From Triads to Tools: A Comprehensive Review of the Expanding Roles of G-Triplex Structures
Abstract
1. Introduction
2. Structural Basis of G-Triplex DNA and RNA
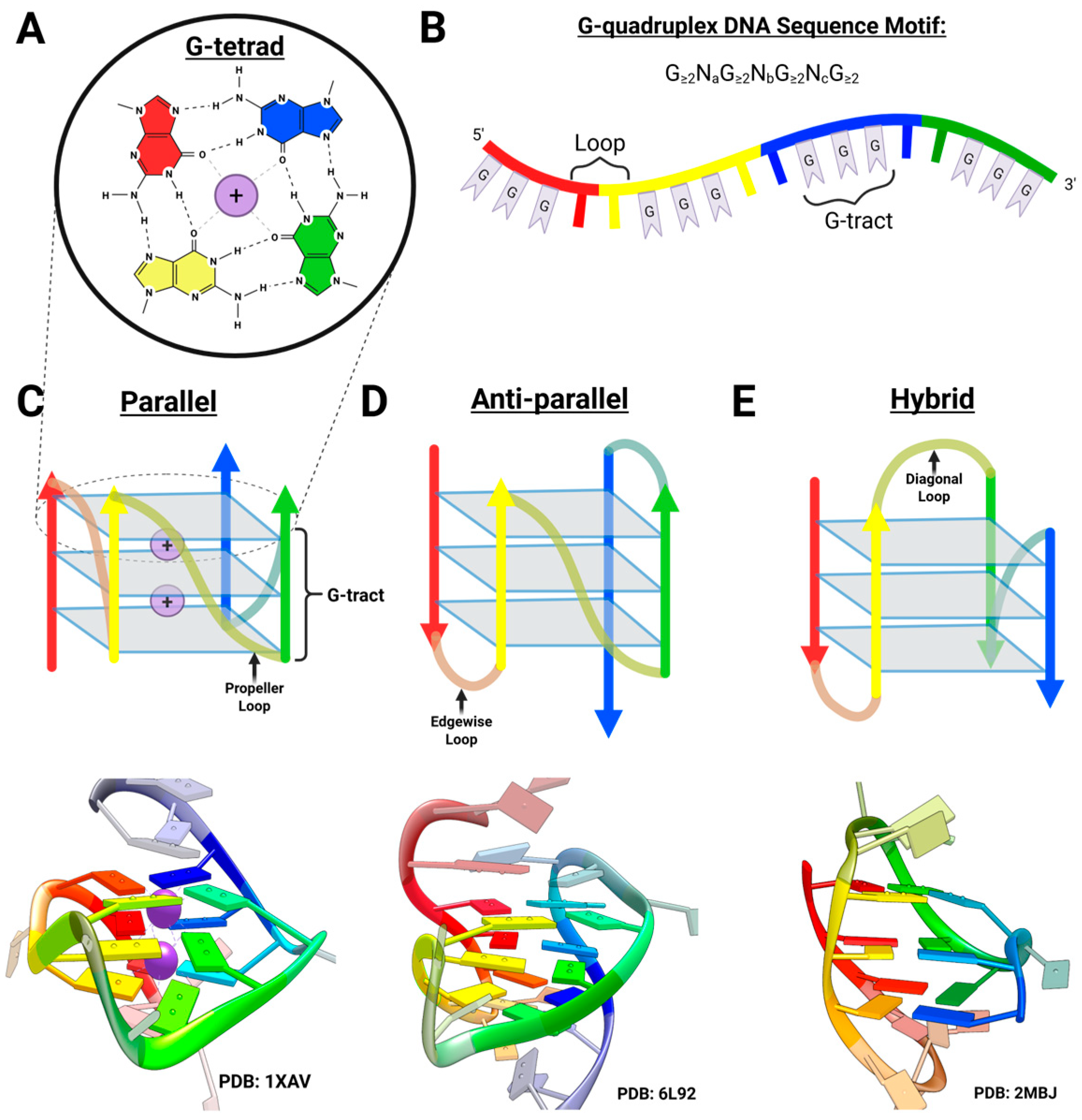
2.1. Overview of G-Quadruplex DNA
2.2. G-Triplex DNA as a Telomeric Folding Intermediate
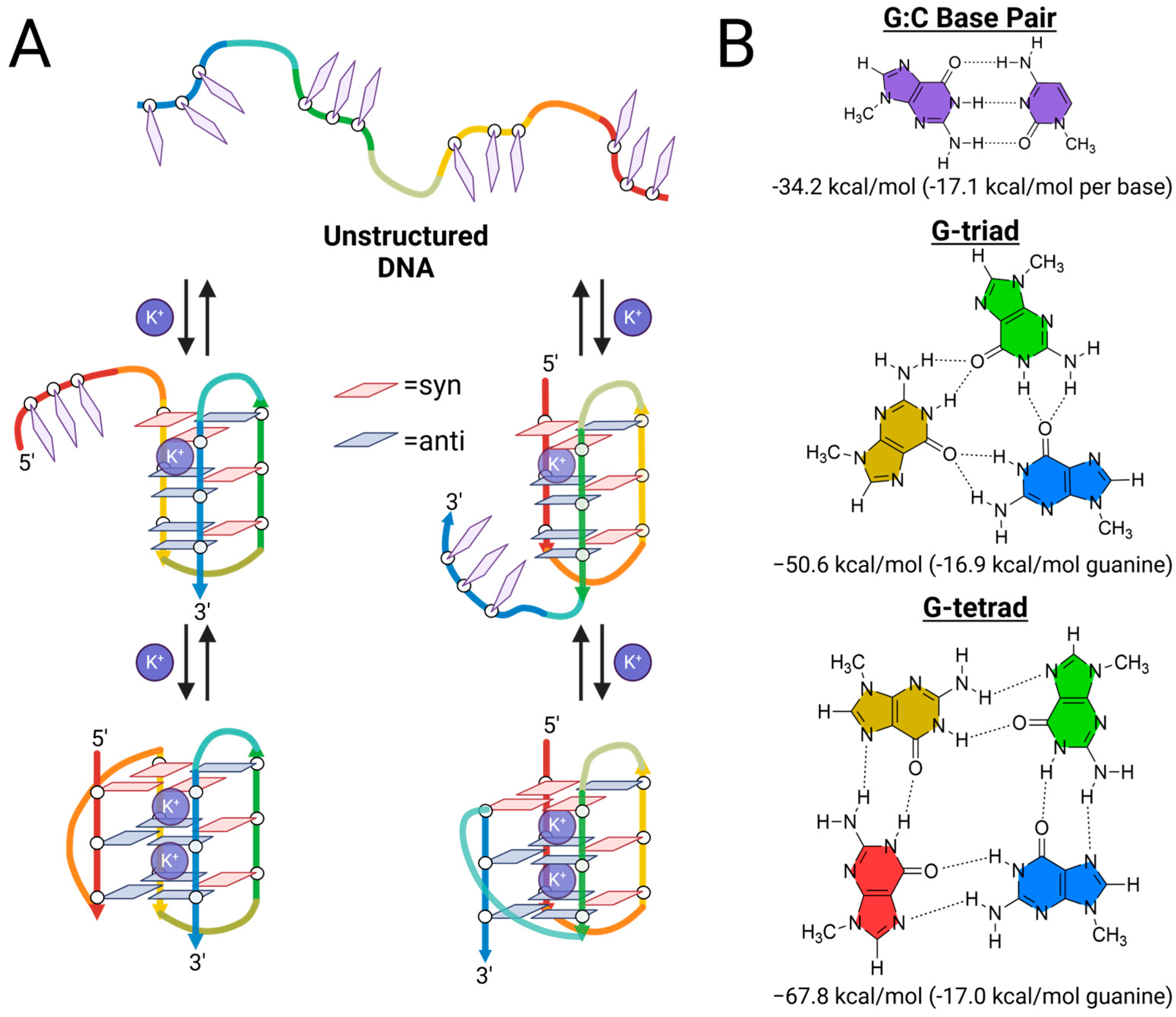
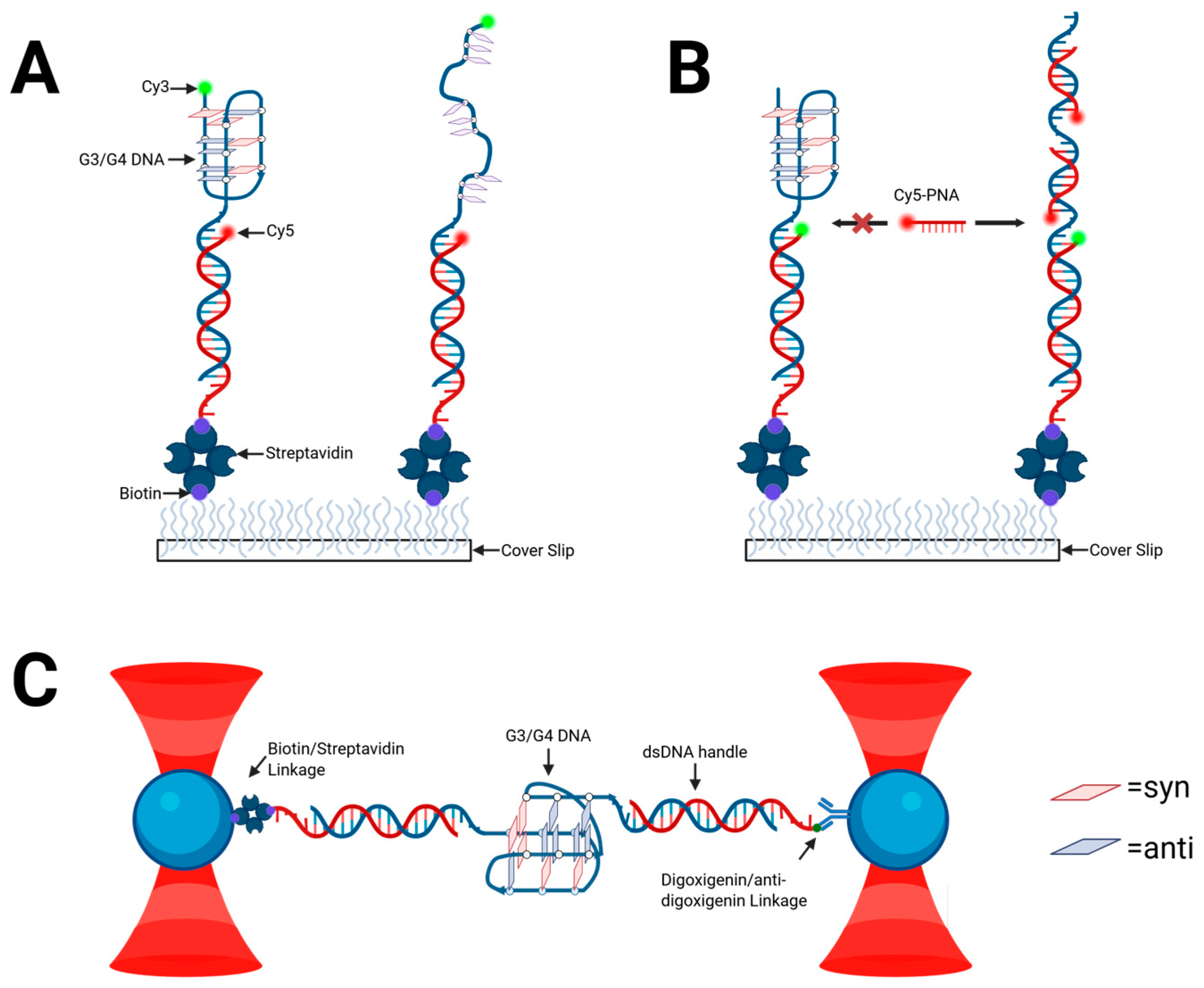
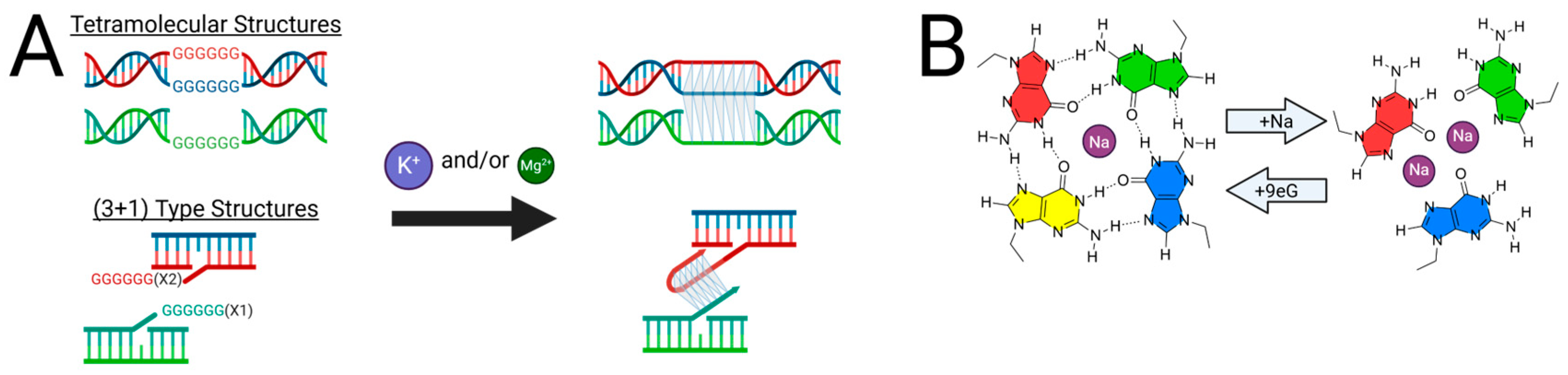
2.3. Structural Characterization of G-Triplex DNA
2.4. Further Evidence for G-Triad Formation
2.5. Other Putative G-Triplex Sequences
2.6. G-Triplex Formation in RNA
2.7. Environmental Conditions and Sequence Contexts That Promote G-Triplex Formation
| Oligonucleotide | Ion (mM) | Tm (°C) | Technique |
|---|---|---|---|
| TBA (G4) | K+ (80) | 53.0 ± 0.5 | DSC [63] |
| K+ (100) | 48.9 | UV [30] | |
| Na+ (100) | 22.7 | UV [30] | |
| Mg2+ (100) | 21.2 | UV [30] | |
| Ca2+ (100) | 54.4 | UV [30] | |
| T1 (G3) | K+ (80) | 33.5 ± 1.0 | CD [45] |
| K+ (100) | 26.4 | UV [30] | |
| Na+ (100) | undetected | UV [30] | |
| Mg2+ (100) | 20.7 | UV [30] | |
| Ca2+ (100) | 24.2 | UV [30] | |
| Hum21 (G4) 2 | K+ (100) | 60.1 | UV [30] |
| Na+ (100) | 58.4 | UV [30] | |
| Mg2+ (100) | 39.4 | UV [30] | |
| Ca2+ (100) | 54.9 | UV [30] | |
| Tel15 (G3) | K+ (100) | 43.2 | UV [30] |
| K+ (110) | 40–60 3 | CD, DSC [27] | |
| Na+ (100) | 32.6 | UV [30] | |
| Mg2+ (100) | 31.2 | UV [30] | |
| Ca2+ (100) | 47.7 | UV [30] | |
| Tel18 (G3) | Na+ (100) | 55 ± 1.0 | CD [22] |
| CatG4 (G4) | K+ (20) | 62.8 | CD [64] |
| G31 (G3) | K+ (50) | 60 | CD [51] |
| NH4+ (300) | 51.2 (±0.3) | CD [62] | |
| myc2345 (G4) 4 | K+ (20) | 75 | CD [65] |
| cMYC-G3 | K+ (20) | 38 | UV [54] |
| Ca2+ (7.5) | 51.0 (±0.7) | UV [54] |
2.8. Structural Polymorphism and Challenges in the Study of G-Triplex Structures
3. Biological Functions of G-Triplexes
3.1. G-Triplex and Regulatory Proteins
3.2. Interaction of Ligands with the G-Triplex
3.3. Catalytic Activity of G3 DNA
4. Applications of G-Triplexes
4.1. G-Triplex Function in Biosensor Development
4.2. G-Triplex Nucleic Acid Detection Assays
4.3. G-Triplex Protein Detection and Enzyme Activity Assays
4.4. G-Triplex for Small-Molecule Detection
4.5. G-Triplex-Based Metal Ion Detection
5. Conclusions and Future Directions
6. 2025 Advances in G-Triplex and Related Structures
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Saini, N.; Zhang, Y.; Usdin, K.; Lobachev, K.S. When Secondary Comes First-The Importance of Non-Canonical DNA Structures. Biochimie 2013, 95, 117–123. [Google Scholar] [CrossRef]
- Wang, G.; Vasquez, K.M. Dynamic Alternative DNA Structures in Biology and Disease. Nat. Rev. Genet. 2023, 24, 211–234. [Google Scholar] [CrossRef]
- Bansal, A.; Kaushik, S.; Kukreti, S. Non-Canonical DNA Structures: Diversity and Disease Association. Front. Genet. 2022, 13, 959258. [Google Scholar] [CrossRef]
- Pandya, N.; Bhagwat, S.R.; Kumar, A. Regulatory Role of Non-Canonical DNA Polymorphisms in Human Genome and Their Relevance in Cancer. Biochim. Et Biophys. Acta (BBA)—Rev. Cancer 2021, 1876, 188594. [Google Scholar] [CrossRef]
- Bachurin, S.S.; Kletskii, M.E.; Burov, O.N.; Bibov, M.Y.; Dobaeva, N.M.; Berezovskiy, D.P. Oligonucleotides-Transformers for Molecular Biology and Nanoengineering. Gene 2022, 820, 146277. [Google Scholar] [CrossRef]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix Formation by Guanylic Acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I.J.; Blackburn, E.H. Telomeric DNA Oligonucleotides Form Novel Intramolecular Structures Containing Guanine-Guanine Base Pairs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Patel, D.J. Solution Structure of a Parallel-Stranded G-Quadruplex DNA. J. Mol. Biol. 1993, 234, 1171–1183. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. G-Quadruplexes in Promoters throughout the Human Genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Mergny, J.L. Shape Matters: Size-Exclusion HPLC for the Study of Nucleic Acid Structural Polymorphism. Nucleic Acids Res. 2014, 42, e149. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Thompson, B.; Cathers, B.E.; Salazar, M.; Kerwin, S.M.; Trent, J.O.; Jenkins, T.C.; Neidle, S.; Hurley, L.H. Inhibition of Human Telomerase by a G-Quadruplex-Interactive Compound. J. Med. Chem. 1997, 40, 2113–2116. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct Evidence for a G-Quadruplex in a Promoter Region and Its Targeting with a Small Molecule to Repress c-MYC Transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed]
