Molecular Networking in Cosmetic Analysis: A Review of Non-Targeted Profiling for Safety Hazards and Bioactive Compounds
Abstract
1. Introduction
2. MS/MS-Based MN
2.1. Principles of MN
2.2. Development of MN
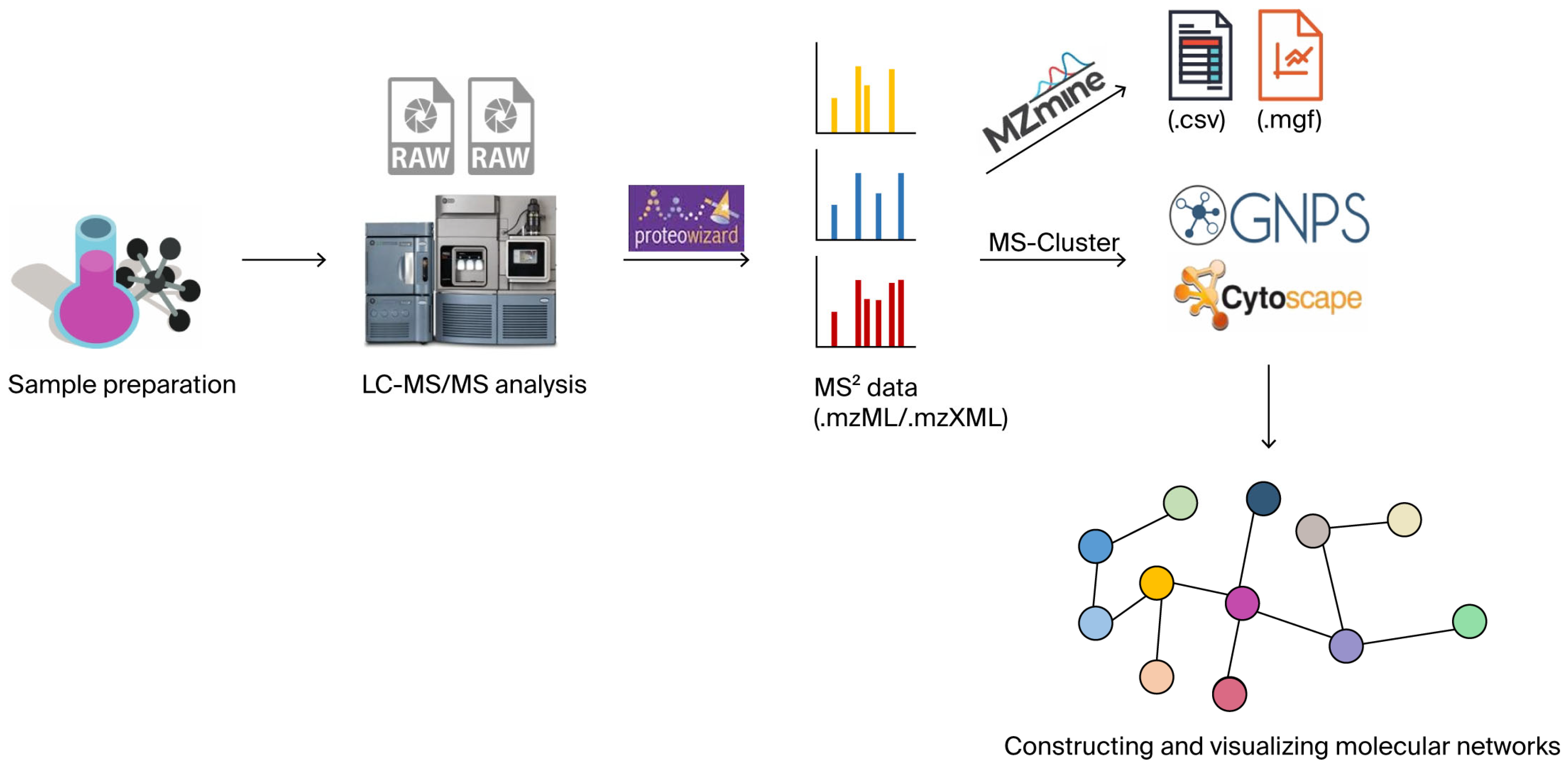
2.3. Molecular Network Construction Process
- (1)
- Sample preparation, including extraction and purification. Matrix interferences are removed using liquid–liquid extraction (LLE), a sample pretreatment method that is quick, easy, cheap, effective, rugged, and safe (QuEChERS), and solid-phase extraction (SPE), while quality control samples are prepared to ensure the reliability of the data.
- (2)
- MS/MS data acquisition in a data-dependent mode using LC-MS/MS or LC-HRMS with different collision energy gradients to cover compounds of varying stability.
- (3)
- Raw data are converted to mzML/mzXML formats using tools such as ProteoWizard v3.0.23246, then imported into MZmine 3, MS-DIAL 4, or equivalent platforms for chromatographic peak detection and peak list alignment. These operations yield a comprehensive feature table (.csv format) containing mass-to-charge ratios (m/z), retention times, peak areas, and cross-sample correlation metrics, alongside representative processed MS2 spectral files (.mgf format) [60].
- (4)
- The finalized feature table (.csv) and processed spectral files (.mgf) are then co-submitted to GNPS, where the platform autonomously constructs molecular networks based on MS2 spectral similarity thresholds.
