Conjugation of Short-Chain Fatty Acids to Bicyclic-Amines for Analysis by Liquid Chromatography Tandem Mass Spectroscopy
Abstract
1. Introduction
2. Results
2.1. Selection of Amine Types for Conjugation
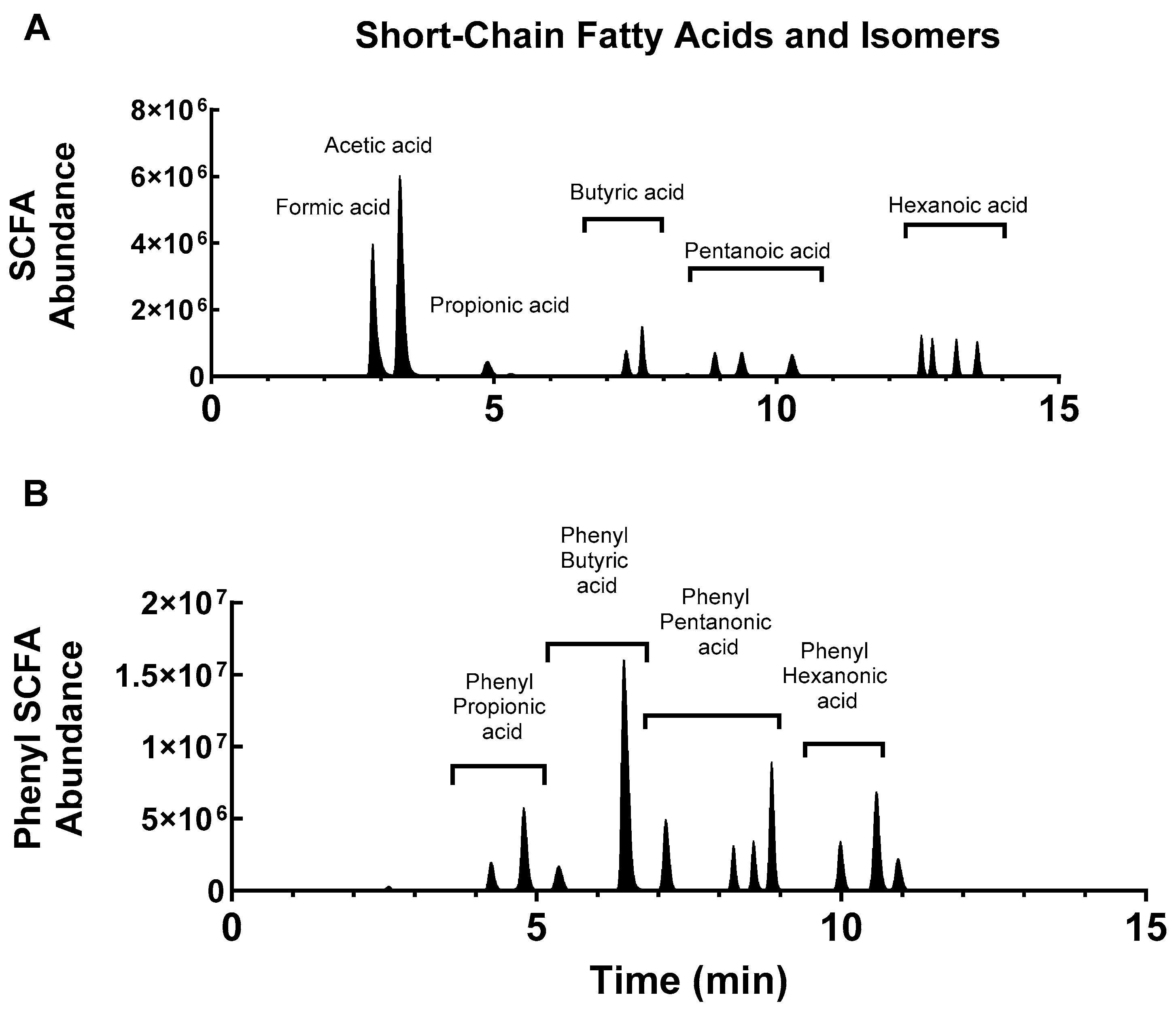
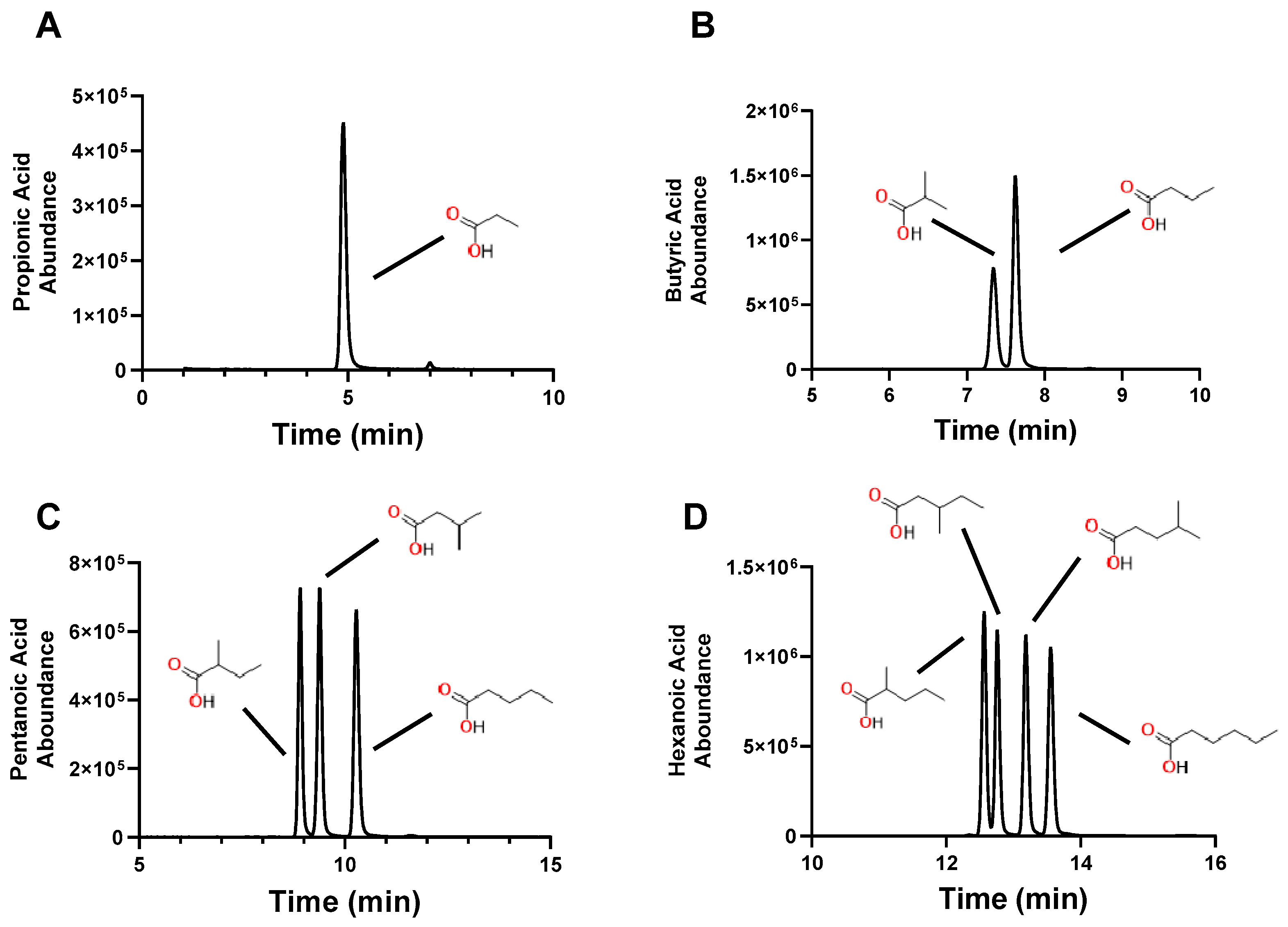
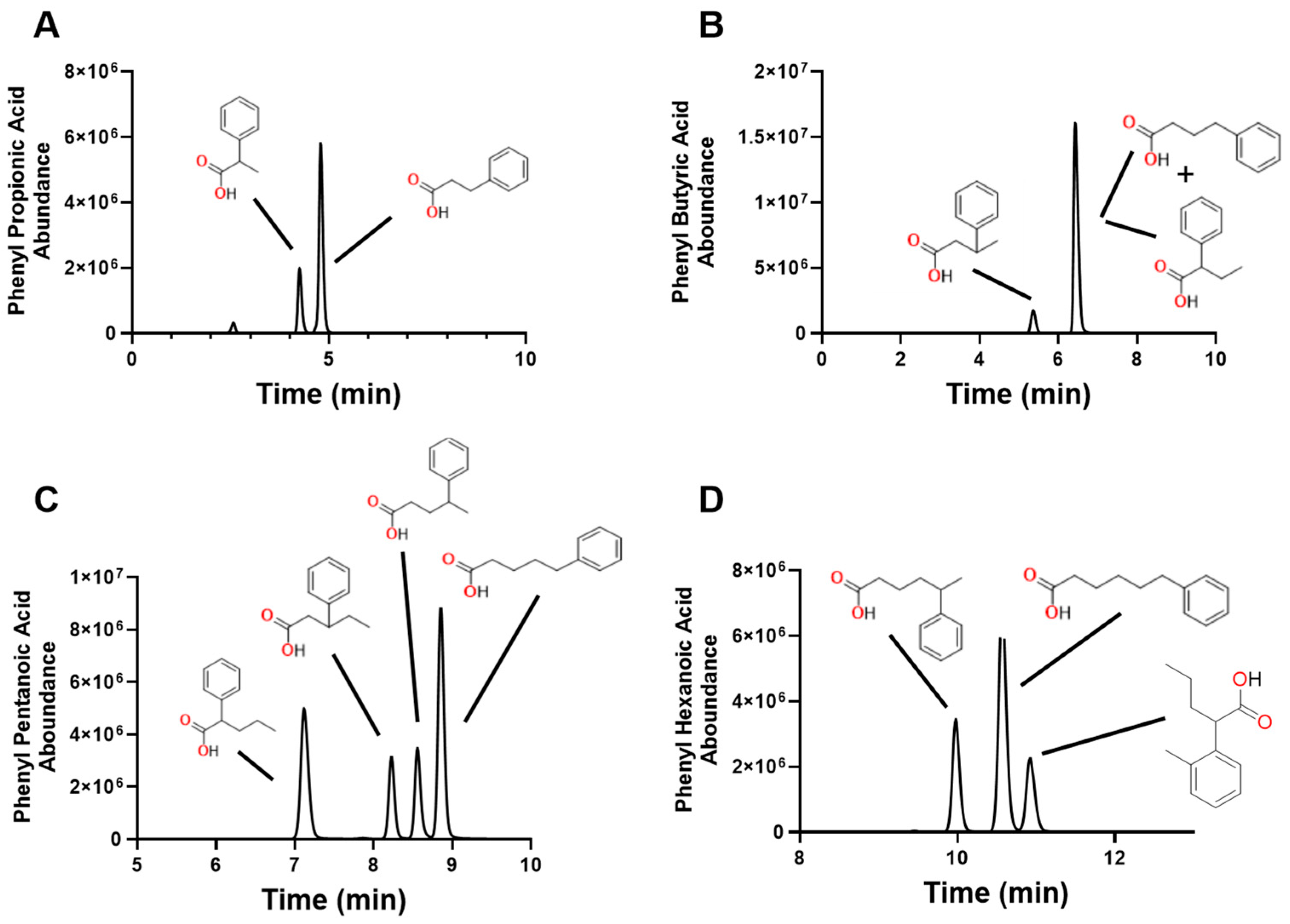
2.2. Conjugation of 4PyBA to SCFA
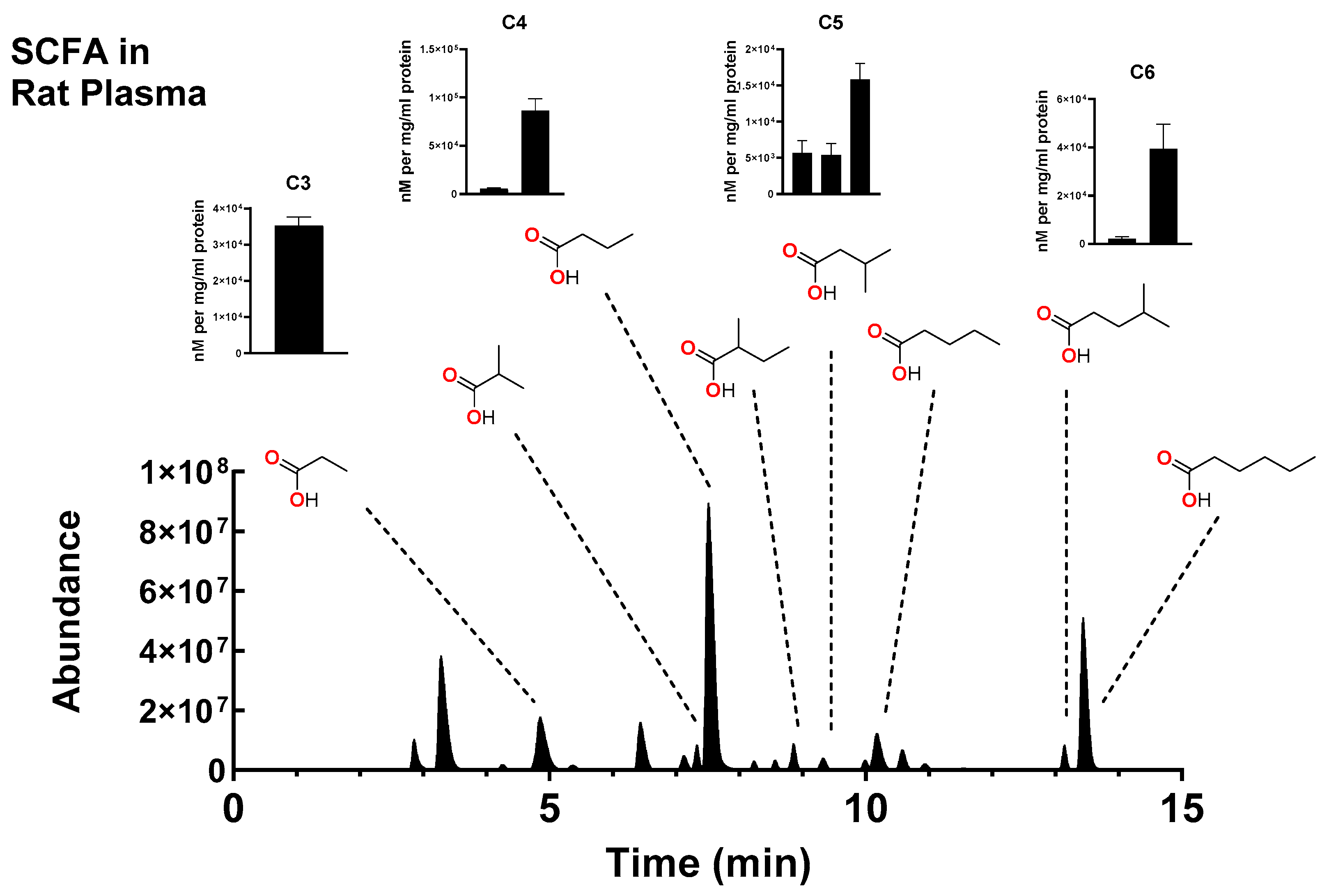
2.3. Measurement of SCFA in Biological Samples
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Conjugation Reaction
4.3. LC-MS/MS
4.4. Screening of SCFA Conjugation to 27 Different Amines
4.5. Extraction of SCFAs from Plasma and Tissue
4.6. Extraction and Conjugation
4.7. Data Analysis
5. Conclusions
6. Patents
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CID | collision-induced dissociation |
| EtOH | ethanol |
| ESI | electrospray ionization |
| MS | mass spectroscopy |
| MeCN | acetonitrile |
| MeOH | methanol |
| LC-MS/MS | liquid chromatography tandem mass spectroscopy |
| Q | quadrupole (configuration of a single linear accelerator in MS/MS) |
| SRM | selective reaction monitor (or single ion monitoring, multiple ion monitoring) |
| SCFAs | short-chain fatty acids |
| 4PyBA | 4-(pyrrolidin-1-ylmethyl) benzylamine |
| MALDI | matrix-assisted laser desorption/ionization |
References
- De Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Fusco, W.; Lorenzo, M.B.; Cintoni, M.; Porcari, S.; Rinninella, E.; Kaitsas, F.; Lener, E.; Mele, M.C.; Gasbarrini, A.; Collado, M.C.; et al. Short-Chain Fatty-Acid-Producing Bacteria: Key Components of the Human Gut Microbiota. Nutrients 2023, 15, 2211. [Google Scholar] [CrossRef]