- Seenisamy, J.; Rezler, E.M.; Powell, T.J.; Tye, D.; Gokhale, V.; Joshi, C.S.; Siddiqui-Jain, A.; Hurley, L.H. The Dynamic Character of the G-Quadruplex Element in the c-MYC Promoter and Modification by TMPyP4. J. Am. Chem. Soc. 2004, 126, 8702–8709. [Google Scholar] [CrossRef]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly Prevalent Putative Quadruplex Sequence Motifs in Human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef]
- Eddy, J.; Maizels, N. Gene Function Correlates with Potential for G4 DNA Formation in the Human Genome. Nucleic Acids Res. 2006, 34, 3887–3896. [Google Scholar] [CrossRef]
- Carvalho, J.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-Quadruplex, Friend or Foe: The Role of the G-Quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef]
- Sanchez-Martin, V.; Soriano, M.; Garcia-Salcedo, J.A. Quadruplex Ligands in Cancer Therapy. Cancers 2021, 13, 3156. [Google Scholar] [CrossRef] [PubMed]
- Antonacci, C.; Chaires, J.B.; Sheardy, R.D. Biophysical Characterization of the Human Telomeric (TTAGGG)4 Repeat in a Potassium Solution. Biochemistry 2007, 46, 4654–4660. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.D.; Chaires, J.B. Kinetics and Mechanism of K+- and Na+-Induced Folding of Models of Human Telomeric DNA into G-Quadruplex Structures. Nucleic Acids Res. 2008, 36, 4191–4203. [Google Scholar] [CrossRef]
- Okamoto, K.; Sannohe, Y.; Mashimo, T.; Sugiyama, H.; Terazima, M. G-Quadruplex Structures of Human Telomere DNA Examined by Single Molecule FRET and BrG-Substitution. Bioorganic Med. Chem. 2008, 16, 6873–6879. [Google Scholar] [CrossRef]
- Mashimo, T.; Yagi, H.; Sannohe, Y.; Rajendran, A.; Sugiyama, H. Folding Pathways of Human Telomeric Type-1 and Type-2 G-Quadruplex Structures. J. Am. Chem. Soc. 2010, 132, 14910–14918. [Google Scholar] [CrossRef]
- Koirala, D.; Mashimo, T.; Sannohe, Y.; Yu, Z.; Mao, H.; Sugiyama, H. Intramolecular Folding in Three Tandem Guanine Repeats of Human Telomeric DNA. Chem. Commun. 2012, 48, 2006–2008. [Google Scholar] [CrossRef]
- Bončina, M.; Lah, J.; Prislan, I.; Vesnaver, G. Energetic Basis of Human Telomeric DNA Folding into G-Quadruplex Structures. J. Am. Chem. Soc. 2012, 134, 9657–9663. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.D.; Buscaglia, R.; Chaires, J.B. Populated Intermediates in the Thermal Unfolding of the Human Telomeric Quadruplex. J. Am. Chem. Soc. 2012, 134, 16834–16844. [Google Scholar] [CrossRef] [PubMed]
- An, N.; Fleming, A.M.; Burrows, C.J. Interactions of the Human Telomere Sequence with the Nanocavity of the α-Hemolysin Ion Channel Reveal Structure-Dependent Electrical Signatures for Hybrid Folds. J. Am. Chem. Soc. 2013, 135, 8562–8570. [Google Scholar] [CrossRef]
- Pérez-Arnáiz, C.; Busto, N.; Leal, J.M.; García, B. New Microsecond Intramolecular Reactions of Human Telomeric DNA in Solution. RSC Adv. 2016, 6, 39204–39208. [Google Scholar] [CrossRef]
- Caterino, M.; Virgilio, A.; Esposito, V.; Petraccone, L.; Galeone, A.; Giancola, C. G-Triplex Stability in Human Telomeric DNA with Epigenetic Modification/Oxidative Damage to Thymine. J. Therm. Anal. Calorim. 2018, 134, 1253–1259. [Google Scholar] [CrossRef]
- Koirala, D.; Ghimire, C.; Bohrer, C.; Sannohe, Y.; Sugiyama, H.; Mao, H. Long-Loop G-Quadruplexes Are Misfolded Population Minorities with Fast Transition Kinetics in Human Telomeric Sequences. J. Am. Chem. Soc. 2013, 135, 2235–2241. [Google Scholar] [CrossRef]
- Li, W.; Hou, X.M.; Wang, P.Y.; Xi, X.G.; Li, M. Direct Measurement of Sequential Folding Pathway and Energy Landscape of Human Telomeric G-Quadruplex Structures. J. Am. Chem. Soc. 2013, 135, 6423–6426. [Google Scholar] [CrossRef]
- Jiang, H.X.; Cui, Y.; Zhao, T.; Fu, H.W.; Koirala, D.; Punnoose, J.A.; Kong, D.M.; Mao, H. Divalent Cations and Molecular Crowding Buffers Stabilize G-Triplex at Physiologically Relevant Temperatures. Sci. Rep. 2015, 5, 9255. [Google Scholar] [CrossRef]
- Mitra, J.; Makurath, M.A.; Ngo, T.T.M.; Troitskaia, A.; Chemla, Y.R.; Ha, T. Extreme Mechanical Diversity of Human Telomeric DNA Revealed by Fluorescence-Force Spectroscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 8350–8359. [Google Scholar] [CrossRef]
- Hou, X.M.; Fu, Y.B.; Wu, W.Q.; Wang, L.; Teng, F.Y.; Xie, P.; Wang, P.Y.; Xi, X.G. Involvement of G-Triplex and G-Hairpin in the Multi-Pathway Folding of Human Telomeric G-Quadruplex. Nucleic Acids Res. 2017, 45, 11401–11412. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.M.; Li, H.; You, J.; Li, W.; Wang, P.Y.; Li, M.; Dou, S.X.; Xi, X.G. Folding Dynamics of Parallel and Antiparallel G-Triplexes under the Influence of Proximal DNA. J. Phys. Chem. B 2018, 122, 9499–9506. [Google Scholar] [CrossRef]
- Wu, W.Q.; Zhang, M.L.; Song, C.P. A Comprehensive Evaluation of a Typical Plant Telomeric G-Quadruplex (G4) DNA Reveals the Dynamics of G4 Formation, Rearrangement, and Unfolding. J. Biol. Chem. 2020, 295, 5461–5469. [Google Scholar] [CrossRef] [PubMed]