- (5)
- After constructing the molecular network on GNPS, the graph file is exported (e.g., .graphml) and the analysis results are visualized using Cytoscape v3.10.2 software to refine network topology and annotate nodes/edges.
- (6)
- Using the GNPS data platform for molecular network analysis, structural analogs of known and undiscovered compounds are inferred and identified based on topological relationships between molecular nodes.
3. Computational Framework, Integration Challenges, and Future Perspectives of MN
3.1. Algorithmic Foundations of MN
3.2. Challenges and Strategies for Integrating Diverse Datasets
3.3. Integration Prospects with Deep Learning Methods
4. Applications of MN in Cosmetic Raw Material Exploration and Risk Substance Detection
4.1. Analysis of Naturally Active Ingredients
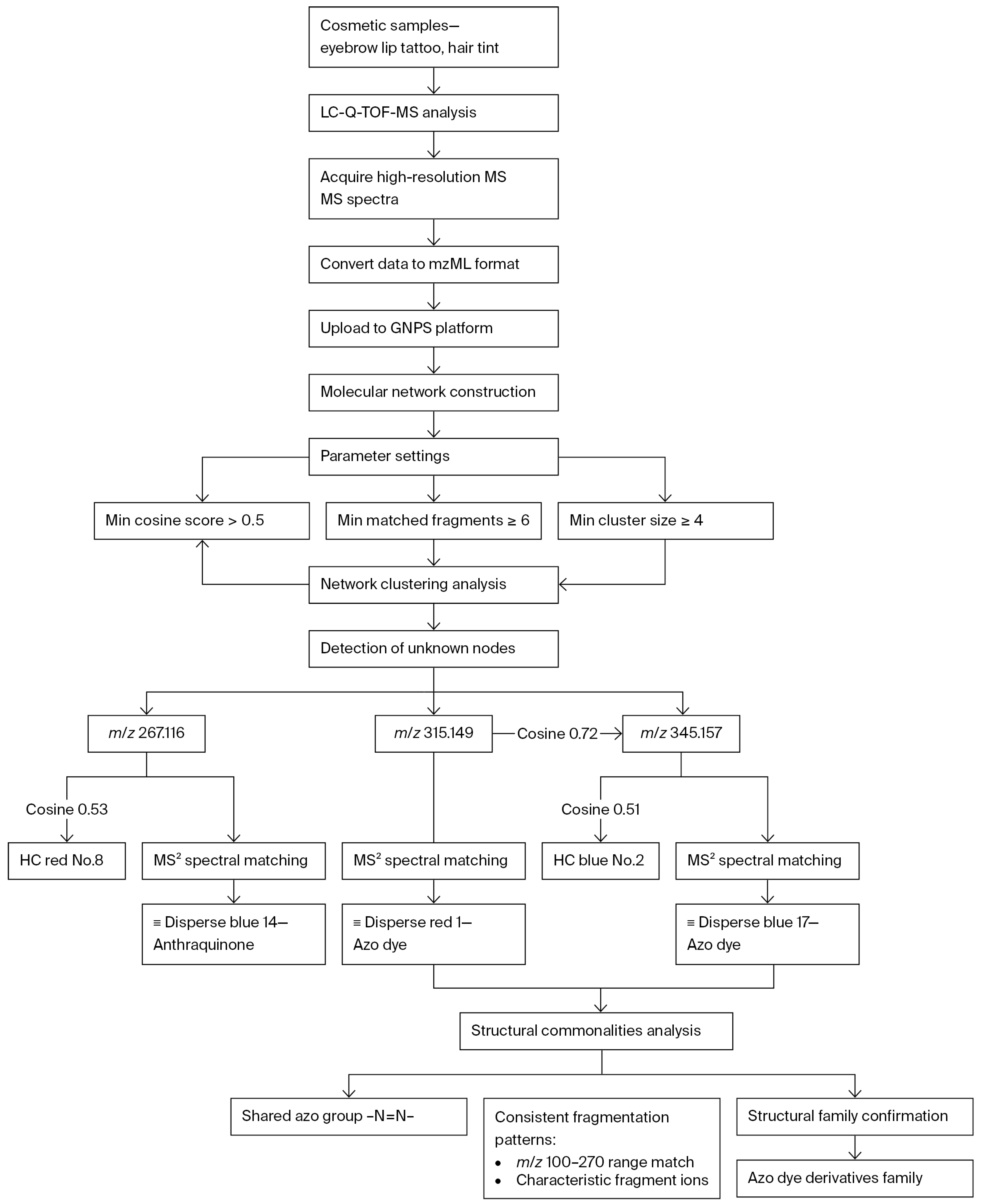
4.2. Identification of Prohibited Ingredients and Risk Substances
5. Challenges and Optimization Strategies
5.1. Challenges in MS Data Quality
5.2. Methodological Limitations of MN for Structurally Modified Adulterants
5.3. Matrix Interference in Cosmetic Analysis
5.4. Limitations of Spectral Databases in Cosmetic MN
- -
- Polymeric substances resulting from recent synthesis, utilized in formulations;
- -
- Specific natural products derived from rare plant species Existing databases frequently contain an insufficient amount of data regarding these compounds.