- Li, M.; Van Esch, B.C.A.M.; Wagenaar, G.T.M.; Garssen, J.; Folkerts, G.; Henricks, P.A.J. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef]
- Li, M.; Van Esch, B.C.A.M.; Wagenaar, G.T.M.; Garssen, J.; Folkerts, G.; Henricks, P.A.J. Pro- and anti-inflammatory effects of short chain fatty acids on immune and endothelial cells. Eur. J. Pharmacol. 2018, 831, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Rastall, R.; Diez-Municio, M.; Forssten, S.; Hamaker, B.; Meynier, A.; Moreno, F.J.; Respondek, F.; Stahl, B.; Venema, K.; Wiese, M. Structure and function of non-digestible carbohydrates in the gut microbiome. Benef. Microbes 2022, 13, 95–168. [Google Scholar] [CrossRef]
- Chen, S.-J.; Chen, C.-C.; Liao, H.-Y.; Lin, Y.-T.; Wu, Y.-W.; Liou, J.-M.; Wu, M.-S.; Kuo, C.-H.; Lin, C.-H. Association of Fecal and Plasma Levels of Short-Chain Fatty Acids With Gut Microbiota and Clinical Severity in Patients With Parkinson Disease. Neurology 2022, 98, e848–e858. [Google Scholar] [CrossRef]
- Guo, Y.; Zou, J.; Xu, X.; Zhou, H.; Sun, X.; Wu, L.; Zhang, S.; Zhong, X.; Xiong, Z.; Lin, Y.; et al. Short-chain fatty acids combined with intronic DNA methylation of HIF3A: Potential predictors for diabetic cardiomyopathy. Clin. Nutr. 2021, 40, 3708–3717. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Y.F.; Miao, J.; Zheng, R.F.; Li, J.Y. Short-chain fatty acids: Important components of the gut-brain axis against AD. Biomed. Pharmacother. 