- Kodikara, S.G.; Merkel, K.J.; Haas, S.J.; Shiekh, S.; Balci, H. Detecting Secondary Structure Formation with FRET-PAINT. Analyst 2023, 19, 4655–4658. [Google Scholar] [CrossRef]
- Zhang, M.L.; Xu, Y.P.; Kumar, A.; Zhang, Y.; Wu, W.Q. Studying the Potassium-Induced G-Quadruplex DNA Folding Process Using Microscale Thermophoresis. Biochemistry 2019, 58, 3955–3959. [Google Scholar] [CrossRef]
- Šponer, J.; Bussi, G.; Stadlbauer, P.; Kührová, P.; Banáš, P.; Islam, B.; Haider, S.; Neidle, S.; Otyepka, M. Folding of Guanine Quadruplex Molecules–Funnel-like Mechanism or Kinetic Partitioning? An Overview from MD Simulation Studies. Biochim. Et Biophys. Acta—Gen. Subj. 2017, 1861, 1246–1263. [Google Scholar] [CrossRef]
- Stadlbauer, P.; Mlýnský, V.; Krepl, M.; Šponer, J. Complexity of Guanine Quadruplex Unfolding Pathways Revealed by Atomistic Pulling Simulations. J. Chem. Inf. Model. 2023, 63, 4716–4731. [Google Scholar] [CrossRef] [PubMed]
- Stadlbauer, P.; Trantírek, L.; Cheatham, T.E.; Koča, J.; Šponer, J. Triplex Intermediates in Folding of Human Telomeric Quadruplexes Probed by Microsecond-Scale Molecular Dynamics Simulations. Biochimie 2014, 105, 22–35. [Google Scholar] [CrossRef]
- Bian, Y.; Tan, C.; Wang, J.; Sheng, Y.; Zhang, J.; Wang, W. Atomistic Picture for the Folding Pathway of a Hybrid-1 Type Human Telomeric DNA G-Quadruplex. PLoS Comput. Biol. 2014, 10, e1003562. [Google Scholar] [CrossRef]
- Kim, E.; Yang, C.; Pak, Y. Free-Energy Landscape of a Thrombin-Binding DNA Aptamer in Aqueous Environment. J. Chem. Theory Comput. 2012, 8, 4845–4851. [Google Scholar] [CrossRef]
- Sun, L.; Jin, H.; Zhao, X.; Liu, Z.; Guan, Y.; Yang, Z.; Zhang, L.; Zhang, L. Unfolding and Conformational Variations of Thrombin-Binding DNA Aptamers: Synthesis, Circular Dichroism and Molecular Dynamics Simulations. ChemMedChem 2014, 9, 993–1001. [Google Scholar] [CrossRef]
- Yang, C.; Jang, S.; Pak, Y. Multiple Stepwise Pattern for Potential of Mean Force in Unfolding the Thrombin Binding Aptamer in Complex with Sr2+. J. Chem. Phys. 2011, 135, 225104. [Google Scholar] [CrossRef]
- Zgarbová, M.; Šponer, J.; Otyepka, M.; Cheatham, T.E.; Galindo-Murillo, R.; Jurečka, P. Refinement of the Sugar–Phosphate Backbone Torsion Beta for AMBER Force Fields Improves the Description of Z- and B-DNA. J. Chem. Theory Comput. 2015, 11, 5723–5736. [Google Scholar] [CrossRef] [PubMed]
- Limongelli, V.; De Tito, S.; Cerofolini, L.; Fragai, M.; Pagano, B.; Trotta, R.; Cosconati, S.; Marinelli, L.; Novellino, E.; Bertini, I.; et al. The G-Triplex DNA. Angew. Chem.—Int. Ed. 2013, 52, 2269–2273. [Google Scholar] [CrossRef]
- Cerofolini, L.; Amato, J.; Giachetti, A.; Limongelli, V.; Novellino, E.; Parrinello, M.; Fragai, M.; Randazzo, A.; Luchinat, C. G-Triplex Structure and Formation Propensity. Nucleic Acids Res. 2014, 42, 13393–13404. [Google Scholar] [CrossRef]
- Frelih, T.; Wang, B.; Plavec, J.; Šket, P. Pre-Folded Structures Govern Folding Pathways of Human Telomeric G-Quadruplexes. Nucleic Acids Res. 2020, 48, 2189–2197. [Google Scholar] [CrossRef]
- Rajendran, A.; Endo, M.; Hidaka, K.; Sugiyama, H. Direct and Single-Molecule Visualization of the Solution-State Structures of G-Hairpin and G-Triplex Intermediates. Angew. Chem.—Int. Ed. 2014, 53, 4107–4112. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Xie, L.; Li, D.; Shen, H.; Li, C.; Xu, W. Interconversion between Guanine Quartets and Triads on the Au(111) Surface. Chem. Commun. (Camb. Engl.) 2022, 58, 3198–3201. [Google Scholar] [CrossRef] [PubMed]
- Heddi, B.; Martín-Pintado, N.; Serimbetov, Z.; Kari, T.M.A.; Phan, A.T. G-Quadruplexes with (4n − 1) Guanines in the G-Tetrad Core: Formation of a G-Triad·water Complex and Implication for Small-Molecule Binding. Nucleic Acids Res. 2016, 44, 910–916. [Google Scholar] [CrossRef]
- Zhou, H.; Wu, Z.-F.; Han, Q.-J.; Zhong, H.-M.; Peng, J.-B.; Li, X.; Fan, X.-L. Stable and Label-Free Fluorescent Probe Based on G-Triplex DNA and Thioflavin T. Anal. Chem. 2018, 90, 3220–3226. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Han, X.; Xia, L.; Kong, R.-M.; Qu, F. A G-Triplex Based Molecular Beacon for Label-Free Fluorescence “Turn-on” Detection of Bleomycin. Analyst 2018, 143, 5474–5480. [Google Scholar] [CrossRef]
- Zhao, L.L.; Cao, T.; Zhou, Q.Y.; Zhang, X.H.; Zhou, Y.L.; Yang, L.J.; Zhang, X.X. The Exploration of a New Stable G-Triplex DNA and Its Novel Function in Electrochemical Biosensing. Anal. Chem. 2019, 91, 10731–10737. [Google Scholar] [CrossRef]