5.5. Optimization Strategies for MN in Cosmetic Analysis
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| BBMN | Blocks-based molecular network |
| BMN | Bioactive molecular network |
| CAS | Chemical Abstracts Service |
| CLMN | Classic molecular networking |
| DPPH | 2,2-Diphenyl-1-picrylhydrazyl |
| FBMN | Feature-based molecular networking |
| FRAP | Ferric reducing antioxidant power assay |
| GC-MS | Gas chromatography-mass spectrometry |
| GNPS | Global Natural Product Social Molecular Networking |
| HILIC | Hydrophilic interaction liquid chromatography |
| HPLC | High-performance liquid chromatography |
| HPLC-MS/MS | High-performance liquid chromatography–tandem mass spectrometry |
| HRESIMS | High-resolution electrospray ionization mass spectrometry |
| HRMS | High-resolution mass spectrometry |
| IIMN | Ion identity molecular networking |
| LC-MS/MS | Liquid chromatography–tandem mass spectrometry |
| LC-Q-TOF-MS | Liquid chromatography-quadrupole-time-of-flight-mass spectrometry |
| MeOH | Methanol |
| MN | Molecular networking |
| MSH | Melanocyte-stimulating hormone |
| MS/MS | Tandem mass spectrometry |
| MS1 | First-stage mass spectrometry |
| MS2 | Second-stage mass spectrometry |
| NIST | The National Institute of Standards and Technology |
| NMR | Nuclear magnetic resonance |
| ROS | Reactive oxygen species |
| QuEChERS | Quick, easy, cheap, effective, rugged and safe |
| TOF | Time of flight |
References
- Alves, A.; Sousa, E.; Kijjoa, A.; Pinto, M. Marine-derived compounds with potential use as cosmeceuticals and nutricosmetics. Molecules 2020, 25, 2536. [Google Scholar] [CrossRef]
- Ferrier, M.; Gémin, M.-P.; Malinowska, M.A.; Abdallah, C.; Magot, F.; Birer-Williams, C.; Giglioli-Guivarc’h, N.; Hano, C.; Lanoue, A. Metabolomics applications in natural cosmetics: Addressing the new challenges of bio-sourced ingredients. In Phytochemistry Reviews; Springer Nature: Berlin/Heidelberg, Germany, 2024. [Google Scholar] [CrossRef]
- Ekiert, H.; Klimek-Szczykutowicz, M.; Rzepiela, A.; Klin, P.; Szopa, A. Artemisia species with high biological values as a potential source of medicinal and cosmetic raw materials. Molecules 2022, 27, 6427. [Google Scholar] [CrossRef] [PubMed]
- Mondello, A.; Salomone, R.; Mondello, G. Exploring circular economy in the cosmetic industry: Insights from a literature review. Environ. Impact Assess. Rev. 2024, 105, 107443. [Google Scholar] [CrossRef]
- Nurkolis, F. Marine bioactives: Pioneering sustainable solutions for advanced cosmetics and therapeutics. Pharmacol. Res. 2025, 218, 107868. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Qu, H.; Ning, X.; Cui, S.; Cao, J. Utilizing cost-effective determination techniques to authenticate cosmetics. Appl. Sci. 2024, 14, 3198. [Google Scholar] [CrossRef]
- Chen, D.; Chen, Y.; Zhang, Y.; Du, J.; Xiao, H.; Yang, Z.; Xu, J. Multi-class analysis of 100 drug residues in cosmetics using high-performance liquid chromatography-quadrupole time-of-flight high-resolution mass spectrometry. Talanta 2024, 266, 124954. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, M.; Fu, X.; Qian, J.; Wang, M.; Tan, G. Novel hapten design, highly sensitive monoclonal antibody production, and immunoassay development for rapid screening of illegally added chloramphenicol in cosmetics. J. Immunol. Methods 2024, 525, 113604. [Google Scholar] [CrossRef]
- Jian, P.; Muhammad, T.; Wei, A.; Wu, B.; Zhou, T. A membrane-protected micro-solid-phase extraction method based on molecular imprinting and its application to the determination of local anesthetics in cosmetics. J. Sep. Sci. 2022, 45, 2675–2686. [Google Scholar] [CrossRef]
- Castiñeira-Landeira, A.; Gomez-Feas, A.; Carro, A.M.; Dagnac, T.; Almeida, P.J.; Llompart, M. Novel gas-diffusion microextraction followed by gas chromatography coupled to tandem mass spectrometry methodology for the determination of fragrance allergens in cosmetic products. Adv. Sample Prep. 2025, 14, 100187. [Google Scholar] [CrossRef]
- Lian, X.-H.; Wang, C.; Meng, X.-S.; Bai, H.; Sun, X.-J.; Xue, H.-Y.; Ma, Q. Determination of 10 kinds of caine-type prohibited ingredients in cosmetics by ultra-performance liquid chromatography-differential mobility spectrometry-mass spectrometry. Chin. J. Anal. Chem. 2019, 47, 756–764. [Google Scholar] [CrossRef]
- Yang, P.P.; Huang, W.; Li, L.X.; Liu, H. Determination of new glucocorticoid called clobetasol acetate in cosmetics by ultra performance liquid chromatography-tandem mass spectrometry. Chin. J. Chromatogr. 2023, 41, 250–256. [Google Scholar] [CrossRef]
