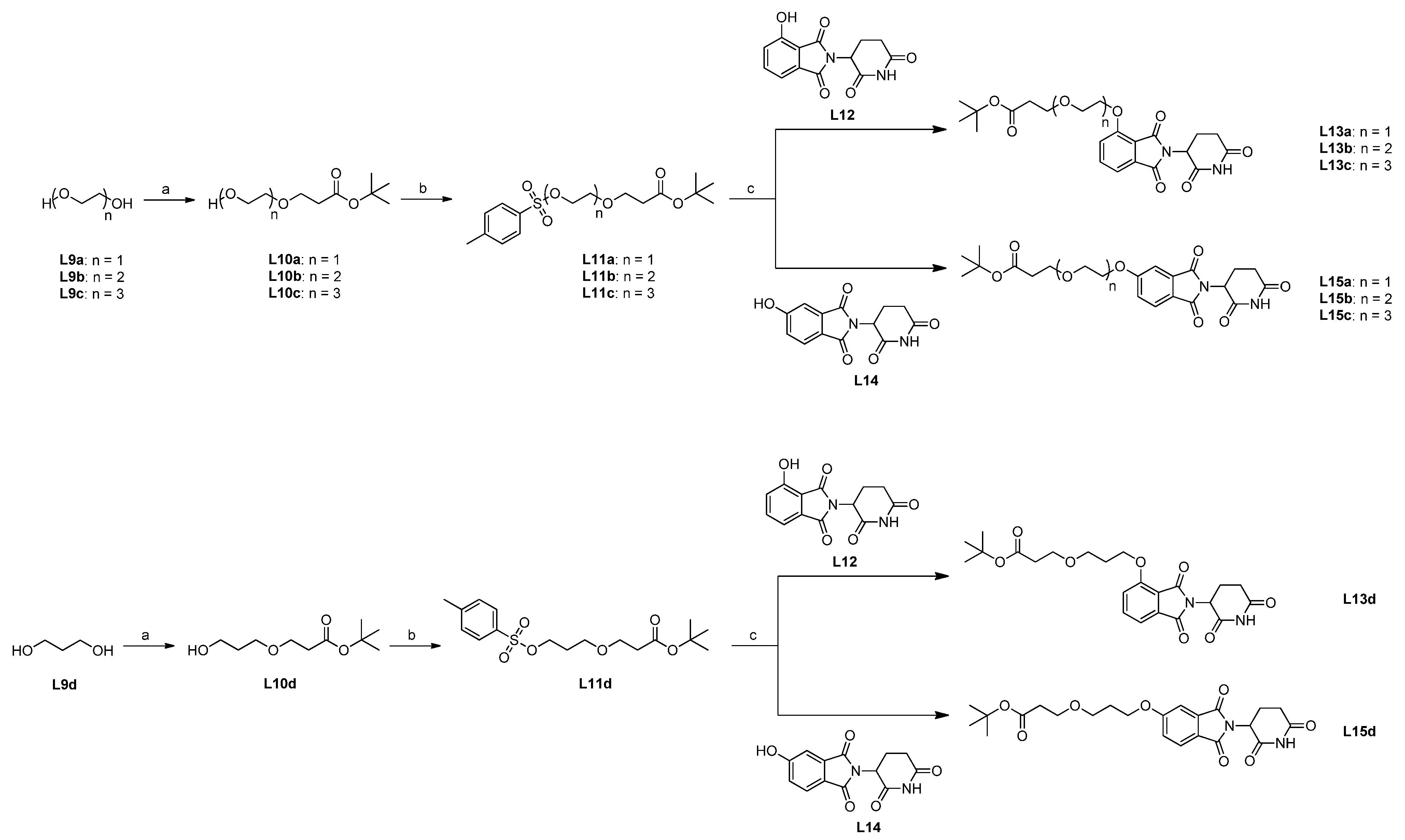
2.4. Synthesis of Candidate CK1δ PROTACs Containing PEG Linker Motifs
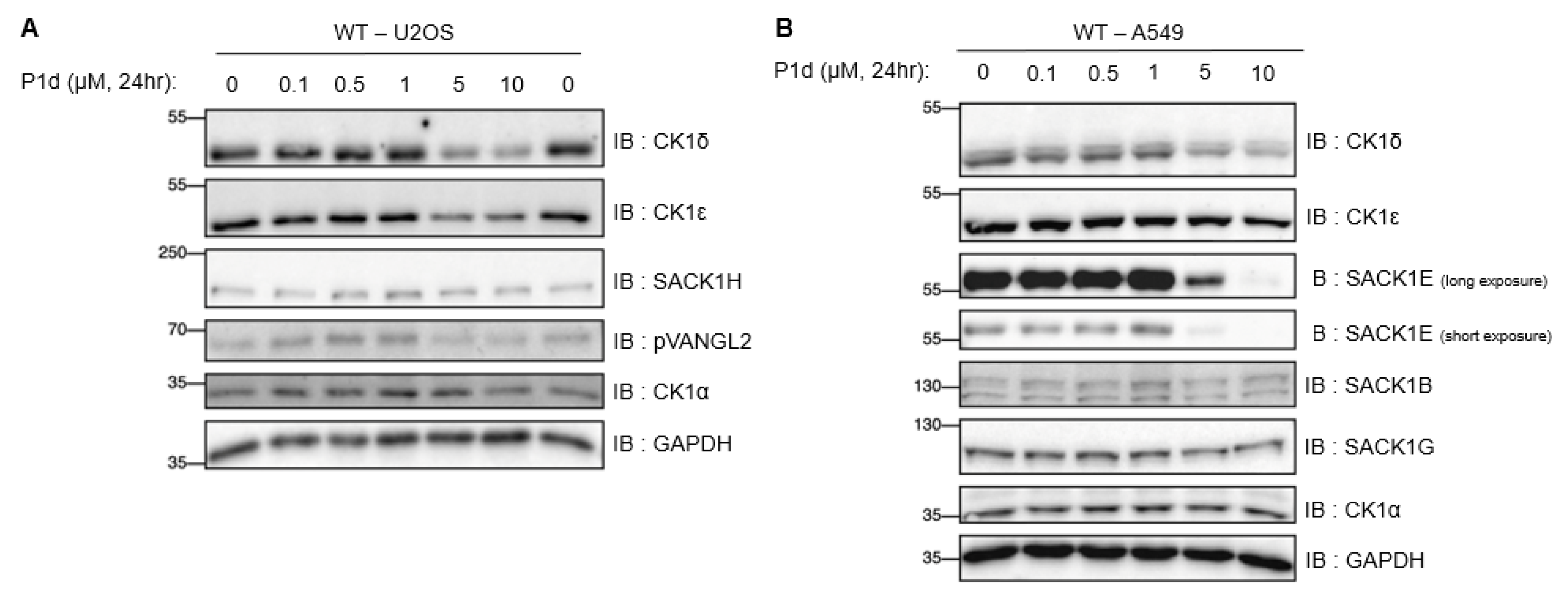
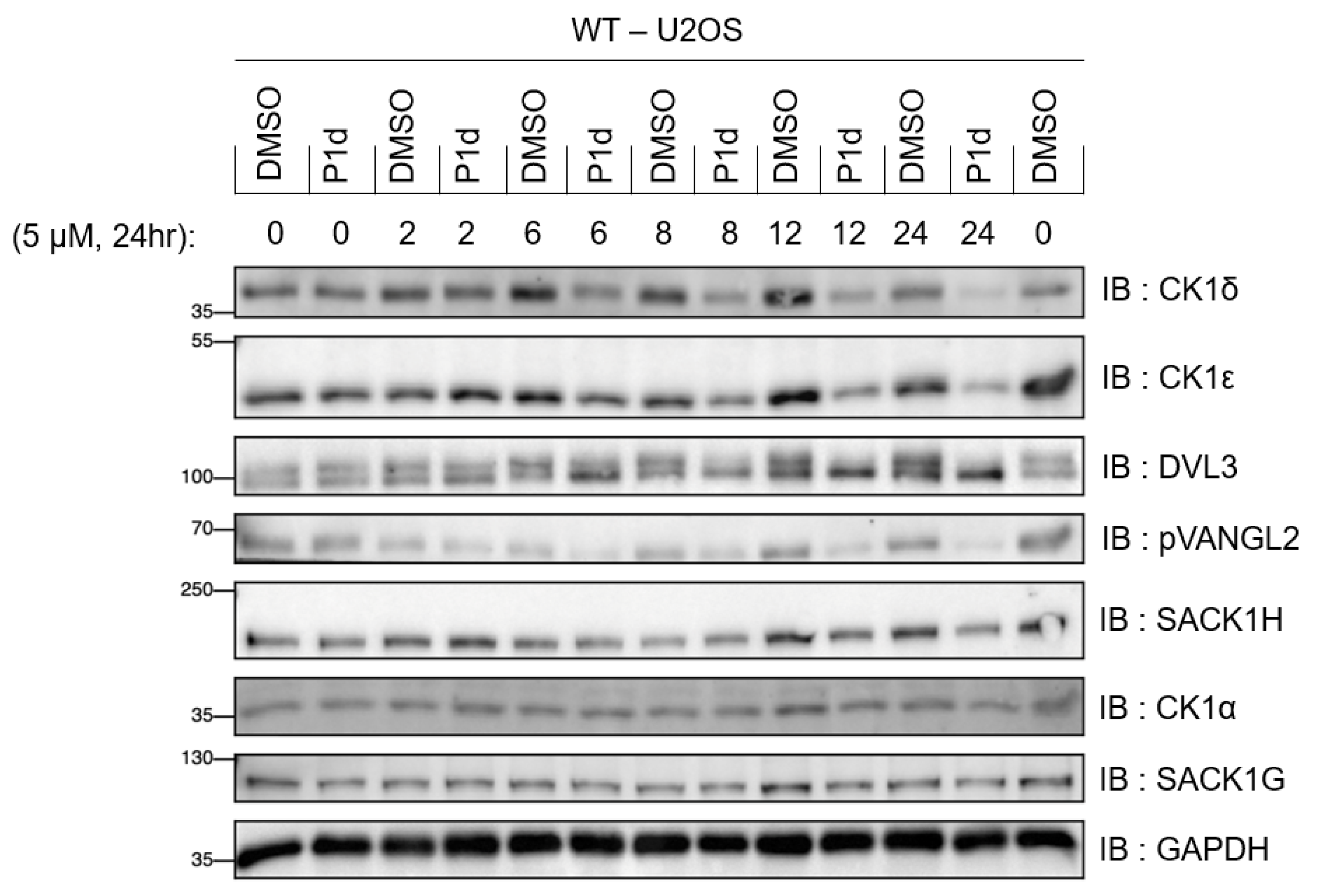
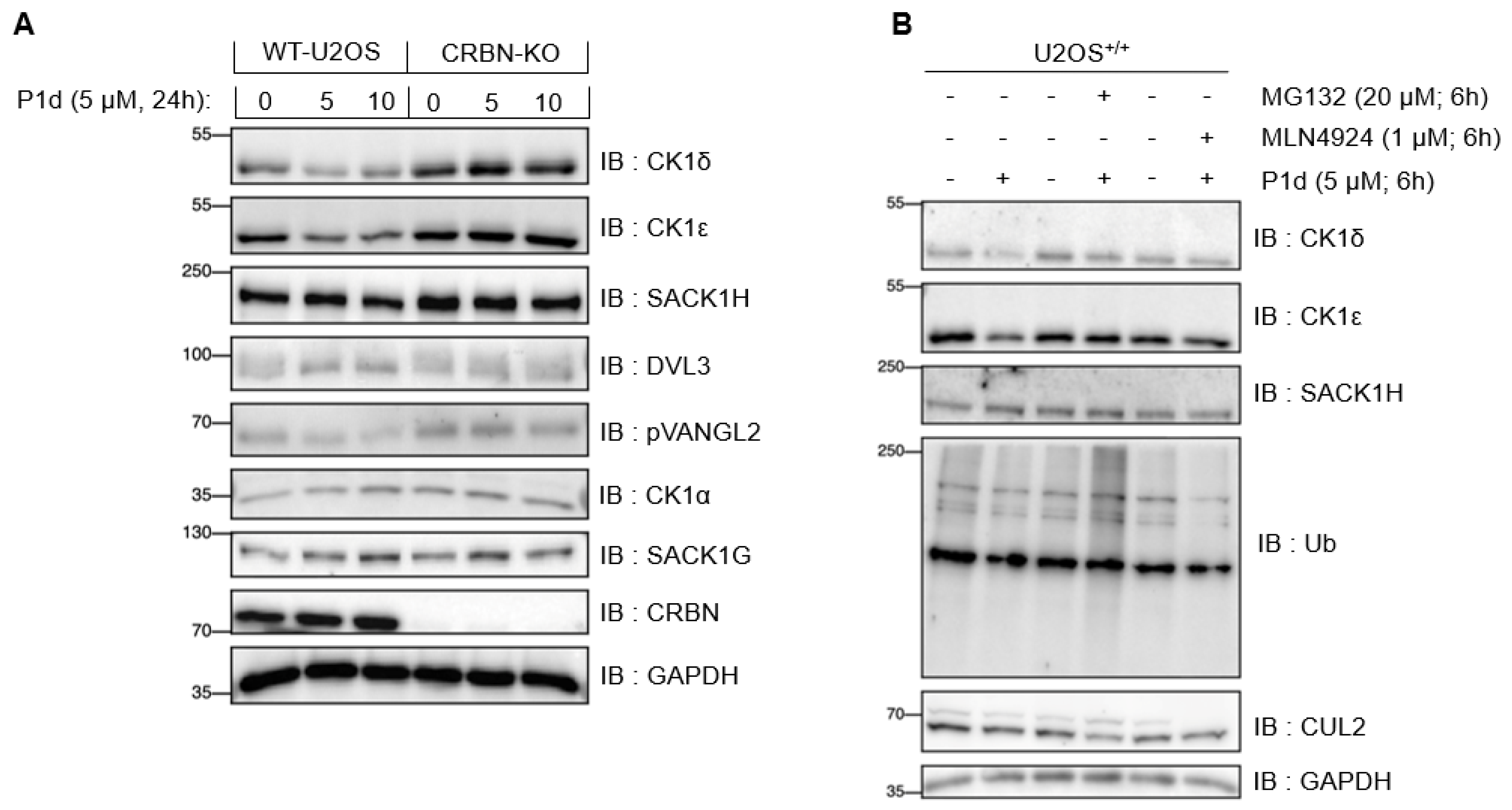
2.5. Biological Results
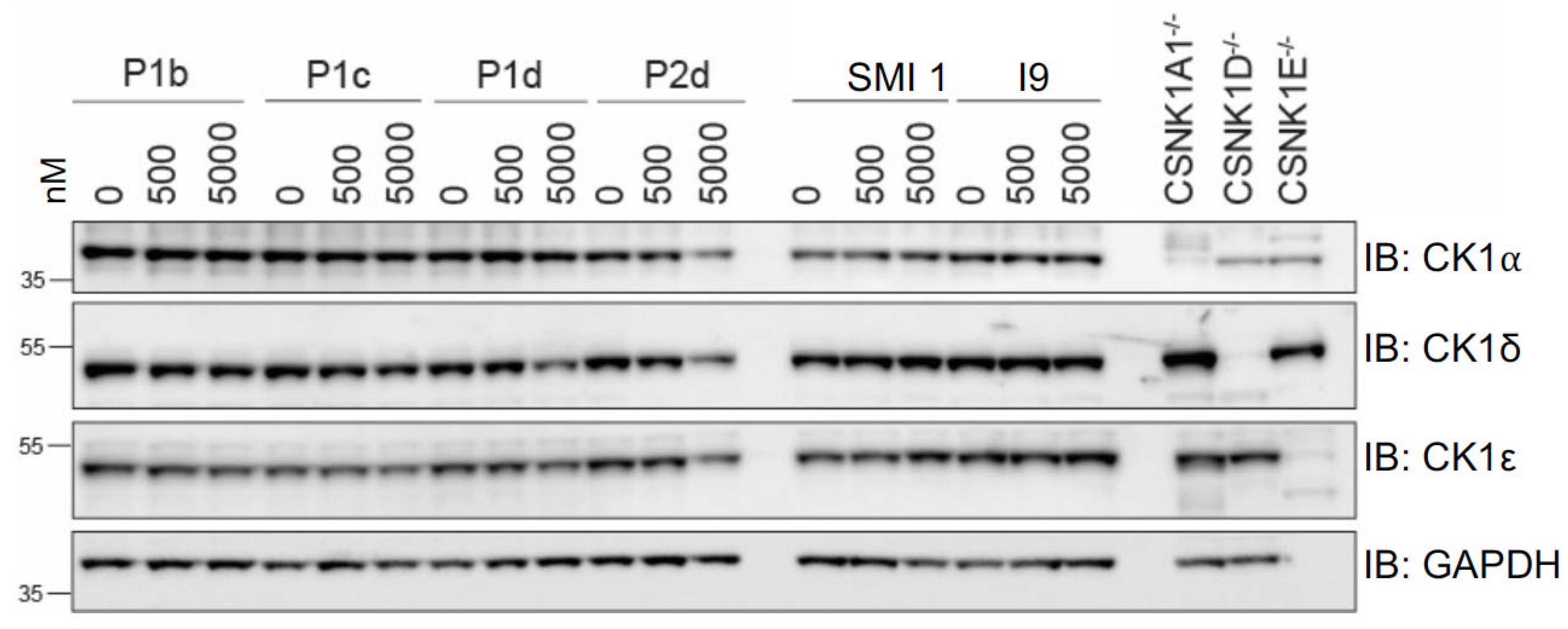
2.5.1. Differential Degradation of CK1δ/ε by Different PROTACs
2.5.2. PROTACs as Inhibitors of CK1δ/ε
2.5.3. Extended Dose–Response Analysis of CK1δ/ε Degradation by Best Degrader P1d Across Multiple Cells
2.5.4. Kinetic Analysis of CK1δ/ε Degradation by P1d
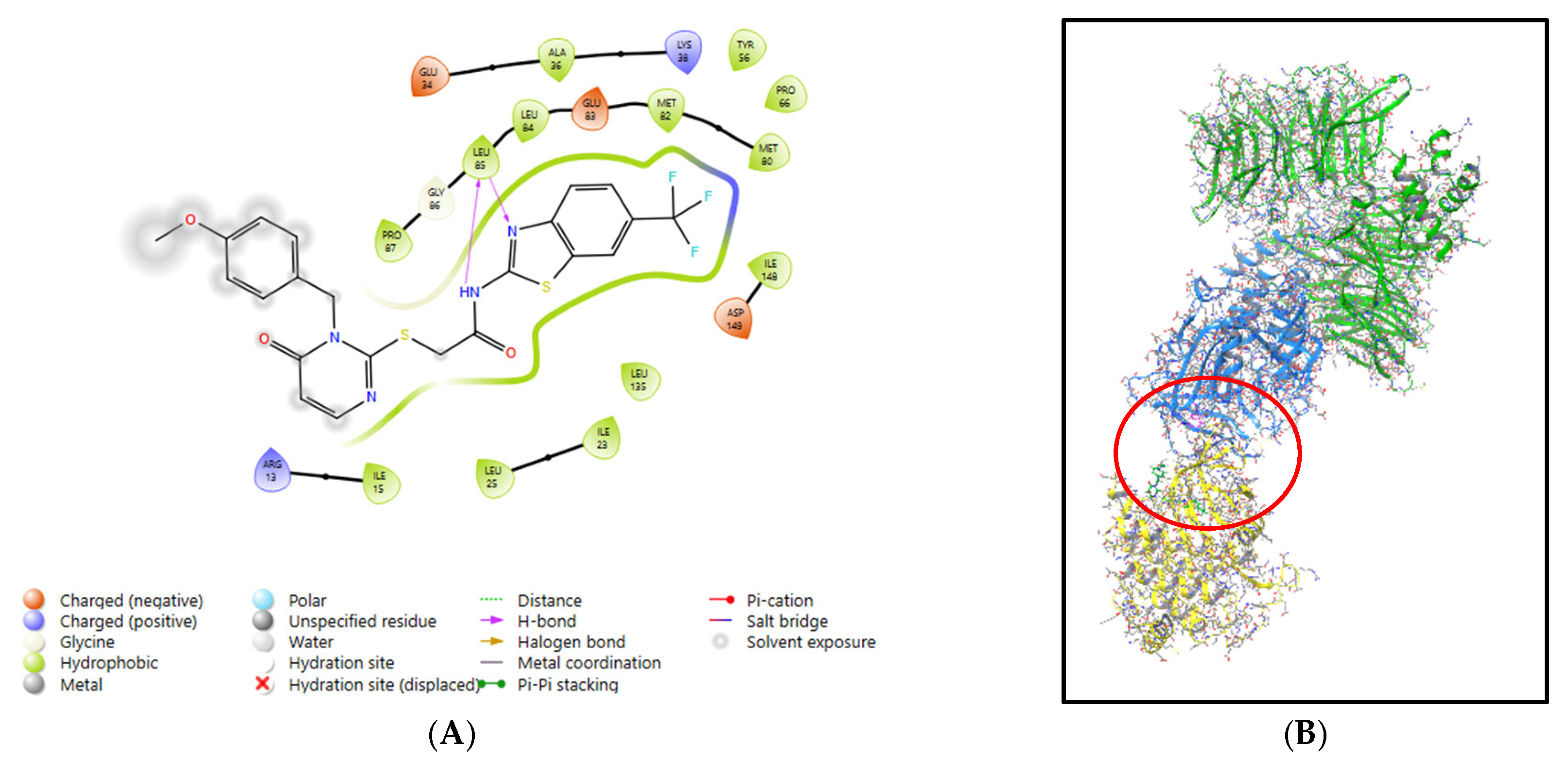
2.5.5. Establishing Mode of Action for P1d
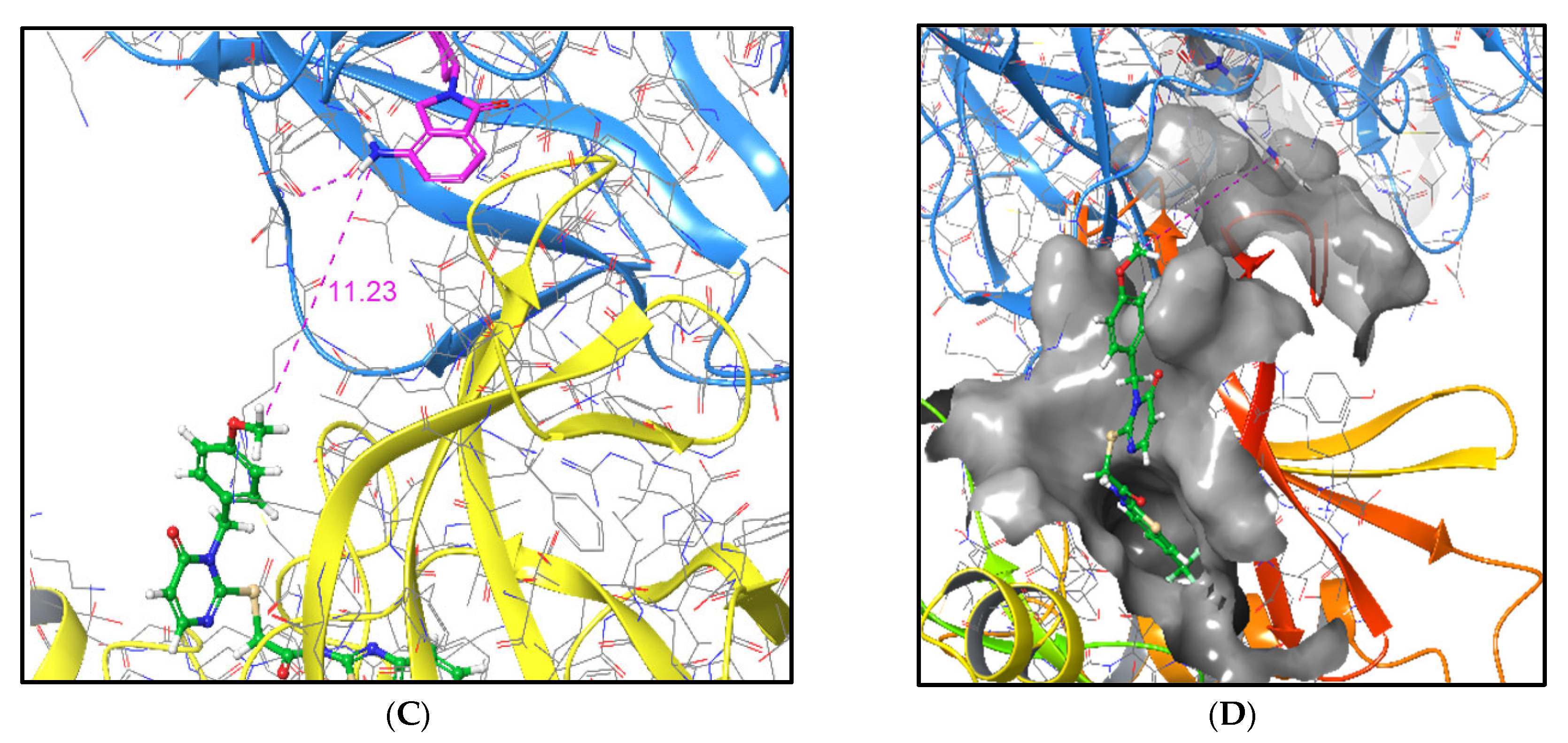
2.5.6. Correlation Between Computed and Experimental Linker Dimensions
3. Materials and Methods
3.1. Molecular Modelling
3.2. Chemistry
3.2.1. General Information
3.2.2. Synthesis of Compound I3
3.2.3. Synthesis of Compound I6
3.2.4. Synthesis of Compound I7
3.2.5. Synthesis of Compound I8
3.2.6. Synthesis of Compound I9
3.2.7. The General Procedure for the Synthesis of Linker Compounds L2a–d
3.2.8. The General Procedure for the Synthesis of Linker Compounds L3a–d
3.2.9. The General Procedure for the Synthesis of Linker Compounds L4a–d
3.2.10. The General Procedure for the Synthesis of Compounds L6a–d
3.2.11. The General Procedure for the Synthesis of Compounds L8a–d
3.2.12. The General Procedure for the Synthesis of Linker Compounds L10a–d
