Synthesis of First Copper Metal Complex of C=C Extended Curcuminoid Analogue: Structure, β-Cyclodextrin Association, and Biological Properties
Abstract
1. Introduction
2. Results
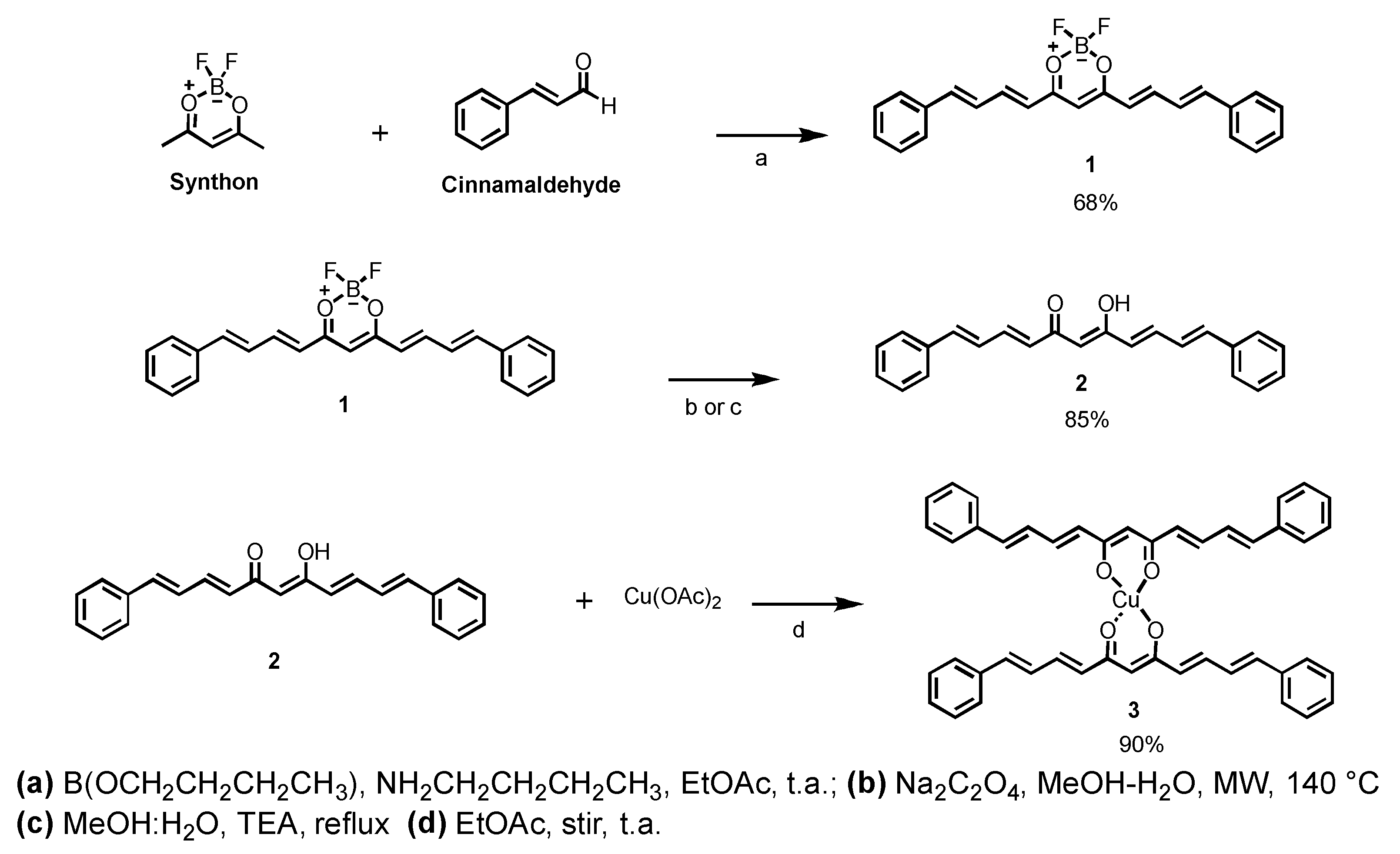
2.1. Synthesis
2.2. Nuclear Magnetic Resonance
2.3. Electronic Paramagnetic Resonance
2.4. Mass Spectrometry
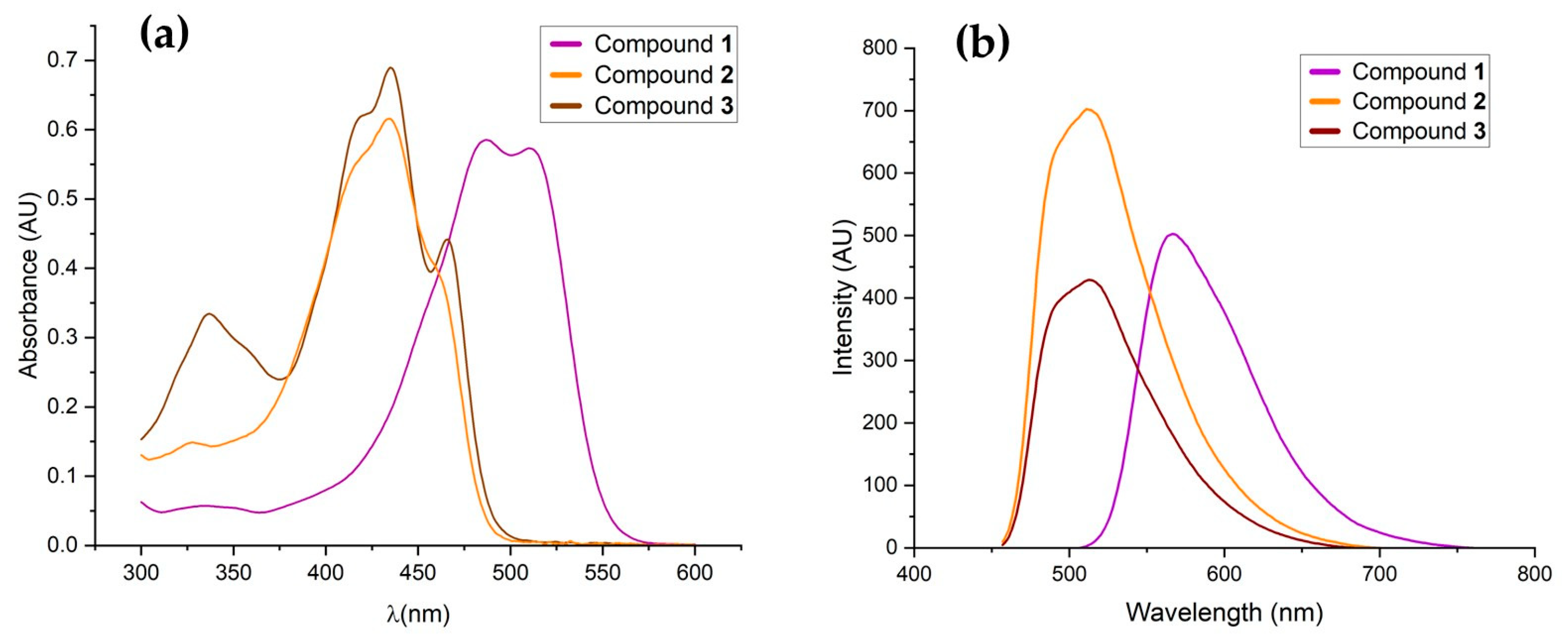
2.5. Optical Properties
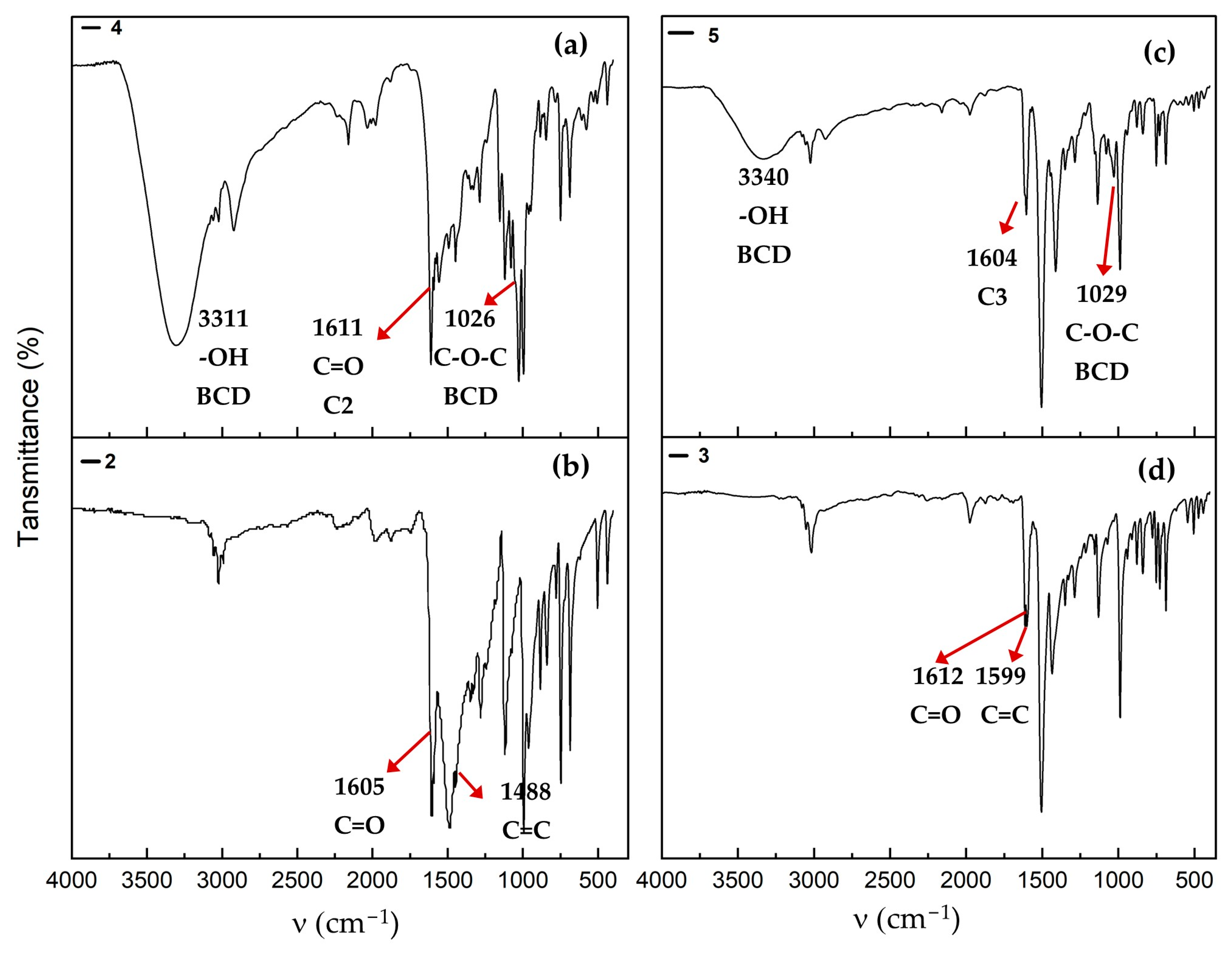
2.6. Infrared-FT Spectroscopy
2.7. Single-Crystal X-Ray Diffraction
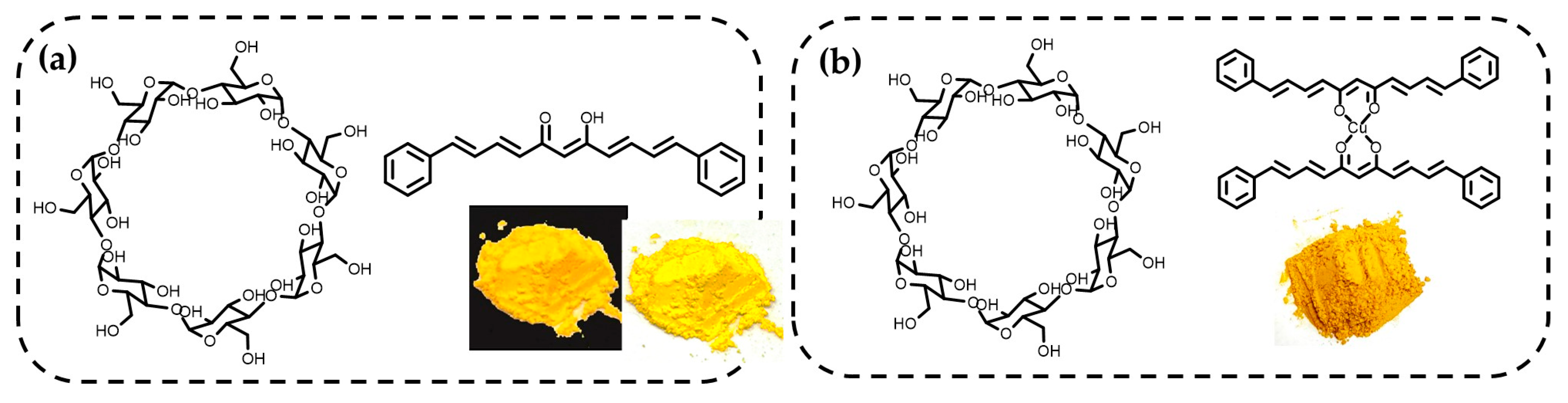
2.8. Improvement of Water Solubility Through Association with β-Cyclodextrin (BCD) Formation and Biological Activity Evaluation
2.8.1. Nuclear Magnetic Resonance of the Association Complexes with BCD
2.8.2. UV-Vis and Infrared Spectroscopy of Association Complexes with BCD
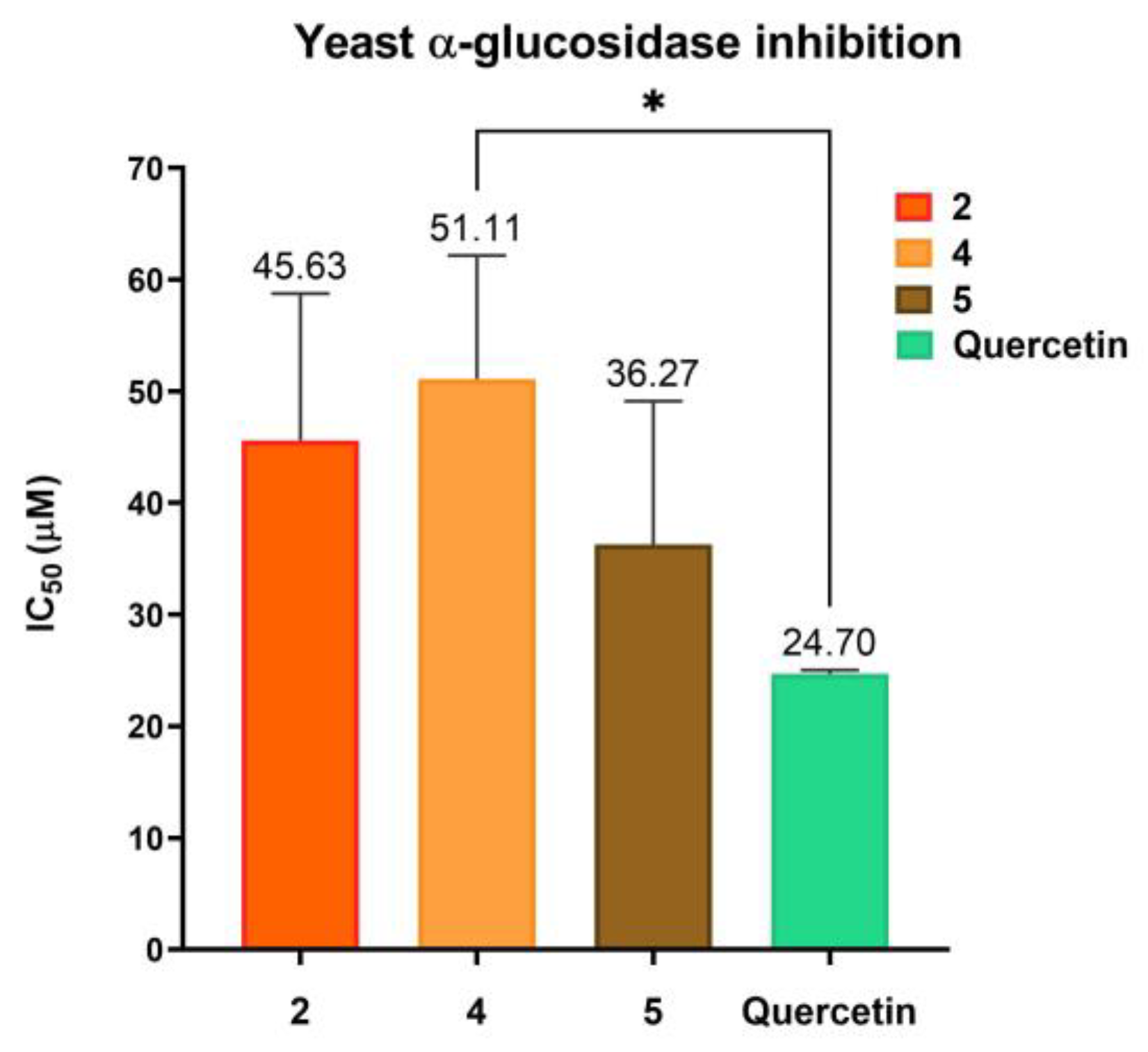
2.9. Biological Activity
3. Discussion
4. Materials and Methods
4.1. Analytical Determinations
4.2. Synthetic Procedures
4.3. Preparation of Association Complexes with β-Cyclodextrin
4.4. Antiproliferative Activity
4.5. Inhibition of Yeast α-Glucosidase
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NOESY | Nuclear Overhauser Effect Spectroscopy |
| NMR | Nuclear magnetic resonance |
| FT-IR | Fourier Transform Infrared spectroscopy |
| UV-Vis | Ultraviolet–Visible spectroscopy |
| BDC | β-Cyclodextrin |
| ORTEP | Oak Ridge Thermal Ellipsoid Plot |
| RMS | Root Mean Square |
| CCDC | Cambridge Crystallographic Data Centre |
| MS | Mass Spectrometry |
References
- Mir, R.H.; Mohi-ud-din, R.; Mir, P.A.; Shah, A.J.; Banday, N.; Sabreen, S.; Maqbool, M.; Jan, R.; Shafi, N.; Masoodi, M.H. Chapter 9—Curcumin as a Privileged Scaffold Molecule for Various Biological Targets in Drug Development. In Studies in Natural Products Chemistry; Atta-ur-Rahman, Ed.; Elsevier: Amsterdam, The Netherlands, 2022; Volume 73, pp. 405–434. ISBN 1572-5995. [Google Scholar]