- Tang, C.; Liang, W.; Xia, Z.; Ye, J.; Liang, H.; Cai, J.; Tan, J.; Xie, Q. Determination of polyfluoroalkyl substances in cosmetic products using dispersed liquid-liquid extraction coupled with ULC-MS/MS. Anal. Methods 2023, 15, 6727–6737. [Google Scholar] [CrossRef]
- Dalmaz, A.; Sivrikaya Özak, S. Rapid and eco-friendly microextraction procedure based on green hydrophobic deep eutectic solvent for simultaneous determination of five sex hormones from cosmetic samples. Microchem. J. 2024, 207, 111979. [Google Scholar] [CrossRef]
- Shang, Y.; Meng, X.; Liu, J.; Song, N.; Zheng, H.; Han, C.; Ma, Q. Applications of mass spectrometry in cosmetic analysis: An overview. J. Chromatogr. A 2023, 1705, 464175. [Google Scholar] [CrossRef] [PubMed]
- Serb, A.F.; Georgescu, M.; Onulov, R.; Novaconi, C.R.; Sisu, E.; Bolocan, A.; Sandu, R.E. Mass-spectrometry-based research of cosmetic ingredients. Molecules 2024, 29, 1336. [Google Scholar] [CrossRef] [PubMed]
- Celeiro, M.; Garcia-Jares, C.; Llompart, M.; Lores, M. Recent advances in sample preparation for cosmetics and personal care products analysis. Molecules 2021, 26, 4900. [Google Scholar] [CrossRef]
- Abedi, G.; Talebpour, Z.; Jamechenarboo, F. The survey of analytical methods for sample preparation and analysis of fragrances in cosmetics and personal care products. TrAC Trends Anal. Chem. 2018, 102, 41–59. [Google Scholar] [CrossRef]
- Sun, J.; Xue, G.X.; Gong, X.; Zhang, Z.P.; Xu, J.; Chen, L.; Cao, L.; Feng, Y.L.; Zhang, Y.J. Rapid determination of 54 dye components in hair dyes by liquid chromatography coupled to quadrupole orbitrap high-resolution mass spectrometry. Anal. Methods 2024, 16, 7341–7351. [Google Scholar] [CrossRef]
- Hu, B.; Li, L.; Ding, X.; Liu, H.; Huang, W.; Lü, W.; Li, X. Rapid determination of 87 prohibited ingredients in cosmetics by ultra performance liquid chromatography-tandem mass spectrometry. Chin. J. Chromatogr. 2024, 42, 38–51. [Google Scholar] [CrossRef]
- Jian, L.; Han, J.; Wen, H.; Shen, Y.; Zhang, K.; Yu, L.; Zheng, R.; Peng, X.; Zhao, L.; Sun, C. Rapid determination of 111 anti-infective drugs possibly added in cosmetics using high-performance liquid chromatography-tandem mass spectrometry with scheduled multiple reaction monitoring. Rapid Commun. Mass Spectrom. 2024, 38, e9778. [Google Scholar] [CrossRef]
- Rahman, M.S.; Yoshida, N.; Hanafusa, M.; Matsuo, A.; Zhu, S.; Stub, Y.; Takahashi, C.; Tsuboi, H.; Matsushita, R.; Maekawa, K.; et al. Screening and quantification of undeclared PGF(2α) analogs in eyelash-enhancing cosmetic serums using LC-MS/MS. J. Pharm. Biomed. Anal. 2022, 219, 114940. [Google Scholar] [CrossRef] [PubMed]
- Schettino, L.; García-Juan, A.; Fernández-Lozano, L.; Benedé, J.L.; Chisvert, A. Trace determination of prohibited acrylamide in cosmetic products by vortex-assisted reversed-phase dispersive liquid-liquid microextraction and liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2023, 1687, 463651. [Google Scholar] [CrossRef] [PubMed]
- Azorín, C.; Benedé, J.L.; Chisvert, A.; Salvador, A. Trace determination of tetrahydrocannabinol (THC) in cosmetic products by stir bar sorptive dispersive microextraction followed by liquid chromatography-tandem mass spectrometry. Talanta 2023, 253, 123934. [Google Scholar] [CrossRef] [PubMed]
- Han, K.-M.; Kim, Y.K.; Seo, S.; Kim, J.H.; Lee, J.H.; Kim, H.I.; Cho, S. Analysis of 13 banned colorants in cosmetics via liquid chromatographic and mass spectrometric techniques. Appl. Sci. 2023, 13, 5967. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wu, X.; Lu, Y. Characterization and determination of benvitimod, an unknown risk substance in cosmetics, using nuclear magnetic resonance spectroscopy and LC-MS/MS. J. Sep. Sci. 2022, 45, 3652–3662. [Google Scholar] [CrossRef]
- Bălan, S.A.; Bruton, T.A.; Harris, K.; Hayes, L.; Leonetti, C.P.; Mathrani, V.C.; Noble, A.E.; Phelps, D.S.C. The total mass of per- and polyfluoroalkyl substances (PFASs) in California cosmetics. Environ. Sci. Technol. 2024, 58, 12101–12112. [Google Scholar] [CrossRef]
- Lai, Y.H.; Wang, Y.S. Advances in high-resolution mass spectrometry techniques for analysis of high mass-to-charge ions. Mass Spectrom. Rev. 2023, 42, 2426–2445. [Google Scholar] [CrossRef]