- Das, M.K.; Williams, E.P.; Myhre, M.W.; David, W.M.; Kerwin, S.M. Calcium-Dependent Chemiluminescence Catalyzed by a Truncated c-MYC Promoter G-Triplex DNA. Molecules 2024, 29, 4457. [Google Scholar] [CrossRef]
- Zhao, L.-L.; Gu, Y.-X.; Dong, J.-H.; Li, X.-T.; Pan, H.-Y.; Xue, C.-Y.; Liu, Y.; Zhou, Y.-L.; Zhang, X.-X. New G-Triplex DNA Dramatically Activates the Fluorescence of Thioflavin T and Acts as a Turn-On Fluorescent Sensor for Uracil-DNA Glycosylase Activity Detection. Anal. Chem. 2024, 96, 8458–8466. [Google Scholar] [CrossRef]
- Wang, L.; Xu, Y.P.; Bai, D.; Shan, S.W.; Xie, J.; Li, Y.; Wu, W.Q. Insights into the Structural Dynamics and Helicase-Catalyzed Unfolding of Plant RNA G-Quadruplexes. J. Biol. Chem. 2022, 298, 102165. [Google Scholar] [CrossRef]
- Campanile, M.; Improta, R.; Esposito, L.; Platella, C.; Oliva, R.; Del Vecchio, P.; Winter, R.; Petraccone, L. Experimental and Computational Evidence of a Stable RNA G-Triplex Structure at Physiological Temperature in the SARS-CoV-2 Genome. Angew. Chem.—Int. Ed. 2024, 63, e202415448. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Wang, B.; Lo, K.W.; Tsang, C.M.; Kwok, C.K. Pre-Defined Stem-Loop Structure Library for the Discovery of L-RNA Aptamers That Target RNA G-Quadruplexes. Angew. Chem.—Int. Ed. 2024, 64, e202417247. [Google Scholar] [CrossRef] [PubMed]
- Ugrina, M.; Burkhart, I.; Müller, D.; Schwalbe, H.; Schwierz, N. RNA G-Quadruplex Folding Is a Multi-Pathway Process Driven by Conformational Entropy. Nucleic Acids Res. 2024, 52, 87–100. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef]
- Hardin, C.C.; Watson, T.; Corregan, M.; Bailey, C. Cation-Dependent Transition between the Quadruplex and Watson-Crick Hairpin Forms of d(CGCG3GCG). Biochemistry 1992, 31, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fu, B.; Wang, J.; Long, Y.; Zhang, X.; Peng, S.; Guo, P.; Tian, T.; Zhou, X. Novel Amplex Red Oxidases Based on Noncanonical DNA Structures: Property Studies and Applications in MicroRNA Detection. Anal. Chem. 2014, 86, 2925–2930. [Google Scholar] [CrossRef]
- Pagano, B.; Martino, L.; Randazzo, A.; Giancola, C. Stability and Binding Properties of a Modified Thrombin Binding Aptamer. Biophys. J. 2008, 94, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-K.; Kuo, C.-J.; Chen, L.-C. Synthetic Multivalent DNAzymes for Enhanced Hydrogen Peroxide Catalysis and Sensitive Colorimetric Glucose Detection. Anal. Chim. Acta 2015, 856, 96–102. [Google Scholar] [CrossRef]
- Hatzakis, E.; Okamoto, K.; Yang, D. Thermodynamic Stability and Folding Kinetics of the Major G-Quadruplex and Its Loop Isomers Formed in the Nuclease Hypersensitive Element in the Human c-Myc Promoter: Effect of Loops and Flanking Segments on the Stability of Parallel-Stranded Intramolecular G-Quadruplexes. Biochemistry 2010, 49, 9152–9160. [Google Scholar] [CrossRef]
- Zhao, Z.; Lin, F.; Ye, H.; Huang, R.; Xu, X. Effects of Modified-Guanosine on the Stability of G-Triplex. Tetrahedron Lett. 2016, 57, 5321–5325. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Muraoka, T.; Kinbara, K.; Sugimoto, N. G-Quadruplexes with Tetra(Ethylene Glycol)-Modified Deoxythymidines Are Resistant to Nucleases and Inhibit HIV-1 Reverse Transcriptase. ChemBioChem 2016, 17, 1399–1402. [Google Scholar] [CrossRef]
- Samanta, P.K.; Pati, S.K. Understanding the Unfolding Mechanism of Human Telomeric G-Quadruplex Using Steered Molecular Dynamics Simulation. Indian J. Chem.—Sect. A Inorg. Phys. Theor. Anal. Chem. 2017, 56A, 907–912. [Google Scholar]
- Zhang, Q.; Yang, T.; Zheng, G.; Gao, H.; Yan, C.; Zheng, X.; Zhou, X.; Shao, Y. Characterization of Intermolecular G-Quadruplex Formation over Intramolecular G-Triplex for DNA Containing Three G-Tracts. Analyst 2020, 145, 4254–4259. [Google Scholar] [CrossRef] [PubMed]
- Dahan, D.; Tsirkas, I.; Dovrat, D.; Sparks, M.A.; Singh, S.P.; Galletto, R.; Aharoni, A. Pif1 Is Essential for Efficient Replisome Progression through Lagging Strand G-Quadruplex DNA Secondary Structures. Nucleic Acids Res. 2018, 46, 11847–11857. [Google Scholar] [CrossRef]
- Hou, X.M.; Wu, W.Q.; Duan, X.L.; Liu, N.N.; Li, H.H.; Fu, J.; Dou, S.X.; Li, M.; Xi, X.G. Molecular Mechanism of G-Quadruplex Unwinding Helicase: Sequential and Repetitive Unfolding of G-Quadruplex by Pif1 Helicase. Biochem. J. 2015, 466, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Badie, S.; Escandell, J.M.; Bouwman, P.; Carlos, A.R.; Thanasoula, M.; Gallardo, M.M.; Suram, A.; Jaco, I.; Benitez, J.; Herbig, U.; et al. BRCA2 Acts as a RAD51 Loader to Facilitate Telomere Replication and Capping. Nat. Struct. Mol. Biol. 2010, 17, 1461–1469. [Google Scholar] [CrossRef]
- Lee, J.; Sung, K.; Joo, S.Y.; Jeong, J.H.; Kim, S.K.; Lee, H. Dynamic Interaction of BRCA2 with Telomeric G-Quadruplexes Underlies Telomere Replication Homeostasis. Nat. Commun. 2022, 13, 3396. [Google Scholar] [CrossRef]