3.2.13. The General Procedure for the Synthesis of Linker Compounds L11a–d
3.2.14. The General Procedure for the Synthesis of Compounds L13a–d
3.2.15. The General Procedure for the Synthesis of Compounds L15a–d
3.2.16. The General Procedure for the Synthesis of PROTAC Compounds P1a–d, P2a, P2d, P3a–d, and P4a–d
3.3. Biology
3.3.1. Cell Culture, Drug Treatment, and Lysis
3.3.2. SDS-PAGE and Immunoblotting
3.3.3. In Vitro Kinase Assays for IC50 Determination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CK1 | Casein Kinase 1 |
| PROTAC | Proteolysis Targeting Chimera |
| PEG | Polyethylene Glycol |
| MOA | Mode of Action |
| SMI | Small Molecule Inhibitor |
| POI | Protein of Interest |
| UPS | Ubiquitin Proteasome System |
| APC | Adenomatous Polyposis Coli |
| AXIN | Axis Inhibition Protein |
| GSK3 protein | Glycogen Synthase Kinase 3 Protein |
| HRII | Hydrophobic Region II |
| ATP | Adenosine Triphosphate |
| TPD | Targeted Protein Degradation |
| IWP | Inhibitor of Wnt Production |
| CRBN | Cereblon |
| p-MeO | Para-Methoxy |
| SAR | Structure–Activity Relationship |
| TEA | Triethylamine |
| DMSO | Dimethylsulfoxide |
| DMF | Dimethylformamide |
| equiv. | Equivalent |
| TFA | Trifluororacetic Acid |
| HATU | Hexafluorophosphate Azabenzotriazole Tetramethyl Uronium |
| DIPEA | N, N-Diisopropylethylamine |
| EDC | 1-Ethyl-3-(3-dimethylaminopropyl) Carbodiimide |
| HOBt | Hydroxybenzotriazole |
| DMAP | 4-Dimethylaminopyridine |