- Shang, W.; Wei, G.; Li, H.; Zhao, G.; Wang, D. Advances in high-resolution mass spectrometry-based metabolomics: Applications in food analysis and biomarker discovery. J. Agric. Food Chem. 2025, 73, 3305–3325. [Google Scholar] [CrossRef]
- Aceña, J.; Stampachiacchiere, S.; Pérez, S.; Barceló, D. Advances in liquid chromatography-high-resolution mass spectrometry for quantitative and qualitative environmental analysis. Anal. Bioanal. Chem. 2015, 407, 6289–6299. [Google Scholar] [CrossRef]
- Guo, Z.; Huang, S.; Wang, J.; Feng, Y.L. Recent advances in non-targeted screening analysis using liquid chromatography-high resolution mass spectrometry to explore new biomarkers for human exposure. Talanta 2020, 219, 121339. [Google Scholar] [CrossRef]
- Szabo, D.; Falconer, T.M.; Fisher, C.M.; Heise, T.; Phillips, A.L.; Vas, G.; Williams, A.J.; Kruve, A. Online and offline prioritization of chemicals of interest in suspect screening and non-targeted screening with high-resolution mass spectrometry. Anal. Chem. 2024, 96, 3707–3716. [Google Scholar] [CrossRef]
- Mandal, V.; Ajabiya, J.; Khan, N.; Tekade, R.K.; Sengupta, P. Advances and challenges in non-targeted analysis: An insight into sample preparation and detection by liquid chromatography-mass spectrometry. J. Chromatogr. A 2024, 1737, 465459. [Google Scholar] [CrossRef] [PubMed]
- Quinn, R.A.; Nothias, L.F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular networking as a drug discovery, drug metabolism, and precision medicine strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Ollivier, S.; Fanuel, M.; Rogniaux, H.; Ropartz, D. Molecular networking of high-resolution tandem ion mobility spectra: A structurally relevant way of organizing data in glycomics? Anal. Chem. 2021, 93, 10871–10878. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.; Seo, H.; Kim, G.B.; Hong, J.; Yoo, H.H. MS-based molecular networking of designer drugs as an approach for the detection of unknown derivatives for forensic and doping applications: A case of NBOME derivatives. Anal. Chem. 2019, 91, 5483–5488. [Google Scholar] [CrossRef]
- Baskiyar, S.; Ren, C.; Heck, K.L.; Hall, A.M.; Gulfam, M.; Packer, S.; Seals, C.D.; Calderón, A.I. Bioactive natural products identification using automation of molecular networking software. J. Chem. Inf. Model. 2022, 62, 6378–6385. [Google Scholar] [CrossRef]
- Le Daré, B.; Ferron, P.J.; Allard, P.M.; Clément, B.; Morel, I.; Gicquel, T. New insights into quetiapine metabolism using molecular networking. Sci. Rep. 2020, 10, 19921. [Google Scholar] [CrossRef]
- Le Daré, B.; Allard, S.; Couette, A.; Allard, P.M.; Morel, I.; Gicquel, T. Comparison of illicit drug seizures products of natural origin using a molecular networking approach. Int. J. Toxicol. 2022, 41, 108–114. [Google Scholar] [CrossRef]
- Oberleitner, D.; Schmid, R.; Schulz, W.; Bergmann, A.; Achten, C. Feature-based molecular networking for identification of organic micropollutants including metabolites by non-target analysis applied to riverbank filtration. Anal. Bioanal. Chem. 2021, 413, 5291–5300. [Google Scholar] [CrossRef]
- Le Daré, B.; Ferron, P.J.; Couette, A.; Ribault, C.; Morel, I.; Gicquel, T. In vivo and in vitro α-amanitin metabolism studies using molecular networking. Toxicol. Lett. 2021, 346, 1–6. [Google Scholar] [CrossRef]
- Zhao, W.; Yan, T.; Huang, X.; Zhang, Y. Analysis of steroidal glycoalkaloids and their metabolites in Solanum nigrum fruits based on liquid chromatography-tandem mass spectrometry and molecular networking. J. Sep. Sci. 2023, 46, e2200804. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, J.; Mao, H.; You, W.; Chen, J.; Xu, H.; Wu, J.; Gong, Y.; Guo, L.; Liu, T.; et al. Structural annotation, semi-quantification and toxicity prediction of pyrrolizidine alkaloids from functional food: In silico and molecular networking strategy. Food Chem. Toxicol. 2023, 176, 113738. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Pang, X.; Gu, Z.; Guo, Z.; Xin, Y.; Zhang, L. Rapidly analyzing of ingredients during chewing and processing of areca nut using feature-based molecular networking. Food Chem. 2023, 410, 135205. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.R.; Wang, M.; Nothias, L.F.; van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef]