- Yang, H.; Zeng, F.; Luo, Y.; Zheng, C.; Ran, C.; Yang, J. Curcumin Scaffold as a Multifunctional Tool for Alzheimer’s Disease Research. Molecules 2022, 27, 3879. [Google Scholar] [CrossRef]
- Li, S.; Yuan, W.; Deng, G.; Wang, P.; Yang, P.; Aggarwal, B.B. Chemical Composition and Product Quality Control of Turmeric (Curcuma longa L.). Pharm. Crops 2011, 5, 28–54. [Google Scholar] [CrossRef]
- Inoue, K.; Nomura, C.; Ito, S.; Nagatsu, A.; Hino, T.; Oka, H. Purification of Curcumin, Demethoxycurcumin, and Bisdemethoxycurcumin by High-Speed Countercurrent Chromatography. J. Agric. Food Chem. 2008, 56, 9328–9336. [Google Scholar] [CrossRef]
- Huang, C.; Lu, H.-F.; Chen, Y.-H.; Chen, J.-C.; Chou, W.-H.; Huang, H.-C. Curcumin, Demethoxycurcumin, and Bisdemethoxycurcumin Induced Caspase-Dependent and –Independent Apoptosis via Smad or Akt Signaling Pathways in HOS Cells. BMC Complement. Med. Ther. 2020, 20, 68. [Google Scholar] [CrossRef]
- Ramsewak, R.S.; DeWitt, D.L.; Nair, M.G. Cytotoxicity, Antioxidant and Anti-Inflammatory Activities of Curcumins I–III from Curcuma Longa. Phytomedicine 2000, 7, 303–308. [Google Scholar] [CrossRef]
- Guo, L.Y.; Cai, X.F.; Lee, J.J.; Kang, S.S.; Shin, E.M.; Zhou, H.Y.; Jung, J.W.; Kim, Y.S. Comparison of Suppressive Effects of Demethoxycurcumin and Bisdemethoxycurcumin on Expressions of Inflammatory Mediators In Vitro and In Vivo. Arch. Pharm. Res. 2008, 31, 490–496. [Google Scholar] [CrossRef]
- Hung, S.-J.; Hong, Y.-A.; Lin, K.-Y.; Hua, Y.-W.; Kuo, C.-J.; Hu, A.; Shih, T.-L.; Chen, H.-P. Efficient Photodynamic Killing of Gram-Positive Bacteria by Synthetic Curcuminoids. Int. J. Mol. Sci. 2020, 21, 9024. [Google Scholar] [CrossRef] [PubMed]
- Den Hartogh, D.J.; Gabriel, A.; Tsiani, E. Antidiabetic Properties of Curcumin II: Evidence from In Vivo Studies. Nutrients 2019, 12, 58. [Google Scholar] [CrossRef] [PubMed]