| MeCN | Acetonitrile |
| RP-HPLC | Reversed-Phase High-Performance Liquid Chromatography |
| MS | Mass Spectrometry |
| HR-MS | High-Resolution Mass Spectrometry |
| NMR | Nuclear Magnetic Resonance |
| EA | Ethyl Acetate |
| MeOH | Methanol |
References
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Li, K.; Crews, C.M. PROTACs: Past, present and future. Chem. Soc. Rev. 2022, 51, 5214–5236. [Google Scholar] [CrossRef]
- Nalawansha, D.A.; Crews, C.M. PROTACs: An Emerging Therapeutic Modality in Precision Medicine. Cell Chem. Biol. 2020, 8, 998–1014. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Kleiger, G.; Mayor, T. Perilous journey: A tour of the ubiquitin-proteasome system. Trends Cell Biol. 2014, 6, 352–359. [Google Scholar] [CrossRef]
- Nandi, D.; Tahiliani, P.; Kumar, A.; Chandu, D. The ubiquitin-proteasome system. J. Biosci. 2006, 31, 137–155. [Google Scholar] [CrossRef]
- Paiva, S.-L.; Da Silva, S.R.; de Araujo, E.D.; Gunning, P.T. Regulating the Master Regulator: Controlling Ubiquitination by Thinking Outside the Active Site. J. Med. Chem. 2018, 2, 405–421. [Google Scholar] [CrossRef]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef]
- Jiang, Y.; Beaudet, A.L. Human disorders of ubiquitination and proteasomal degradation. Curr. Opin. Pediatr. 2004, 4, 419–426. [Google Scholar] [CrossRef]
- Paiva, S.-L.; Crews, C.M. Targeted protein degradation: Elements of PROTAC design. Curr. Opin. Chem. Biol. 2019, 50, 111–119. [Google Scholar] [CrossRef]