- Joo, S.Y.; Sung, K.; Lee, H. Balancing Act: BRCA2’s Elaborate Management of Telomere Replication through Control of G-Quadruplex Dynamicity. BioEssays 2024, 46, 2300229. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative Visualization of DNA G-Quadruplex Structures in Human Cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Johnson, S.A.; Paul, T.; Sanford, S.L.; Schnable, B.L.; Detwiler, A.C.; Thosar, S.A.; Van Houten, B.; Myong, S.; Opresko, P.L. BG4 Antibody Can Recognize Telomeric G-Quadruplexes Harboring Destabilizing Base Modifications and Lesions. Nucleic Acids Res. 2024, 52, 1763–1778. [Google Scholar] [CrossRef]
- Abram, S.B.; Marušič, M.; Živković, M.L.; Brčić, J.; Plavec, J. Non-Canonical Structures in Promoter Modulate Gene Expression in Escherichia coli. Croat. Chem. Acta 2018, 91, 163–170. [Google Scholar] [CrossRef]
- Teng, F.Y.; Hou, X.M.; Fan, S.H.; Rety, S.; Dou, S.X.; Xi, X.G. Escherichia coli DNA Polymerase I Can Disrupt G-Quadruplex Structures during DNA Replication. FEBS J. 2017, 284, 4051–4065. [Google Scholar] [CrossRef]
- Li, T.; Hu, R.; Xia, J.; Xu, Z.; Chen, D.; Xi, J.; Liu, B.F.; Zhu, J.; Li, Y.; Yang, Y.; et al. G-Triplex: A New Type of CRISPR-Cas12a Reporter Enabling Highly Sensitive Nucleic Acid Detection. Biosens. Bioelectron. 2021, 187, 113292. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Endo, M.; Hidaka, K.; Teulade-Fichou, M.P.; Mergny, J.L.; Sugiyama, H. Small Molecule Binding to a G-Hairpin and a G-Triplex: A New Insight into Anticancer Drug Design Targeting G-Rich Regions. Chem. Commun. 2015, 51, 9181–9184. [Google Scholar] [CrossRef]
- Ma, D.L.; Lu, L.; Lin, S.; He, B.; Leung, C.H. A G-Triplex Luminescent Switch-on Probe for the Detection of Mung Bean Nuclease Activity. J. Mater. Chem. B 2015, 3, 348–352. [Google Scholar] [CrossRef]
- Amato, J.; Pagano, A.; Cosconati, S.; Amendola, G.; Fotticchia, I.; Iaccarino, N.; Marinello, J.; De Magis, A.; Capranico, G.; Novellino, E.; et al. Discovery of the First Dual G-Triplex/G-Quadruplex Stabilizing Compound: A New Opportunity in the Targeting of G-Rich DNA Structures. Biochim. Et Biophys. Acta-Gen. Subj. 2017, 1861, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Bonnat, L.; Dautriche, M.; Saidi, T.; Revol-Cavalier, J.; Dejeu, J.; Defrancq, E.; Lavergne, T. Scaffold Stabilization of a G-Triplex and Study of Its Interactions with G-Quadruplex Targeting Ligands. Org. Biomol. Chem. 2019, 17, 8726–8736. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Zheng, X.; Yang, T.; Zhang, Q.; Yan, C.; Zhou, X.; Shao, Y. A pH-Triggered G-Triplex Switch with K+ Tolerance. Chem. Commun. 2020, 56, 7349–7352. [Google Scholar] [CrossRef]
- Gao, H.; Peng, S.; Yan, C.; Zhang, Q.; Zheng, X.; Yang, T.; Wang, D.; Zhou, X.; Shao, Y. Stimuli-Responsive and Reversible Nanoassemblies of G-Triplexes. ChemBioChem 2022, 23, e202100587. [Google Scholar] [CrossRef]
- Kataoka, Y.; Fujita, H.; Endoh, T.; Sugimoto, N.; Kuwahara, M. Effects of Modifying Thioflavin T at the N3-Position on Its G4 Binding and Fluorescence Emission. Molecules 2020, 25, 4936. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Wu, Z.; Li, X.; Hu, X.; Wu, J.; You, Z.; Qiu, J. Comparison of Double-Stranded DNA at the 5′ and 3′ Ends of the G-Triplex and Its Application in the Detection of Hg(II). Int. J. Mol. Sci. 2024, 25, 8159. [Google Scholar] [CrossRef]
- Wen, C.J.; Wen, C.J.; Gong, J.Y.; Gong, J.Y.; Zheng, K.W.; He, Y.D.; Zhang, J.Y.; Hao, Y.H.; Tan, Z.; Tan, Z. Targeting Nucleic Acids with a G-Triplex-to-G-Quadruplex Transformation and Stabilization Using a Peptide-PNA G-Tract Conjugate. Chem. Commun. 2020, 56, 6567–6570. [Google Scholar] [CrossRef]
- Xu, X.W.; Mao, W.X.; Lin, F.; Hu, J.L.; He, Z.Y.; Weng, X.C.; Wang, C.J.; Zhou, X. Enantioselective Diels-Alder Reactions Using a G-Triplex DNA-Based Catalyst. Catal. Commun. 2016, 74, 16–18. [Google Scholar] [CrossRef]
- Kankia, B. Quadruplex-Templated and Catalyzed Ligation of Nucleic Acids. ChemBioChem 2021, 22, 1261–1267. [Google Scholar] [CrossRef]
- Wang, S.; Fu, B.; Peng, S.; Zhang, X.; Tian, T.; Zhou, X. The G-Triplex DNA Could Function as a New Variety of DNA Peroxidase. Chem. Commun. 2013, 49, 7920–7922. [Google Scholar] [CrossRef]
- Kosman, J.; Juskowiak, B. Peroxidase-Mimicking DNAzymes for Biosensing Applications: A Review. Anal. Chim. Acta 2011, 707, 7–17. [Google Scholar] [CrossRef]
- Chen, J.; Cheng, M.; Wang, J.; Dehui, Q.; Monchaud, D.; Mergny, J.-L.; Ju, H.; Zhou, J. The Catalytic Properties of DNA G-quadruplexes Rely on Their Structural Integrity. Chin. J. Catal. 2021, 42, 1102–1107. [Google Scholar] [CrossRef]