- Amalraj, A.; Pius, A.; Gopi, S.; Gopi, S. Biological Activities of Curcuminoids, Other Biomolecules from Turmeric and Their Derivatives—A Review. J. Tradit. Complement. Med. 2017, 7, 205–233. [Google Scholar] [CrossRef]
- Zhang, H.A.; Kitts, D.D. Turmeric and Its Bioactive Constituents Trigger Cell Signaling Mechanisms That Protect Against Diabetes and Cardiovascular Diseases. Mol. Cell. Biochem. 2021, 476, 3785–3814. [Google Scholar] [CrossRef]
- Bhagat, K.K.; Cheke, R.S.; Gavali, V.D.; Kharkar, P.S.; Arote, N.D. A Brief Review on Metal-Curcumin Complexes: Synthesis Approaches and Their Pharmaceutical Applications. Discov. Chem. 2025, 2, 119. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Pan, H.; Li, L.; Yu, Y.; Liu, B. An Insight into the In Vivo Imaging Potential of Curcumin Analogues as Fluorescence Probes. Asian J. Pharm. Sci. 2021, 16, 419–431. [Google Scholar] [CrossRef]
- Bai, G.; Yu, C.; Cheng, C.; Hao, E.; Wei, Y.; Mu, X.; Jiao, L. Syntheses and Photophysical Properties of BF2 Complexes of Curcumin Analogues. Org. Biomol. Chem. 2014, 12, 1618–1626. [Google Scholar] [CrossRef]
- Xu, G.; Wang, J.; Liu, T.; Wang, M.; Zhou, S.; Wu, B.; Jiang, M. Synthesis and Crystal Structure of a Novel Copper (II) Complex of Curcumin-Type and Its Application in In Vitro and In Vivo Imaging. J. Mater. Chem. B 2014, 2, 3659–3666. [Google Scholar] [CrossRef] [PubMed]
- Festa, R.A.; Thiele, D.J. Copper: An Essential Metal in Biology. Curr. Biol. 2011, 21, R877–R883. [Google Scholar] [CrossRef]
- Esmaeili, L.; Perez, M.G.; Jafari, M.; Paquin, J.; Ispas-Szabo, P.; Pop, V.; Andruh, M.; Byers, J.; Mateescu, M.A. Copper Complexes for Biomedical Applications: Structural Insights, Antioxidant Activity and Neuron Compatibility. J. Inorg. Biochem. 2019, 192, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, J.; Riaz, M.A. Biological Potential of Copper Complexes: A Review. Turk. J. Chem. 2022, 46, 595–623. [Google Scholar] [CrossRef]