- An, S.; Fu, L. Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. EBioMedicine 2018, 36, 553–562. [Google Scholar] [CrossRef]
- Berndsen, C.E.; Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307. [Google Scholar] [CrossRef]
- Schreiber, A.; Peter, M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim. Biophys. Acta 2014, 1, 163–181. [Google Scholar] [CrossRef]
- Alabi, S.B.; Crews, C.M. Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J. Biol. Chem. 2021, 296, 100647. [Google Scholar] [CrossRef]
- Bondeson, D.P.; Mares, A.; Smith, I.E.D.; Ko, E.; Campos, S.; Miah, A.H.; Mulholland, K.E.; Routly, N.; Buckley, D.L.; Gustafson, J.L.; et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. [Google Scholar] [CrossRef]
- Zagidullin, A.; Milyukov, V.; Rizvanov, A.; Bulatov, E. Novel approaches for the rational design of PROTAC linkers. Explor. Target. Anti-Tumor Ther. 2020, 1, 381–390. [Google Scholar] [CrossRef]
- Troup, R.I.; Fallan, C.; Baud, M.G.J. Current strategies for the design of PROTAC linkers: A critical review. Explor. Target. Anti-Tumor Ther. 2020, 1, 273–312. [Google Scholar] [CrossRef]
- Poongavanam, V.; Peintner, S.; Abeje, Y.; Kölling, F.; Meibom, D.; Erdelyi, M.; Kihlberg, J. Linker-Determined Folding and Hydrophobic Interactions Explain a Major Difference in PROTAC Cell Permeability. ACS Med. Chem. Lett. 2025, 16, 681–687. [Google Scholar] [CrossRef]