- Bai, Y.; Xu, P.; Li, S.; Wang, D.; Zhang, K.; Zheng, D.; Yue, D.; Zhang, G.; He, S.; Li, Y.; et al. Signal Amplification Strategy of DNA Self-Assembled Biosensor and Typical Applications in Pathogenic Microorganism Detection. Talanta 2024, 272, 125759. [Google Scholar] [CrossRef]
- Gul, I.; Raheem, M.A.; Reyad-ul-Ferdous, M.; Yuan, X.; Chen, Z.; Lv, C.; Chen, M.; Ji, J.; Wu, D.; Zhao, Q.; et al. State-of-the-Art Signal Amplification Strategies for Nucleic Acid and Non-Nucleic Acid Biosensors. Sens. Actuators Rep. 2025, 9, 100268. [Google Scholar] [CrossRef]
- Lee, J.E.; Pack, S.P. Recent Progress in DNA Biosensors: Target-Specific and Structure-Guided Signal Amplification. Biosensors 2025, 15, 476. [Google Scholar] [CrossRef]
- Wu, Z.; Zhou, H.; He, J.; Li, M.; Ma, X.; Xue, J.; Li, X.; Fan, X. G-Triplex Based Molecular Beacon with Duplex-Specific Nuclease Amplification for the Specific Detection of microRNA. Analyst 2019, 144, 5201–5206. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, S.; Zou, R.; Chen, C.; Cai, C. An Enzyme-Free Probe Based on G-Triplex Assisted by Silver Nanocluster Pairs for Sensitive Detection of microRNA-21. Microchim. Acta 2021, 188, 55. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.L.; Pan, H.Y.; Zhang, X.X.; Zhou, Y.L. Ultrasensitive Detection of microRNA Based on a Homogeneous Label-Free Electrochemical Platform Using G-Triplex/Methylene Blue as a Signal Generator. Anal. Chim. Acta 2020, 1116, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Chen, F.; Jiang, M.; Guo, Q.; Wang, Y.; Wang, J.; Zhang, D.W. A Homogeneous Label-Free Electrochemical microRNA Biosensor Coupling With G-Triplex/Methylene Blue Complex and λ-Exonuclease-Assisted Recycling Amplification. Front. Chem. 2021, 9, 753253. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, T.; Xue, D.; Yang, H.; Sui, Z.; Yuan, X.; Xu, J. An Allosteric Palindromic Hairpin Probe Based Dual-Mode Interactive Strand Displacement Amplification Enables Robust miRNA Biosensing. Chem. Commun. 2024, 60, 2910–2913. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, C.; He, Y.; Gao, J.; Li, W.; Cheng, L.; Sun, F.; Xia, P.; Wang, Q. A Generic and Non-Enzymatic Electrochemical Biosensor Integrated Molecular Beacon-like Catalyzed Hairpin Assembly Circuit with MOF@Au@G-Triplex/Hemin Nanozyme for Ultrasensitive Detection of miR-721. Biosens. Bioelectron. 2022, 203, 114051. [Google Scholar] [CrossRef]
- Li, R.; Liu, Q.; Jin, Y.; Li, B. G-Triplex/Hemin DNAzyme: An Ideal Signal Generator for Isothermal Exponential Amplification Reaction-Based Biosensing Platform. Anal. Chim. Acta 2019, 1079, 139–145. [Google Scholar] [CrossRef]
- Li, R.; Liu, Q.; Jin, Y.; Li, B. Sensitive Colorimetric Determination of microRNA Let-7a through Rolling Circle Amplification and a Peroxidase-Mimicking System Composed of Trimeric G-Triplex and Hemin DNAzyme. Mikrochim. Acta 2020, 187, 139. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zou, R.; Xiang, L.; Chen, C.; Cai, C. Engineering a Label- and Enzyme-Free Detection of HIV-DNA on a Cyclic DNA Self-Assembling Strategy Using G-Triplexes as the Signal Reporter. Microchem. J. 2020, 155, 104656. [Google Scholar] [CrossRef]
- Yi, G.; Duan, Q.; Yan, Q.; Huang, Y.; Zhang, W.; Zhao, S. Polymerase/Nicking Enzyme Powered Dual-Template Multi-Cycle G-Triplex Machine for HIV-1 Determination. Anal. Sci. 2021, 37, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Yang, B.; Shi, L.; Tang, Q.; Wang, J.; Liu, W.; Li, B.; Jin, Y. Label-Free and Portable Detection of HIV-DNA by a Handheld Luminometer. Anal. Chim. Acta 2024, 1304, 342553. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Duan, Q.; Huang, Y.; Guo, J.; Zhong, L.; Wang, H.; Yi, G. Symmetric Exponential Amplification Reaction-Based DNA Nanomachine for the Fluorescent Detection of Nucleic Acids. RSC Adv. 2019, 9, 41305–41310. [Google Scholar] [CrossRef]
- Gao, J.; Liu, Q.; Liu, W.; Jin, Y.; Li, B. Comparative Evaluation and Design of a G-Triplex/Thioflavin T-Based Molecular Beacon. Analyst 2021, 146, 2567–2573. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, X.; Pan, L.; Yang, S.; Chen, X.; Wang, F.; Yang, D.; Li, M.; Wang, P. A CRISPR-Cas12a-Based Assay for One-Step Preamplification-Free Detection of Viral DNA. Sens. Actuators B Chem. 2024, 399, 134813. [Google Scholar] [CrossRef]
- Liu, Q.; Hou, L.; Zhang, Y.; Liu, M.; Jin, Y.; Li, B. Improving Efficiency of Entropy-Driven DNA Amplification Biosensing through Producing Two Label-Free Signal Strands in One Cycle. Anal. Chim. Acta 2022, 1232, 340484. [Google Scholar] [CrossRef]