- Rani, J.J.; Roy, S. Recent Development of Copper (II) Complexes of Polypyridyl Ligands in Chemotherapy and Photodynamic Therapy. ChemMedChem 2023, 18, e202200652. [Google Scholar] [CrossRef] [PubMed]
- Mani, A.; Feng, T.; Gandioso, A.; Vinck, R.; Notaro, A.; Gourdon, L.; Burckel, P.; Saubaméa, B.; Blacque, O.; Cariou, K.; et al. Structurally Simple Osmium(II) Polypyridyl Complexes as Photosensitizers for Photodynamic Therapy in the Near Infrared. Angew. Chem. 2023, 135, e202218347. [Google Scholar] [CrossRef]
- Shoair, A.F.; El-Bindary, A.A.; El-Ghamaz, N.A.; Rezk, G.N. Synthesis, Characterization, DNA Binding and Antitumor Activities of Cu(II) Complexes. J. Mol. Liq. 2018, 269, 619–638. [Google Scholar] [CrossRef]
- Choroba, K.; Machura, B.; Erfurt, K.; Casimiro, A.R.; Cordeiro, S.; Baptista, P.V.; Fernandes, A.R. Copper(II) Complexes with 2,2′:6′,2″-Terpyridine Derivatives Displaying Dimeric Dichloro−μ–Bridged Crystal Structure: Biological Activities from 2D and 3D Tumor Spheroids to In Vivo Models. J. Med. Chem. 2024, 67, 5813–5836. [Google Scholar] [CrossRef]
- Ngece, K.; Khwaza, V.; Paca, A.M.; Aderibigbe, B.A. The Antimicrobial Efficacy of Copper Complexes: A Review. Antibiotics 2025, 14, 516. [Google Scholar] [CrossRef] [PubMed]
- Meza-Morales, W.; Machado-Rodriguez, J.C.; Alvarez-Ricardo, Y.; Obregón-Mendoza, M.A.; Nieto-Camacho, A.; Toscano, R.A.; Soriano-García, M.; Cassani, J.; Enríquez, R.G. A New Family of Homoleptic Copper Complexes of Curcuminoids: Synthesis, Characterization and Biological Properties. Molecules 2019, 24, 910. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, M.; Binaeizadeh, M.R.; Iraji, A.; Larijani, B.; Saeedi, M.; Mahdavi, M. A Review on α-Glucosidase Inhibitory Activity of First Row Transition Metal Complexes: A Futuristic Strategy for Treatment of Type 2 Diabetes. RSC Adv. 2022, 12, 12011–12052. [Google Scholar] [CrossRef] [PubMed]