- Cyrus, K.; Wehenkel, M.; Choi, E.-Y.; Han, H.-J.; Lee, H.; Swanson, H.; Kim, K.-B. Impact of linker length on the activity of PROTACs. Mol. Biosyst. 2011, 7, 359–364. [Google Scholar] [CrossRef]
- Knippschild, U.; Krüger, M.; Richter, J.; Xu, P.; García-Reyes, B.; Peifer, C.; Halekotte, J.; Bakulev, V.; Bischof, J. The CK1 Family: Contribution to Cellular Stress Response and Its Role in Carcinogenesis. Front. Oncol. 2014, 4, 96. [Google Scholar] [CrossRef]
- Narasimamurthy, R.; Hunt, S.R.; Lu, Y.; Fustin, J.-M.; Okamura, H.; Partch, C.L.; Forger, D.B.; Kim, J.K.; Virshup, D.M. CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. Proc. Natl. Acad. Sci. USA 2018, 23, 5986–5991. [Google Scholar] [CrossRef]
- Sinnberg, T.; Wang, J.; Sauer, B.; Schittek, B. Casein kinase 1α has a non-redundant and dominant role within the CK1 family in melanoma progression. BMC Cancer 2016, 16, 594. [Google Scholar] [CrossRef]
- Fulcher, L.J.; Sapkota, G.P. Functions and regulation of the serine/threonine protein kinase CK1 family: Moving beyond promiscuity. Biochem. J. 2020, 23, 4603–4621. [Google Scholar] [CrossRef]
- Gybeľ, T.; Čada, Š.; Klementová, D.; Schwalm, M.P.; Berger, B.-T.; Šebesta, M.; Knapp, S.; Bryja, V. Splice variants of CK1α and CK1α-like: Comparative analysis of subcellular localization, kinase activity, and function in the Wnt signaling pathway. J. Biol. Chem. 2024, 7, 107407. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 1, 9–26. [Google Scholar] [CrossRef]