2024, 175, 116601. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Chen, T.; Shi, L.; Wang, D.; Tang, D. Regulatory role of short-chain fatty acids in inflammatory bowel disease. Cell Commun. Signal 2022, 20, 64. [Google Scholar] [CrossRef]
- Pohl, K.; Moodley, P.; Dhanda, A. The effect of increasing intestinal short-chain fatty acid concentration on gut permeability and liver injury in the context of liver disease: A systematic review. J. Gastroenterol. Hepatol. 2022, 37, 1498–1506. [Google Scholar] [CrossRef] [PubMed]
- Dicks, L.M.T. How important are fatty acids in human health and can they be used in treating diseases? Gut Microbes 2024, 16, 2420765. [Google Scholar] [CrossRef]
- Michaudel, C.; Sokol, H. The Gut Microbiota at the Service of Immunometabolism. Cell Metab. 2020, 32, 514–523. [Google Scholar] [CrossRef]
- Woting, A.; Blaut, M. The Intestinal Microbiota in Metabolic Disease. Nutrients 2016, 8, 202. [Google Scholar] [CrossRef]
- Chadchan, S.B.; Popli, P.; Ambati, C.R.; Tycksen, E.; Han, S.J.; E Bulun, S.; Putluri, N.; Biest, S.W.; Kommagani, R. Gut microbiota-derived short-chain fatty acids protect against the progression of endometriosis. Life Sci. Alliance 2021, 4, e202101224. [Google Scholar] [CrossRef]
- Filippone, A.; Lanza, M.; Campolo, M.; Casili, G.; Paterniti, I.; Cuzzocrea, S.; Esposito, E. Protective effect of sodium propionate in Abeta(1-42)-induced neurotoxicity and spinal cord trauma. Neuropharmacology 2020, 166, 107977. [Google Scholar] [CrossRef] [PubMed]