- Wang, C.; Li, J.; Wang, P. The Effect of Adjacent Double-Strand DNA on the G-Triplex-ThT Complex Fluorescence Intensity Enhancement and Its Application in TNOS and Hg2+ Detection. Talanta 2023, 252, 123884. [Google Scholar] [CrossRef]
- Tang, Q.; Li, Z.; Li, J.; Chen, H.; Yan, H.; Deng, J.; Liu, L. PCR-Free, Label-Free, and Centrifugation-Free Diagnosis of Multiplex Antibiotic Resistance Genes by Combining mDNA-Au@Fe3O4 from Heating Dry and DNA Concatamers with G-Triplex. Anal. Chem. 2023, 96, 292–300. [Google Scholar] [CrossRef]
- You, S.; Chen, L.; Li, D.; Yang, W.; Chen, L. Construction of a Novel “Self-Regenerative” Electrochemical Biosensor Based on Metal–Organic Frameworks and Its Application to the Detection of Mycoplasma Ovine Pneumonia. Bioelectrochemistry 2023, 152, 108409. [Google Scholar] [CrossRef]
- Dong, J.-H.; Zhang, R.-H.; Zhao, L.-L.; Xue, C.-Y.; Pan, H.-Y.; Zhong, X.-Y.; Zhou, Y.-L.; Zhang, X.-X. Identification and Quantification of Locus-Specific 8-Oxo-7,8-Dihydroguanine in DNA at Ultrahigh Resolution Based on G-Triplex-Assisted Rolling Circle Amplification. Anal. Chem. 2024, 96, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Chen, X.; Xu, Z.; Li, T.; Zhao, S.; Hu, R.; Zhu, J.; Li, Y.; Yang, Y.; Liu, M. Telomere G-Triplex Lights up Thioflavin T for RNA Detection: New Wine in an Old Bottle. Anal. Bioanal. Chem. 2022, 414, 6149–6156. [Google Scholar] [CrossRef]
- Liu, Q.; Sun, X.; Liu, M.; Jin, Y.; Li, B. G-Triplex Molecular Beacon—based Fluorescence Biosensor for Sensitive Detection of Small Molecule-Protein Interaction via Exonuclease III—assisted Recycling Amplification. Sens. Actuators B Chem. 2020, 310, 127804. [Google Scholar] [CrossRef]
- Xue, J.; Yi, J.; Zhou, H. Label-Free Fluorescence Molecular Beacon Probes Based on G-Triplex DNA and Thioflavin T for Protein Detection. Molecules 2021, 26, 2962. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Che, B.; Dai, H. A New G-Triplex-Based Strategy for Sensitivity Enhancement of the Detection of Endonuclease Activity and Inhibition. RSC Adv. 2021, 11, 28008–28013. [Google Scholar] [CrossRef] [PubMed]
- Que, H.Y.; Yan, X.Y.; Guo, B.; Ma, H.M.; Wang, T.; Liu, P.; Gan, X.F.; Yan, Y.R. Terminal Deoxynucleotidyl Transferase and Rolling Circle Amplification Induced G-Triplex Formation: A Label-Free Fluorescent Strategy for DNA Methyltransferase Activity Assay. Sens. Actuators B-Chem. 2019, 291, 394–400. [Google Scholar] [CrossRef]
- Zhu, W.; Li, Z.; Dai, L.; Yang, W.; Li, Y. Label-Free Fluorescence Detection of Alkaline Phosphatase Activity Using a G-Triplex Based Dumbbell-Shaped Probe. Anal. Sci. 2023, 39, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Wang, L.; Liang, Y.; Wang, Z.; Zhang, W.; Zhao, Q.; Yang, X.; Jiang, Y. G-Triplex/Hemin DNAzyme Mediated Colorimetric Aptasensor for Escherichia coli O157:H7 Detection Based on Exonuclease III-Assisted Amplification and Aptamers-Functionalized Magnetic Beads. Talanta 2024, 269, 125457. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Li, Y.-T.; Zhang, Y.; Xiao, M.-M.; Zhang, Z.; Liu, Y.; Zhang, Z.-Y.; Zhang, G.-J. Plug-and-Play Smart Transistor Bio-Chips Implementing Point-of-Care Diagnosis of AMI with Modified CRISPR/Cas12a System. Biosens. Bioelectron. 2024, 246, 115909. [Google Scholar] [CrossRef]
- Chen, F.; Fu, X.; Meng, Y.; Jiang, M.; Wang, J.; Zhou, Y.L.; Zhang, D.W. A Novel Miniaturized Homogeneous Label-Free Electrochemical Biosensing Platform Combining Integrated Microelectrode and Functional Nucleic Acids. Anal. Chim. Acta 2021, 1158, 338415. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, C.; Wei, Q.; Song, Y.; Chen, P.; Wang, L.; Yang, X.; Chen, X. A Sensitive Aptasensor Based on Rolling Circle Amplification and G-Rich ssDNA/Terbium (III) Luminescence Enhancement for Ofloxacin Detection in Food. Talanta 2021, 235, 122783. [Google Scholar] [CrossRef]
- Wang, C.; Li, J. Fluorescence Method for Kanamycin Detection Based on the Conversion of G-Triplex and G-Quadruplex. Anal. Bioanal. Chem. 2021, 413, 7073–7080. [Google Scholar] [CrossRef]
- Liu, Z.J.; Liang, Y.Q.; Li, J.Y.; Wu, B.; Huang, C.; Liu, Y.W.; Zhang, C.Z.; Yang, Y.; Cai, N.Q.; Chen, J.Y.; et al. Engineered Aptamer-Derived Fluorescent Aptasensor: The Label-Free, Single-Step, Rapid Detection of Vancomycin in Clinical Samples. Small 2024, 21, 2407799. [Google Scholar] [CrossRef]
- Fan, Y.; Wen, J.; Li, J.; Yang, X.; Zhang, L.; Zhang, Z. Structure-switching Aptasensors for Sensitive Detection of Ochratoxin A. Luminescence 2023, 38, 1678–1685. [Google Scholar] [CrossRef]
- Wen, J.; Fan, Y.Y.; Li, J.; Yang, X.W.; Zhang, X.X.; Zhang, Z.Q. A G-Triplex and G-Quadruplex Concatemer-Enhanced Fluorescence Probe Coupled with Hybridization Chain Reaction for Ultrasensitive Aptasensing of Ochratoxin A. Anal. Chim. Acta 2023, 1272, 341503. [Google Scholar] [CrossRef]