- Bryja, V.; Bernatík, O. Dishevelled at the Crossroads of Pathways. In Wnt Signaling in Development and Disease: Molecular Mechanisms and Biological Functions; Wiley: Hoboken, NJ, USA, 2014; p. 207. [Google Scholar] [CrossRef]
- Xu, P.; Ianes, C.; Gärtner, F.; Liu, C.; Burster, T.; Bakulev, V.; Rachidi, N.; Knippschild, U.; Bischof, J. Structure, regulation, and (patho-)physiological functions of the stress-induced protein kinase CK1 delta (CSNK1D). Gene 2019, 715, 144005. [Google Scholar] [CrossRef]
- Mazzoldi, E.L.; Pastò, A.; Ceppelli, E.; Pilotto, G.; Barbieri, V.; Amadori, A.; Pavan, S. Casein Kinase 1 Delta Regulates Cell Proliferation, Response to Chemotherapy and Migration in Human Ovarian Cancer Cells. Front. Oncol. 2019, 9, 1211. [Google Scholar] [CrossRef]
- Cheong, J.K.; Virshup, D.M. Casein kinase 1: Complexity in the family. Int. J. Biochem. Cell Biol. 2011, 4, 465–469. [Google Scholar] [CrossRef]
- Nishiguchi, G.; Caine, E.A.; McGowan, K.; Shi, Z.; Aggarwal, A.; Mayasundari, A.; Price, J.; Yang, L.; Li, Y.; Fu, X.; et al. Structure–Activity Relationship of Potent, Selective, and Orally Bioavailable Molecular Glue Degraders of CK1α. ACS Med. Chem. Lett. 2024, 15, 1692–1698. [Google Scholar] [CrossRef]
- MedchemExpress.com. Available online: https://www.medchemexpress.com/ck1%CE%B1-degrader-1.html (accessed on 6 March 2025).