- Mantzourani, C.; Kokotou, M.G. Liquid Chromatography-Mass Spectrometry (LC-MS) Derivatization-Based Methods for the Determination of Fatty Acids in Biological Samples. Molecules 2022, 27, 5717. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, Q.-L.; Huang, X.-W.; Ma, Z.-H.; Zheng, X.-L.; Li, S.-L.; Duan, H.-N.; Sun, X.-C.; Lin, F.-F.; Zhao, L.-J.; et al. A rapid and convenient derivatization method for quantitation of short-chain fatty acids in human feces by ultra-performance liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2020, 34, e8730. [Google Scholar] [CrossRef] [PubMed]
- Kanemitsu, Y.; Mishima, E.; Maekawa, M.; Matsumoto, Y.; Saigusa, D.; Yamaguchi, H.; Ogura, J.; Tsukamoto, H.; Tomioka, Y.; Abe, T.; et al. Comprehensive and semi-quantitative analysis of carboxyl-containing metabolites related to gut microbiota on chronic kidney disease using 2-picolylamine isotopic labeling LC-MS/MS. Sci. Rep. 2019, 9, 19075. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.-Y.; Wang, C.-Y.; Lee, C.-H.; Kao, H.-L.; Wu, W.-K.; Kuo, C.-H. Development of an Efficient and Sensitive Chemical Derivatization-Based LC-MS/MS Method for Quantifying Gut Microbiota-Derived Metabolites in Human Plasma and Its Application in Studying Cardiovascular Disease. J. Proteome Res. 2021, 20, 3508–3518. [Google Scholar] [CrossRef]
- Song, H.E.; Lee, H.Y.; Kim, S.J.; Back, S.H.; Yoo, H.J. A Facile Profiling Method of Short Chain Fatty Acids Using Liquid Chromatography-Mass Spectrometry. Metabolites 2019, 9, 173. [Google Scholar] [CrossRef] [PubMed]