- Chung, H.H.; Kao, C.Y.; Wang, T.A.; Chu, J.; Pei, J.; Hsu, C.C. Reaction tracking and high-throughput screening of active compounds in combinatorial chemistry by tandem mass spectrometry molecular networking. Anal. Chem. 2021, 93, 2456–2463. [Google Scholar] [CrossRef]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Bandeira, N. Spectral networks: A new approach to de novo discovery of protein sequences and posttranslational modifications. Biotechniques 2007, 42, 687, 689, 691. [Google Scholar] [CrossRef]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar] [CrossRef]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Afoullouss, S.; Balsam, A.; Allcock, A.L.; Thomas, O.P. Optimization of LC-MS(2) data acquisition parameters for molecular networking applied to marine natural products. Metabolites 2022, 12, 245. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.F.; Zhang, X.; Zhu, F.; Huo, Z.Q.; Yao, Q.Q.; Feng, Q.; Liu, Z.; Zhang, G.M.; Yao, J.C.; Liang, H.B. MS/MS-based molecular networking: An efficient approach for natural products dereplication. Molecules 2022, 28, 157. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Qian, Y.; Fan, F.; Zhang, Z.; Zhang, Y.; Yu, Q.; Zhang, X.; Ren, H.; Geng, J.; Liu, H. Revealing specific transformation pattern of sulfonamides during wastewater biological treatment processes by molecular networking nontarget screening. Water Res. 2023, 235, 119895. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.; Petras, D.; Nothias, L.F.; Wang, M.; Aron, A.T.; Jagels, A.; Tsugawa, H.; Rainer, J.; Garcia-Aloy, M.; Dührkop, K.; et al. Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment. Nat. Commun. 2021, 12, 3832. [Google Scholar] [CrossRef]
- Nothias, L.F.; Nothias-Esposito, M.; da Silva, R.; Wang, M.; Protsyuk, I.; Zhang, Z.; Sarvepalli, A.; Leyssen, P.; Touboul, D.; Costa, J.; et al. Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J. Nat. Prod. 2018, 81, 758–767. [Google Scholar] [CrossRef]
- Tchetan, E.; Ortiz, S.; Hughes, K.; Olounladé, P.A.; Laurent, P.; Azando, E.V.B.; Herent, M.-F.; Hounzangbe-Adote, S.M.; Houinato, M.R.B.; Gbaguidi, F.A.; et al. HPLC-LTQ orbitrap mass spectrometry-based molecular networking for identifying anthelmintic molecules in Morinda lucida Benth. S. Afr. J. Bot. 2023, 161, 53–65. [Google Scholar] [CrossRef]
- He, Q.F.; Wu, Z.L.; Li, L.; Sun, W.Y.; Wang, G.Y.; Jiang, R.W.; Hu, L.J.; Shi, L.; He, R.R.; Wang, Y.; et al. Discovery of neuritogenic Securinega alkaloids from Flueggea suffruticosa by a building blocks-based molecular network strategy. Angew. Chem. Int. Ed. Engl. 2021, 60, 19609–19613. [Google Scholar] [CrossRef]
- Allard, P.M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.L. Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Huber, F.; Ridder, L.; Verhoeven, S.; Spaaks, J.H.; Diblen, F.; Rogers, S.; van der Hooft, J.J.J. Spec2Vec: Improved mass spectral similarity scoring through learning of structural relationships. PLoS Comput. Biol. 2021, 17, e1008724. [Google Scholar] [CrossRef]
- Huber, F.; van der Burg, S.; van der Hooft, J.J.J.; Ridder, L. MS2DeepScore: A novel deep learning similarity measure to compare tandem mass spectra. J. Cheminform. 2021, 13, 84. [Google Scholar] [CrossRef]
- Sheng, Y.; Xue, Y.; Wang, J.; Liu, S.; Jiang, Y. Fast screening and identification of illegal adulteration in dietary supplements and herbal medicines using molecular networking with deep-learning-based similarity algorithms. Anal. Bioanal. Chem. 2023, 415, 3285–3293. [Google Scholar] [CrossRef]
- Sheng, Y.; Xue, Y.; Wang, J.; Liu, S.; Jiang, Y. Nontargeted screening method for detection of illicit adulterants in dietary supplements and herbal medicines using UHPLC-QTOF-MS with fine-tuned Spec2Vec-based spectral similarity and chemical classification filter. J. Pharm. Biomed. Anal. 2024, 239, 115877. [Google Scholar] [CrossRef]
- Kowalczyk, S.; Grymel, M.; Bilik, J.; Kula, W.; Wawoczny, A.; Grymel, P.; Gillner, D. Selected plants as sources of natural and active ingredients for cosmetics of the future. Appl. Sci. 2024, 14, 3487. [Google Scholar] [CrossRef]
- Fonseca, S.; Amaral, M.N.; Reis, C.P.; Custódio, L. Marine natural products as innovative cosmetic ingredients. Mar. Drugs 2023, 21, 170. [Google Scholar] [CrossRef]
- Bouissane, L.; Elfardi, Y.; Khatib, S.; Fatimi, A.; Pereira, C.; Cruz-Martins, N. Medicinal plants and their derivatives for skin and hair: A Mediterranean perspective of women care. Arch. Dermatol. Res. 2025, 317, 710. [Google Scholar] [CrossRef] [PubMed]
- Babich, O.; Ivanova, S.; Bakhtiyarova, A.; Kalashnikova, O.; Sukhikh, S. Medicinal plants are the basis of natural cosmetics. Process Biochem. 2025, 154, 35–51. [Google Scholar] [CrossRef]