- Wang, K.; Jiang, M.; Liu, H.; Meng, C.; Li, M.; Lu, H. Discovery of novel co-degradation CK1α and CDK7/9 PROTACs with p53 activation for treating acute myeloid leukemia. Bioorganic Chem. 2024, 147, 107319. [Google Scholar] [CrossRef]
- Haag, A.; Němec, V.; Janovská, P.; Bartošíková, J.; Adhikari, B.; Müller, J.; Schwalm, M.P.; Čada, Š.; Ohmayer, U.; Daub, H.; et al. Development and Discovery of a Selective Degrader of Casein Kinases 1 δ/ε. J. Med. Chem. 2025, 1, 506–530. [Google Scholar] [CrossRef]
- García-Reyes, B.; Witt, L.; Jansen, B.; Karasu, E.; Gehring, T.; Leban, J.; Henne-Bruns, D.; Pichlo, C.; Brunstein, E.; Baumann, U.; et al. Discovery of Inhibitor of Wnt Production 2 (IWP-2) and Related Compounds As Selective ATP-Competitive Inhibitors of Casein Kinase 1 (CK1) δ/ε. J. Med. Chem. 2018, 9, 4087–4102. [Google Scholar] [CrossRef]
- Liu, C.; Witt, L.; Ianes, C.; Bischof, J.; Bammert, M.-T.; Baier, J.; Kirschner, S.; Henne-Bruns, D.; Xu, P.; Kornmann, M.; et al. Newly Developed CK1-Specific Inhibitors Show Specifically Stronger Effects on CK1 Mutants and Colon Cancer Cell Lines. Int. J. Mol. Sci. 2019, 20, 6184. [Google Scholar] [CrossRef]
- Schade, D.; Plowright, A.T. Medicinal Chemistry Approaches to Heart Regeneration. J. Med. Chem. 2015, 24, 9451–9479. [Google Scholar] [CrossRef]
- Chen, B.; Dodge, M.E.; Tang, W.; Lu, J.; Ma, Z.; Fan, C.-W.; Wei, S.; Hao, W.; Kilgore, J.; Williams, N.S.; et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 2009, 5, 100–107. [Google Scholar] [CrossRef]
- Bischof, J.; Leban, J.; Zaja, M.; Grothey, A.; Radunsky, B.; Othersen, O.; Strobl, S.; Vitt, D.; Knippschild, U. 2-Benzamido-N-(1H-benzodimidazol-2-yl)thiazole-4-carboxamide derivatives as potent inhibitors of CK1δ/ε. Amino Acids 2012, 43, 1577–1591. [Google Scholar] [CrossRef]
- Abeje, Y.E.; Wieske, L.H.E.; Poongavanam, V.; Maassen, S.; Atilaw, Y.; Cromm, P.; Lehmann, L.; Erdelyi, M.; Meibom, D.; Kihlberg, J. Impact of Linker Composition on VHL PROTAC Cell Permeability. J. Med. Chem. 2025, 1, 638–657. [Google Scholar] [CrossRef]
- Petzold, G.; Fischer, E.S.; Thomä, N.H. Structural basis of lenalidomide-induced CK1α degradation by the CRL4(CRBN) ubiquitin ligase. Nature 2016, 532, 127–130. [Google Scholar] [CrossRef]
- Hernández-Núñez, E.; Tlahuext, H.; Moo-Puc, R.; Torres-Gómez, H.; Reyes-Martínez, R.; Cedillo-Rivera, R.; Nava-Zuazo, C.; Navarrete-Vazquez, G. Synthesis and in vitro trichomonicidal, giardicidal and amebicidal activity of N-acetamide(sulfonamide)-2-methyl-4-nitro-1H-imidazoles. Eur. J. Med. Chem. 2009, 7, 2975–2984. [Google Scholar] [CrossRef]
- Roopan, S.M.; Khan, F.R.N.; Mandal, B.K. Fe nano particles mediated C–N bond-forming reaction: Regioselective synthesis of 3-[(2-chloroquinolin-3-yl)methyl]pyrimidin-4(3H)ones. Tetrahedron Lett. 2010, 17, 2309–2311. [Google Scholar] [CrossRef]
- Xu, J.; Yadan, J.-C. A New and Convenient Synthesis of Imidazole-2-thiones from Imidazoles. Synlett 1995, 1995, 239–241. [Google Scholar] [CrossRef]
- Schmuck, C.; Rehm, T.; Geiger, L.; Schäfer, M. Synthesis and self-association properties of flexible guanidiniocarbonylpyrrole-carboxylate zwitterions in DMSO: Intra- versus intermolecular ion pairing. J. Org. Chem. 2007, 16, 6162–6170. [Google Scholar] [CrossRef]
- Kim, K.; Lee, D.H.; Park, S.; Jo, S.-H.; Ku, B.; Park, S.G.; Park, B.C.; Jeon, Y.U.; Ahn, S.; Kang, C.H.; et al. Disordered region of cereblon is required for efficient degradation by proteolysis-targeting chimera. Sci. Rep. 2019, 1, 19654. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Shahabi, D. Synthesis of amide derivatives for electron deficient amines and functionalized carboxylic acids using EDC and DMAP and a catalytic amount of HOBt as the coupling reagents. Tetrahedron Lett. 2021, 63, 152719. [Google Scholar] [CrossRef]
- Cromm, P.M.; Samarasinghe, K.T.G.; Hines, J.; Crews, C.M. Addressing Kinase-Independent Functions of Fak via PROTAC-Mediated Degradation. J. Am. Chem. Soc. 2018, 49, 17019–17026. [Google Scholar] [CrossRef]
- Kaucká, M.; Petersen, J.; Janovská, P.; Radaszkiewicz, T.; Smyčková, L.; Daulat, A.M.; Borg, J.-P.; Schulte, G.; Bryja, V. Asymmetry of VANGL2 in migrating lymphocytes as a tool to monitor activity of the mammalian WNT/planar cell polarity pathway. Cell Commun. Signal. CCS 2015, 13, 2. [Google Scholar] [CrossRef]