- Dirir, A.M.; Daou, M.; Yousef, A.F.; Yousef, L.F. A Review of Alpha-Glucosidase Inhibitors from Plants as Potential Candidates for the Treatment of Type-2 Diabetes. Phytochem. Rev. 2022, 21, 1049–1079. [Google Scholar] [CrossRef]
- Gong, L.; Feng, D.; Wang, T.; Ren, Y.; Liu, Y.; Wang, J. Inhibitors of A-amylase and A-glucosidase: Potential Linkage for Whole Cereal Foods on Prevention of Hyperglycemia. Food Sci. Nutr. 2020, 8, 6320–6337. [Google Scholar] [CrossRef]
- Du, Z.; Liu, R.; Shao, W.; Mao, X.; Ma, L.; Gu, L.; Huang, Z.; Chan, A.S.C. α-Glucosidase Inhibition of Natural Curcuminoids and Curcumin Analogs. Eur. J. Med. Chem. 2006, 41, 213–218. [Google Scholar] [CrossRef]
- Mehrabi, M.; Esmaeili, S.; Ezati, M.; Abassi, M.; Rasouli, H.; Nazari, D.; Adibi, H.; Khodarahmi, R. Antioxidant and Glycohydrolase Inhibitory Behavior of Curcumin-Based Compounds: Synthesis and Evaluation of Anti-Diabetic Properties In Vitro. Bioorg. Chem. 2021, 110, 104720. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, J.; Yu, J.; Chen, X.; Zhang, S.; Cai, Y.; Li, L. Curcumin as a Mild Natural A-glucosidase Inhibitor: A Study on Its Mechanism In Vitro. Int. J. Food Sci. Technol. 2022, 57, 2689–2700. [Google Scholar] [CrossRef]
- Arenaza-Corona, A.; Obregón-Mendoza, M.A.; Meza-Morales, W.; Ramírez-Apan, M.T.; Nieto-Camacho, A.; Toscano, R.A.; Pérez-González, L.L.; Sánchez-Obregón, R.; Enríquez, R.G. The Homoleptic Curcumin–Copper Single Crystal (ML2): A Long Awaited Breakthrough in the Field of Curcumin Metal Complexes. Molecules 2023, 28, 6033. [Google Scholar] [CrossRef]
- Cai, L.; Du, H.; Wang, D.; Lyu, H.; Wang, D. Synthesis and Photophysical Properties of Ditrifluoroacetoxyboron Complexes with Curcumin Analogues. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 249, 119297. [Google Scholar] [CrossRef]