- Cas, M.D.; Paroni, R.; Saccardo, A.; Casagni, E.; Arnoldi, S.; Gambaro, V.; Saresella, M.; Mario, C.; La Rosa, F.; Marventano, I.; et al. A straightforward LC-MS/MS analysis to study serum profile of short and medium chain fatty acids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2020, 1154, 121982. [Google Scholar]
- Jaochico, A.; Sangaraju, D.; Shahidi-Latham, S.K. A rapid derivatization based LC-MS/MS method for quantitation of short chain fatty acids in human plasma and urine. Bioanalysis 2019, 11, 741–753. [Google Scholar] [CrossRef]
- McKay, M.J.; Castaneda, M.; Catania, S.; Charles, K.A.; Shanahan, E.; Clarke, S.J.; Engel, A.; Varelis, P.; Molloy, M.P. Quantification of short-chain fatty acids in human stool samples by LC-MS/MS following derivatization with aniline analogues. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2023, 1217, 123618. [Google Scholar] [CrossRef]
- Zeng, M.; Cao, H. Fast quantification of short chain fatty acids and ketone bodies by liquid chromatography-tandem mass spectrometry after facile derivatization coupled with liquid-liquid extraction. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1083, 137–145. [Google Scholar] [CrossRef]
- Nicholson, S.E.; Merrill, D.; Zhu, C.; Burmeister, D.M.; Zou, Y.; Lai, Z.; Darlington, D.N.; Lewis, A.M.; Newton, L.; Scroggins, S.; et al. Polytrauma independent of therapeutic intervention alters the gastrointestinal microbiome. Am. J. Surg. 2018, 216, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wong, P.Y.; Wang, Q.; Wong, H.Y.; Huang, T.; Cui, C.; Zhang, N.; Cheung, W.H.; Wong, R.M.Y. Short-chain fatty acids enhance muscle mass and function through the activation of mTOR signalling pathways in sarcopenic mice. J. Cachexia Sarcopenia Muscle 2024, 15, 2387–2401. [Google Scholar] [CrossRef]