- Li, H.; Ding, X.; An, Q.; Li, W.; Guo, L.; Zheng, Y.; Zhang, D.; Huo, W. A strategy comprehensively and quickly identifies the herbal composition and chemical constituents in Yixishu Lotion by molecular networking. Biomed. Chromatogr. 2025, 39, e70069. [Google Scholar] [CrossRef]
- Hughes, K.; Ho, R.; Greff, S.; Herbette, G.; Filaire, E.; Ranouille, E.; Berthon, J.Y.; Raharivelomanana, P. Feature-based molecular networks identification of bioactive metabolites from three plants of the Polynesian Cosmetopoeia targeting the dermal papilla cells of the hair cycle. Molecules 2021, 27, 105. [Google Scholar] [CrossRef]
- Chambon, M.; Ho, R.; Baghdikian, B.; Herbette, G.; Bun-Llopet, S.S.; Garayev, E.; Raharivelomanana, P. Identification of antioxidant metabolites from five plants (Calophyllum inophyllum, Gardenia taitensis, Curcuma longa, Cordia subcordata, Ficus prolixa) of the Polynesian Pharmacopoeia and Cosmetopoeia for skin care. Antioxidants 2023, 12, 1870. [Google Scholar] [CrossRef]
- Kim, J.G.; Le, T.P.L.; Han, J.S.; Cho, Y.B.; Kwon, H.; Lee, D.; Lee, M.K.; Hwang, B.Y. Bioactive molecular network-guided discovery of dihydro-β-agarofurans from the fruits of Celastrus orbiculatus. Phytochemistry 2022, 203, 113349. [Google Scholar] [CrossRef]
- Kim, J.G.; Han, J.S.; Cho, Y.B.; An, B.K.; Lee, D.; Lee, M.K.; Hwang, B.Y. Molecular networking-guided isolation of melanogenesis inhibitory dihydro-β-agarofuran sesquiterpenoids from Celastrus orbiculatus. Phytochemistry 2025, 229, 114312. [Google Scholar] [CrossRef]
- Zwerger, M.J.; Hammerle, F.; Siewert, B.; Ganzera, M. Application of feature-based molecular networking in the field of algal research with special focus on mycosporine-like amino acids. J. Appl. Phycol. 2023, 35, 1377–1392. [Google Scholar] [CrossRef]
- Masoumifeshani, B.; Abedian Kenari, A.; Sottorff, I.; Crüsemann, M.; Amiri Moghaddam, J. Identification and evaluation of antioxidant and anti-aging peptide fractions from enzymatically hydrolyzed proteins of Spirulina platensis and Chlorella vulgaris. Mar. Drugs 2025, 23, 162. [Google Scholar] [CrossRef] [PubMed]
- Hegazi, N.M.; Salem, M.A.; Saad, H.H.; Aborehab, N.M.; El Bishbishy, M.H.; Ezzat, S.M. Untargeted metabolomics-based molecular networking for chemical characterization of selected Apiaceae fruit extracts in relation to their antioxidant and anti-cellulite potentials. Fitoterapia 2024, 173, 105782. [Google Scholar] [CrossRef] [PubMed]
- Buche, G.; Laffon, M.; Fougère, L.; Destandau, E. Evaluation and comparison of dermo-cosmetic activities of three oak species by targeting antioxidant metabolites and skin enzyme inhibitors. Metabolites 2023, 13, 804. [Google Scholar] [CrossRef] [PubMed]
- Son, S.R.; Kim, K.S.; Jang, D.S.; Lee, S. Caffeoylglucaric and caffeoylquinic acids from Inula japonica leaves and their anti-skin aging effects in TNF-α-induced normal human fibroblast damage. J. Agric. Food Chem. 2025, 73, 13471–13487. [Google Scholar] [CrossRef]
- Eysseric, E.; Beaudry, F.; Gagnon, C.; Segura, P.A. Non-targeted screening of trace organic contaminants in surface waters by a multi-tool approach based on combinatorial analysis of tandem mass spectra and open access databases. Talanta 2021, 230, 122293. [Google Scholar] [CrossRef]
- Casey, J.S.; Jackson, S.R.; Ryan, J.; Newton, S.R. The use of gas chromatography-high resolution mass spectrometry for suspect screening and non-targeted analysis of per- and polyfluoroalkyl substances. J. Chromatogr. A 2023, 1693, 463884. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, T.; Wang, Z.; Liang, W.; Wang, X.; Zhang, X.; Lu, X.; Liu, X.; Zhao, C.; Xu, G. High-resolution mass spectrometry-based suspect and nontarget screening of natural toxins in foodstuffs and risk assessment of dietary exposure. Environ. Pollut. 2025, 365, 125338. [Google Scholar] [CrossRef]
- Woo, I.S.; Kim, Y.K.; Kim, H.I.; Choi, J.D.; Han, K.M. Characterization of banned colorants in cosmetics: A tandem mass-based molecular networking approach. J. Chromatogr. A 2024, 1724, 464928. [Google Scholar] [CrossRef]
- Kim, Y.K.; Woo, I.S.; Park, C.G.; Kim, A.; Choi, J.D.; Son, K.H.; Han, K.M. Green extraction of prostaglandin analogs in cosmetics using deep eutectic solvents and detection via LC-MS/MS. J. Chromatogr. A 2025, 1739, 465516. [Google Scholar] [CrossRef]
| Technology | Core Principle | Advantages | Limitations | Ideal Applications |
|---|---|---|---|---|