- Lyu, H.; Wang, D.; Cai, L.; Wang, D.-J.; Li, X.-M. Synthesis, Photophysical and Solvatochromic Properties of Diacetoxyboron Complexes with Curcumin Derivatives. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 220, 117126. [Google Scholar] [CrossRef]
- Tahay, P.; Parsa, Z.; Zamani, P.; Safari, N. A Structural and Optical Study of Curcumin and Curcumin Analogs. J. Iran. Chem. Soc. 2022, 19, 3177–3188. [Google Scholar] [CrossRef]
- Kazantzis, K.T.; Koutsonikoli, K.; Mavroidi, B.; Zachariadis, M.; Alexiou, P.; Pelecanou, M.; Politopoulos, K.; Alexandratou, E.; Sagnou, M. Curcumin Derivatives as Photosensitizers in Photodynamic Therapy: Photophysical Properties and In Vitro Studies with Prostate Cancer Cells. Photochem. Photobiol. Sci. 2020, 19, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Urošević, M.; Nikolić, L.; Gajić, I.; Nikolić, V.; Dinić, A.; Miljković, V. Curcumin: Biological Activities and Modern Pharmaceutical Forms. Antibiotics 2022, 11, 135. [Google Scholar] [CrossRef]
- Tønnesen, H.H.; Másson, M.; Loftsson, T. Studies of Curcumin and Curcuminoids. XXVII. Cyclodextrin Complexation: Solubility, Chemical and Photochemical Stability. Int. J. Pharm. 2002, 244, 127–135. [Google Scholar] [CrossRef]
- Arruda, T.R.; Marques, C.S.; Soares, N.F.F. Native Cyclodextrins and Their Derivatives as Potential Additives for Food Packaging: A Review. Polysaccharides 2021, 2, 825–842. [Google Scholar] [CrossRef]
- Del Valle, E.M.M. Cyclodextrins and Their Uses: A Review. Process Biochem. 2004, 39, 1033–1046. [Google Scholar] [CrossRef]
- Obregón-Mendoza, M.A.; Meza-Morales, W.; Alvarez-Ricardo, Y.; Estévez-Carmona, M.M.; Enríquez, R.G. High Yield Synthesis of Curcumin and Symmetric Curcuminoids: A “Click” and “Unclick” Chemistry Approach. Molecules 2022, 28, 289. [Google Scholar] [CrossRef]