- Yang, W.; Garrett, L.; Feng, D.; Elliott, G.; Liu, X.; Wang, N.; Wong, Y.M.; Choi, N.T.; Yang, Y.; Gao, B. Wnt-induced Vangl2 phosphorylation is dose-dependently required for planar cell polarity in mammalian development. Cell Res. 2017, 27, 1466–1484. [Google Scholar] [CrossRef]
- Fulcher, L.J.; Bozatzi, P.; Tachie-Menson, T.; Wu, K.Z.L.; Cummins, T.D.; Bufton, J.C.; Pinkas, D.M.; Dunbar, K.; Shrestha, S.; Wood, N.T.; et al. The DUF1669 domain of FAM83 family proteins anchor casein kinase 1 isoforms. Sci. Signal. 2018, 11, eaao2341. [Google Scholar] [CrossRef]
- Glennie, L.; Curnutt, N.; Cartwright, T.; Dunbar, K.; Le Chatelier, B.; Wood, N.T.; Macartney, T.J.; Woo, C.M.; Sapkota, G.P. The contribution of native protein complexes to targeted protein degradation. BioRxiv 2025. preprint. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Mohamadi, F.; Richards, N.G.J.; Guida, W.C.; Liskamp, R.; Lipton, M.; Caufield, C.; Chang, G.; Hendrickson, T.; Still, W.C. Macromodel—An integrated software system for moedling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 1990, 4, 440–467. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 7, 1739–1749. [Google Scholar] [CrossRef]
- Dunbar, K.; Jones, R.A.; Dingwell, K.; Macartney, T.J.; Smith, J.C.; Sapkota, G.P. FAM83F regulates canonical Wnt signalling through an interaction with CK1α. Life Sci. Alliance 2021, 4, e202000805. [Google Scholar] [CrossRef]






| I9 | L8b–c | Solvent | Base (5 equiv.) | Coupling Reagents | Temperature [°C] | Reaction Time | Additive |
|---|---|---|---|---|---|---|---|
| 1 equiv. | 1 equiv. | MeCN | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 1 equiv. | DMSO | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 1 equiv. | DMF | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 1 equiv. | THF | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 1 equiv. | MeCN | DIPEA | EDC, HOBT, DMAP | 25 °C | 48 h | - |
| 1 equiv. | 1 equiv. | MeCN | Potassium carbonate | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 1 equiv. | MeCN | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 1 equiv. | MeCN | TEA | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 1 equiv. | MeCN | DBU | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 1 equiv. | MeCN | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | Mole sieve (3 Å) |
| 2 equiv. | 1 equiv. | MeCN | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 2 equiv. | MeCN | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | - |
| 1 equiv. | 1 equiv. | MeCN | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | Glutarimide (5 equiv.) |
| 1 equiv. | 1 equiv. | MeCN | DIPEA | EDC, HOBT, DMAP | 40 °C | 48 h | Glutarimide (10 equiv.) |
| 1 equiv. | 1 equiv. | MeCN | DIPEA | HATU | 40 °C | 5 h | - |
| 1 equiv. | 1 equiv. | MeCN | Potassium carbonate | HATU | 40 °C | 5 h | - |
| 1 equiv. | 1 equiv. | MeCN | TEA | HATU | 40 °C | 5 h | - |
| 1 equiv. | 1 equiv. | MeCN | DBU | HATU | 40 °C | 5 h | - |
| 1 equiv. | 1 equiv. | MeCN | DIPEA | HATU | 40 °C | 5 h | Mole sieve (3 Å) |
| 1 equiv. | 1 equiv. | MeCN | DIPEA | HATU | 70 °C | 5 h | - |
| PROTAC | Linker Length (Å) 1 |
|---|---|
| P1a | 4.179 |
| P1b | 4.991 |
| P1c | 6.428 |
| P1d | 7.413 |
| P2a | 4.179 |
| P2d | 7.413 |
| P3a | 7.103 |
| P3b | 10.712 |
| P3c | 14.062 |
| P3d | 8.492 |
| P4a | 7.103 |
| P4b | 10.712 |
| P4c | 14.062 |
| P4d | 8.492 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arnold, M.; Thompson, T.; Glennie, L.; Hollnagel, M.; Sapkota, G.; Peifer, C. Design and Synthesis of Novel Candidate CK1δ Proteolysis Targeting Chimeras (PROTACs). Molecules 2025, 30, 4452. https://doi.org/10.3390/molecules30224452
Arnold M, Thompson T, Glennie L, Hollnagel M, Sapkota G, Peifer C. Design and Synthesis of Novel Candidate CK1δ Proteolysis Targeting Chimeras (PROTACs). Molecules. 2025; 30(22):4452. https://doi.org/10.3390/molecules30224452
Chicago/Turabian StyleArnold, Malte, Temi Thompson, Lorraine Glennie, Mattes Hollnagel, Gopal Sapkota, and Christian Peifer. 2025. "Design and Synthesis of Novel Candidate CK1δ Proteolysis Targeting Chimeras (PROTACs)" Molecules 30, no. 22: 4452. https://doi.org/10.3390/molecules30224452
APA StyleArnold, M., Thompson, T., Glennie, L., Hollnagel, M., Sapkota, G., & Peifer, C. (2025). Design and Synthesis of Novel Candidate CK1δ Proteolysis Targeting Chimeras (PROTACs). Molecules, 30(22), 4452. https://doi.org/10.3390/molecules30224452