- Li, Q.; Peng, S.; Chang, Y.; Yang, M.; Wang, D.; Zhou, X.; Shao, Y. A G-Triplex-Based Label-Free Fluorescence Switching Platform for the Specific Recognition of Chromium Species. J. Photochem. Photobiol. A Chem. 2022, 432, 114071. [Google Scholar] [CrossRef]
- Del Mundo, I.M.A.; Cho, E.J.; Dalby, K.N.; Vasquez, K.M. A “light-up” Intercalator Displacement Assay for Detection of Triplex DNA Stabilizers. Chem. Commun. 2020, 56, 1996–1999. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Yang, B.; Shi, L.; Tang, Q.; Wang, J.; Liu, W.; Li, B.; Jin, Y. Peroxidase-Mimicking DNAzymes as Receptors for Label-Free Discriminating Heavy Metal Ions by Chemiluminescence Sensor Arrays. Anal. Chem. 2023, 95, 3486–3492. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.-M.; Ma, L.; Han, X.; Ma, C.; Qu, F.; Xia, L. Hg2+-Mediated Stabilization of G-Triplex Based Molecular Beacon for Label-Free Fluorescence Detection of Hg2+, Reduced Glutathione, and Glutathione Reductase Activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 228, 117855. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, Y.; Si, X.; Li, J.; Duan, Y.; Jiang, W.; Liu, C.; You, J.; Li, Z.; Shen, Q. A Label- and Enzyme-Free Fluorescent Method for the Rapid and Simple Detection of Hg2+ in Cigarettes Using G-Triplexes as the Signal Reporter. Spectroscopy 2020, 35, 41–52. [Google Scholar]
- Li, X.-Y.; Xiao, Y.-L.; Liu, X.; Wang, Y.-Q.; Li, M.-M.; Wang, J.-P. Label-Free and Ultrasensitive Detection of Environmental Lead Ions Based on Spatially Localized DNA Nanomachines Driven by Hyperbranched Hybridization Chain Reaction. J. Hazard. Mater. 2024, 476, 135115. [Google Scholar] [CrossRef]
- Liu, X.; Li, X.; Zhu, Z.; Yao, F.; Kang, X. Polyaniline-Based Hybrid Membrane for Single-Molecule Protein Nanopore Analysis under High Voltage. Biosens. Bioelectron. 2025, 283, 117520. [Google Scholar] [CrossRef]
- Li, T. Unveiling a Novel RNA G-Triplex Structure: Its Function and Potential in CRISPR-Based Diagnostics. Chem. Commun. 2025, 61, 4002–4005. [Google Scholar] [CrossRef]
- Janeček, M.; Kührová, P.; Mlynsky, V.; Stadlbauer, P.; Otyepka, M.; Bussi, G.; Sponer, J.; Banás, P. Computer Folding of Parallel DNA G-Quadruplex: Hitchhiker’s Guide to the Conformational Space. J. Comput. Chem. 2025, 46, e27535. [Google Scholar] [CrossRef]







| Name | Sequence | Origin |
|---|---|---|
| Tel22 | A GGG TTA GGG TTA GGG TTA GGG | Human telomeric sequence |
| Tel21 | TTA GGG TTA GGG TTA GGG TTA | Truncated human telomeric sequence |
| Tel15 | GGG TTA GGG TTA GGG | Truncated human telomeric sequence |
| TBA | GG TT GG TGT GG TT GG | Selex |
| T1 | GG TT GG TGT GG | 3′ truncation of TBA |
| htel1 | TA GGG TTA GGG TTA GGG TTA GGG | Human telomeric sequence |
| CatG4 | T GGG TA GGG C GGG TT GGG AAA | Selex |
| G31 | T GGG TA GGG C GGG | 3′ truncation of CatG4 |
| G31′ | T GGG AA GGG A GGG | Alteration of G31 |
| EAD2 | CT GGG A GGG A GGG A GGG A | Designed sequence |
| G3-A | CT GGG A GGG A GGG A | 3′ truncation of EAD2 |
| cMYCPu27 | TGG GGA GGG T GGG GA GGG T GGG GAA GG | cMYC protooncogene promoter |
| cMYCG3 | T GGG GA GGG T GGG GAA | 5′ truncation of cMYCPu27 |
| G3-F15 | TC GGG AA GGG A GGG | Modification of G3-A |
| Name | Sequence | Origin |
|---|---|---|
| C | C GG C GG C GG C GG | Arabidopsis transcriptome |
| AA | GG AA GG AA GG AA GG | Arabidopsis transcriptome |
| ATR | GGG A GGG AA GGGG AA GGGG | Arabidopsis 5′-UTR |
| SMLX | GGGGG U GGGGGG UUA GGG UUA GGG | Arabidopsis 5′-UTR |
| RG1 | GG CU GG CAAU GG C GG | SARS-CoV-2 |
| TERRA | UA GGG UUA GGG UUA GGG UUA GGG U | Human telomeres |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Myhre, M.W.; Das, M.K.; Williams, E.P.; David, W.M.; Kerwin, S.M. From Triads to Tools: A Comprehensive Review of the Expanding Roles of G-Triplex Structures. Molecules 2025, 30, 4303. https://doi.org/10.3390/molecules30214303
Myhre MW, Das MK, Williams EP, David WM, Kerwin SM. From Triads to Tools: A Comprehensive Review of the Expanding Roles of G-Triplex Structures. Molecules. 2025; 30(21):4303. https://doi.org/10.3390/molecules30214303
Chicago/Turabian StyleMyhre, Mitchell W., Malay Kumar Das, Elizabeth P. Williams, Wendi M. David, and Sean M. Kerwin. 2025. "From Triads to Tools: A Comprehensive Review of the Expanding Roles of G-Triplex Structures" Molecules 30, no. 21: 4303. https://doi.org/10.3390/molecules30214303
APA StyleMyhre, M. W., Das, M. K., Williams, E. P., David, W. M., & Kerwin, S. M. (2025). From Triads to Tools: A Comprehensive Review of the Expanding Roles of G-Triplex Structures. Molecules, 30(21), 4303. https://doi.org/10.3390/molecules30214303