- Weiss, H.; Reichel, J.; Görls, H.; Schneider, K.R.A.; Micheel, M.; Pröhl, M.; Gottschaldt, M.; Dietzek, B.; Weigand, W. Curcuminoid–BF2 Complexes: Synthesis, Fluorescence and Optimization of BF2 Group Cleavage. Beilstein J. Org. Chem. 2017, 13, 2264–2272. [Google Scholar] [CrossRef]
- Bakun, P.; Kucinska, M.; Kobyłka, P.; Kuźmińska, J.; Koczorowski, T.; Mlynarczyk, D.T.; Popenda, L.; Górska, K.; Kasperkowiak, M.; Murias, M.; et al. Morpholinated Curcuminoids against Urinary Bladder Cancer Cells: Synthesis and Anticancer Evaluation. Med. Chem. Res. 2024, 33, 944–963. [Google Scholar] [CrossRef]
- Karunakaran, C.; Balamurugan, M.; Karthikeyan, M. Applications of Electron Paramagnetic Resonance. In Spin Resonance Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2018; pp. 281–347. ISBN 978-0-12-813608-9. [Google Scholar]
- Łabanowska, M.; Bidzińska, E.; Para, A.; Kurdziel, M. EPR Investigation of Cu(II)-Complexes with Nitrogen Derivatives of Dialdehyde Starch. Carbohydr. Polym. 2012, 87, 2605–2613. [Google Scholar] [CrossRef]
- Barik, A.; Mishra, B.; Shen, L.; Mohan, H.; Kadam, R.M.; Dutta, S.; Zhang, H.-Y.; Priyadarsini, K.I. Evaluation of a New Copper(II)–Curcumin Complex as Superoxide Dismutase Mimic and Its Free Radical Reactions. Free Radic. Biol. Med. 2005, 39, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Gál, E.; Nagy, L.C. Photophysical Properties and Electronic Structure of Symmetrical Curcumin Analogues and Their BF2 Complexes, Including a Phenothiazine Substituted Derivative. Symmetry 2021, 13, 2299. [Google Scholar] [CrossRef]
- Feddaoui, I.; Abdelbaky, M.S.M.; García-Granda, S.; Essalah, K.; Ben Nasr, C.; Mrad, M.L. Synthesis, Crystal Structure, Vibrational Spectroscopy, DFT, Optical Study and Thermal Analysis of a New Stannate(IV) Complex Based on 2-Ethyl-6-Methylanilinium (C9H14N)2[SnCl6]. J. Mol. Struct. 2019, 1186, 31–38. [Google Scholar] [CrossRef]