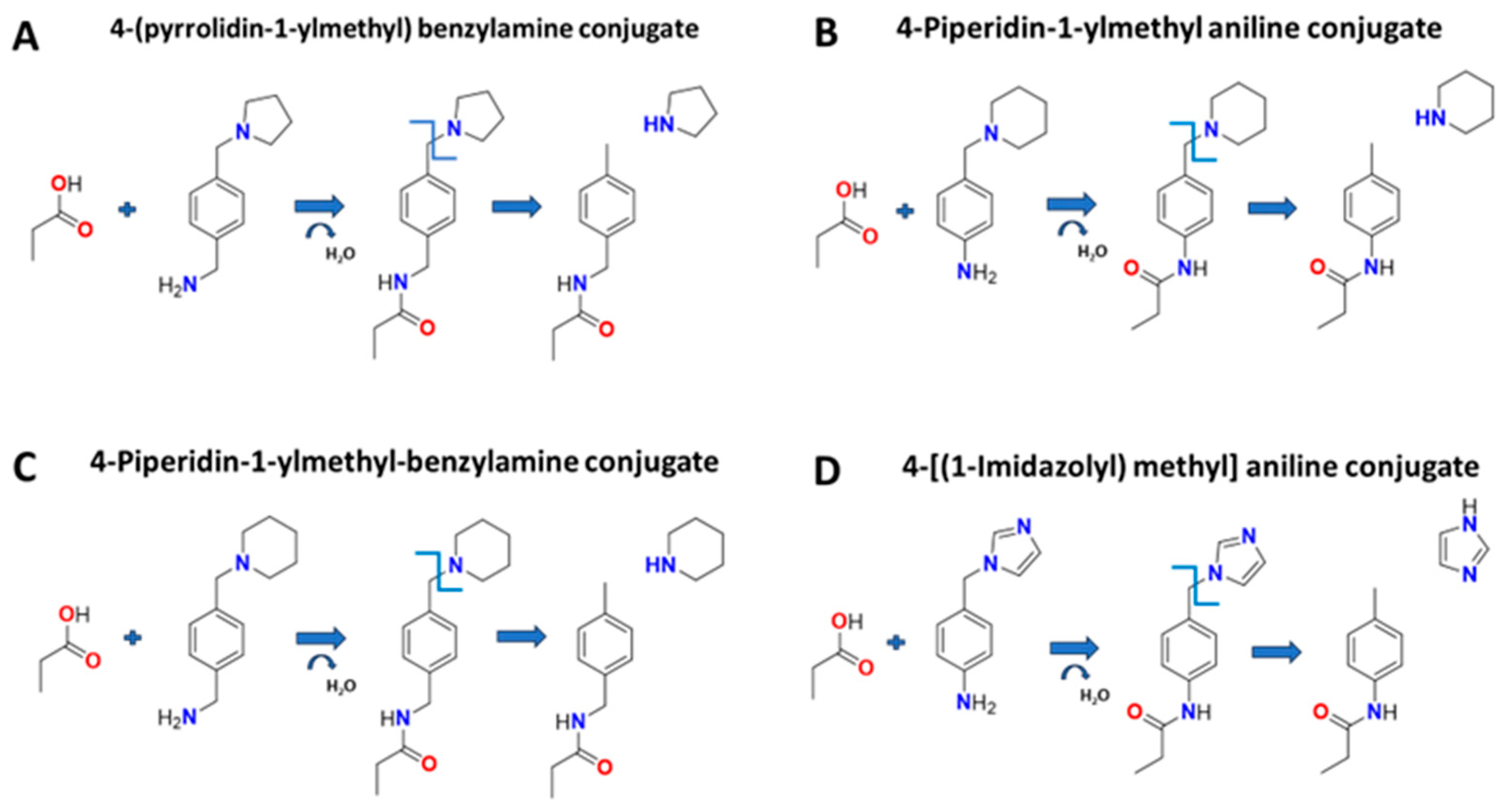
| Amino pyridines | Phenyl amines |
| 2-picolylamine | 3,4-dihyroxybenzylamine |
| 3-picolylamine | O-benzylhydroxylamine |
| Tyramines and tryptamines | Hydrazine |
| 3-methoxytyramine | 3-nitrophenyl hydrazine |
| tyramine | 2,4-dinitrophenyl hydrazine |
| normetanephrine | dansyl hydrazine |
| 5-hydroxy tryptamine (serotonin) | |
| 5-methoxy tryptamine | Phenyl ethyl amines |
| 5-benyloxy tryptamine | 4-methoxyphenethylamine |
| tryptamine | |
| Quinoline | |
| Benzyl Amines | 4-aminomethylquinoline |
| 4-(pyrrolidin-1-ylmethyl) benzylamine * | |
| 4-piperidin-1-ylmethyl-benzylamine * | Anilines |
| 4-(1H-Pyrrol-1-yl) benzylamine | 4-[(1-imidazolyl) methyl] aniline * |
| 3-(piperidin-1-yl) benzylamine | 4-piperidin-1-ylmethyl aniline * |
| 3-phenoxy-benzylamine | 4-(1H-imidazol-1-yl) aniline |
| 3-(pyridin-2-yloxy) benzylamine | 4-(phenylthio) aniline |
| 4-p-tolyloxy-benzylamine |
| (AUC) | Propionic | Butyric | Pentanoic | Hexanoic |
|---|---|---|---|---|
| 4PyBA | 8.44 ± 0.20 | 7.30 ± 0.26 | 4.48 ± 0.12 | 3.59 ± 0.03 |
| 4PpBA | 6.58 ± 0.06 | 6.50 ± 0.03 | 0.47 ± 0.09 | 2.93 ± 0.15 |
| 4PMA | 1.84 ±0.04 | 2.14 ± 0.03 | 1.17 ± 0.01 | 1.45 ± 0.01 |
| 4IMA | 1.02 ± 0.09 | 1.32 ± 0.11 | 0.62 ± 0.04 | 0.51 ± 0.01 |
| Precursor (m/z) | ||||
| 4PyBA | 247.1 | 261.2 | 275.2 | 289.2 |