| CLMN | Clusters compounds via direct comparison of MS2 spectral fragment ion similarity | Simple algorithm; rapid component grouping | Cannot resolve isomers; no quantitative capability; noise-sensitive (low-abundance ions) | Preliminary screening of mixtures |
| FBMN | Integrates RT, ion mobility, isotope patterns, and other features | Improved isomer resolution; enhanced reliability in complex matrices; semi-quantitative analysis | Dependent on LC-MS preprocessing software (e.g., MS-DIAL 4); large data volumes | Fine-scale analysis of complex systems (e.g., plant extracts) |
| IIMN | Correlates different adduct ions (e.g., [M+H]+/[M+Na]+) via chromatographic peak shape correlation | Resolves adduct splitting; enhances annotation propagation; detects ion–ligand complexes | Database-dependent; limited for novel modifications | Multi-adduct systems (e.g., metabolite profiling) |
| BMN | Maps bioactivity data (anti-inflammatory/antimicrobial) onto molecular networks to locate active clusters | Rapid identification of bioactive compounds; guides targeted isolation; links structure to function | Requires additional bioassays; fails with unclear mechanisms | Bioactive ingredient discovery (e.g., cosmetic actives) |
| BBMN | Integrates biosynthetic rules with MN for selective filtering of structural domains | High selectivity for novel scaffolds; simplifies complex datasets; provides visual guidance for new structures | Relies on biosynthetic rule libraries; may miss non-canonical metabolites | Novel scaffold discovery (e.g., microbial secondary metabolites) |
| No. | Study Subject | Identified Compounds | Cosmetic Efficacy | Role of MN in Screening/Identification | Ref. |
|---|---|---|---|---|---|
| 1 | Three Polynesian plants | Glycosylated flavonols, phenolic acids, C-flavonoids, iridoids, secoiridoids | Promoting dermal papilla cell proliferation (hair care) | BMN for identifying bioactive metabolites | [70] |
| 2 | Five Polynesian medicinal plants | Quercetin-O-rhamnoside, rosmarinic acid, curcumin (61 metabolites total) | Antioxidant (anti-photoaging) | LC-MS/MS with MN for identifying seven key phenolic radical scavengers | [71] |
| 3 | Celastrus orbiculatus fruits | 12 novel dihydro-β-agarofuran sesquiterpenes; 15 known compounds | Melanin inhibition (whitening) | BMN for discovering bioactive ingredients | [72,73] |
| 4 | Marine red algae | Mycosporine-like amino acids | UV protection and antioxidant (sunscreen) | UHPLC-HRMS with FBMN and GNPS workflow enabling high-throughput dereplication | [74] |
| 5 | Arthrospira platensis and Chlorella vulgaris | Lys-Val, Val-Arg, Tyr-Phe, Leu-Gly-Leu (8 di/tri-peptides) | Antioxidant and anti-aging | MS-based GNPS networking identifying key bioactive peptides | [75] |
| 6 | Nine Apiaceae fruits | Apigenin and derivatives | Antioxidant and lipogenesis inhibition (anti-cellulite) | UPLC-HRMS with MN for activity screening | [76] |
| 7 | Three French oak extracts | Quercetin derivatives, ellagic acid, procyanidin B2, condensed tannins, flavonol glycosides | Collagenase inhibition and ROS scavenging (anti-aging) | UHPLC-HRMS with MN for activity polyphenol screening | [77] |
| 8 | Inula japonica leaf extract | Inujaponics A-C, caffeoylquinic acids, caffeoylglucuronic acids | MMP-1 inhibition and collagen synthesis (anti-aging) | LC-MS with MN for identifying caffeoylglucaric and caffeoylquinic acids | [78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Li, S.; Wang, J.-S.; Wu, D.; Xu, G.-Q.; Wang, H.-Y. Molecular Networking in Cosmetic Analysis: A Review of Non-Targeted Profiling for Safety Hazards and Bioactive Compounds. Molecules 2025, 30, 3968. https://doi.org/10.3390/molecules30193968
Li L, Li S, Wang J-S, Wu D, Xu G-Q, Wang H-Y. Molecular Networking in Cosmetic Analysis: A Review of Non-Targeted Profiling for Safety Hazards and Bioactive Compounds. Molecules. 2025; 30(19):3968. https://doi.org/10.3390/molecules30193968
Chicago/Turabian StyleLi, Li, Shuo Li, Ji-Shuang Wang, Di Wu, Guang-Qian Xu, and Hai-Yan Wang. 2025. "Molecular Networking in Cosmetic Analysis: A Review of Non-Targeted Profiling for Safety Hazards and Bioactive Compounds" Molecules 30, no. 19: 3968. https://doi.org/10.3390/molecules30193968
APA StyleLi, L., Li, S., Wang, J.-S., Wu, D., Xu, G.-Q., & Wang, H.-Y. (2025). Molecular Networking in Cosmetic Analysis: A Review of Non-Targeted Profiling for Safety Hazards and Bioactive Compounds. Molecules, 30(19), 3968. https://doi.org/10.3390/molecules30193968