| 4PpBA | 261.2 | 275.3 | 289.3 | 303.2 |
| 4PMA | 247.2 | 261.2 | 275.3 | 289.3 |
| 4IMA | 230.2 | 244.2 | 258.2 | 272.2 |
| Product (m/z) | ||||
| 4PyBA | 176.0 | 190.1 | 204.3 | 218.1 |
| 4PpBA | 176.1 | 190.4 | 204.0 | 218.1 |
| 4PMA | 162.1 | 176.0 | 190.0 | 204.1 |
| 4IMA | 162.1 | 176.0 | 190.1 | 204.1 |
| MW Analyte | MW Conjugate-Parent | MDW Conjugate-Daughter | |
|---|---|---|---|
| Propionic acid | 74 | 247 | 176 |
| Butyric acid and C4 isomers | 88 | 261 | 190 |
| Pentanoic acid and C5 isomers | 102 | 275 | 204 |
| Hexanoic acid and C6 isomers | 116 | 289 | 218 |
| Phenyl-propionic acid (2 isomers) | 150 | 323 | 254 |
| Phenyl butyric acid (3 isomers) | 164 | 337 | 266 |
| Phenyl pentanoic acid(4 isomers) | 178 | 351 | 280 |
| Phenyl hexanoic acid (2 isomers) | 192 | 365 | 294 |
| Linear Range (nM) | r2 | LOD (nM) | %CV 1 μM | %CV 100 nM | |
|---|---|---|---|---|---|
| C3 | |||||
| Propionic acid | 1–3333 | 0.9982 | 227 | 1.25 | 4.26 |
| 2 Phenyl Propionic acid | 1–3333 | 0.9994 | 69 | 2.09 | 6.87 |
| 3 Phenyl Propionic acid | 1–3333 | 0.9995 | 117 | 1.95 | 3.44 |
| C4 | |||||
| Butyric acid | 1–1000 | 0.9876 | 187 | 1.46 | 4.61 |
| 2 Methyl Propionic acid | 1–3333 | 0.9993 | 135 | 0.34 | 3.97 |
| 2/4 Phenyl Butyric acid | 1–1000 | 0.9994 | 40 | 2.18 | 6.62 |
| 3 Phenyl Butyric acid | 1–1000 | 0.9993 | 43 | 2.55 | 9.18 |
| C5 | |||||
| Pentanoic acid | 1–1000 | 0.9816 | 229 | 1.48 | 5.71 |
| 2 Methyl Butyric acid | 1–1000 | 0.9970 | 92 | 1.40 | 6.12 |
| 3 Methyl Butyric acid | 1–1000 | 0.9993 | 165 | 1.28 | 6.10 |
| 2 Phenyl Pentanoic acid | 1–1000 | 0.9992 | 46 | 2.34 | 3.96 |
| 3 Phenyl Pentanoic acid | 1–1000 | 0.9993 | 43 | 1.80 | 3.02 |
| 4 Phenyl Pentanoic acid | 1–1000 | 0.9994 | 40 | 2.07 | 4.12 |
| 5 Phenyl Pentanoic acid | 1–1000 | 0.9981 | 51 | 2.00 | 4.26 |
| C6 | |||||
| Hexanoic acid | 1–1000 | 0.9966 | 114 | 1.49 | 4.96 |
| 2 Methyl Pentanoic acid | 1–1000 | 0.9975 | 84 | 1.13 | 5.09 |
| 3 Methyl Pentanoic acid | 1–1000 | 0.9980 | 76 | 0.92 | 5.86 |
| 4 Methyl Pentanoic aci | 1–1000 | 0.9973 | 88 | 1.88 | 5.16 |
| 5 Phenyl Hexanoic acid | 1–1000 | 0.9986 | 63 | 4.79 | 4.33 |
| 6 Phenyl Hexanoic acid | 1–1000 | 0.9992 | 48 | 0.88 | 4.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darlington, D.N. Conjugation of Short-Chain Fatty Acids to Bicyclic-Amines for Analysis by Liquid Chromatography Tandem Mass Spectroscopy. Molecules 2025, 30, 341. https://doi.org/10.3390/molecules30020341
Darlington DN. Conjugation of Short-Chain Fatty Acids to Bicyclic-Amines for Analysis by Liquid Chromatography Tandem Mass Spectroscopy. Molecules. 2025; 30(2):341. https://doi.org/10.3390/molecules30020341
Chicago/Turabian StyleDarlington, Daniel N. 2025. "Conjugation of Short-Chain Fatty Acids to Bicyclic-Amines for Analysis by Liquid Chromatography Tandem Mass Spectroscopy" Molecules 30, no. 2: 341. https://doi.org/10.3390/molecules30020341
APA StyleDarlington, D. N. (2025). Conjugation of Short-Chain Fatty Acids to Bicyclic-Amines for Analysis by Liquid Chromatography Tandem Mass Spectroscopy. Molecules, 30(2), 341. https://doi.org/10.3390/molecules30020341