- Li, S.; Bu, R.; Gou, R.; Zhang, C. Hirshfeld Surface Method and Its Application in Energetic Crystals. Cryst. Growth Des. 2021, 21, 6619–6634. [Google Scholar] [CrossRef]
- Surface Properties|CrystalExplorer. Available online: https://crystalexplorer.net/docs/manual/isosurfaces/properties/ (accessed on 17 July 2025).
- Schneider, H.-J.; Hacket, F.; Rüdiger, V.; Ikeda, H. NMR Studies of Cyclodextrins and Cyclodextrin Complexes. Chem. Rev. 1998, 98, 1755–1786. [Google Scholar] [CrossRef]
- Han, W.; You, J.; Li, H.; Zhao, D.; Nie, J.; Wang, T. Curcuminoid-Based Difluoroboron Dyes as High-Performance Photosensitizers in Long-Wavelength (Yellow and Red) Cationic Photopolymerization. Macromol. Rapid Commun. 2019, 40, 1900291. [Google Scholar] [CrossRef]
- Kaur, K.; Al-Khazaleh, A.K.; Bhuyan, D.J.; Li, F.; Li, C.G. A Review of Recent Curcumin Analogues and Their Antioxidant, Anti-Inflammatory, and Anticancer Activities. Antioxidants 2024, 13, 1092. [Google Scholar] [CrossRef]
- Kuzminska, J.; Szyk, P.; Mlynarczyk, D.T.; Bakun, P.; Muszalska-Kolos, I.; Dettlaff, K.; Sobczak, A.; Goslinski, T.; Jelinska, A. Curcumin Derivatives in Medicinal Chemistry: Potential Applications in Cancer Treatment. Molecules 2024, 29, 5321. [Google Scholar] [CrossRef] [PubMed]
- Sobornova, V.V.; Belov, K.V.; Krestyaninov, M.A.; Khodov, I.A. Influence of Solvent Polarity on the Conformer Ratio of Bicalutamide in Saturated Solutions: Insights from NOESY NMR Analysis and Quantum-Chemical Calculations. Int. J. Mol. Sci. 2024, 25, 8254. [Google Scholar] [CrossRef] [PubMed]
- Leow, P.-C.; Bahety, P.; Boon, C.P.; Lee, C.Y.; Tan, K.L.; Yang, T.; Ee, P.-L.R. Functionalized Curcumin Analogs as Potent Modulators of the Wnt/β-Catenin Signaling Pathway. Eur. J. Med. Chem. 2014, 71, 67–80. [Google Scholar] [CrossRef]
- Lutskii, A.E.; Kotelevskii, N.M.; Osipov, O.A.; Zamaraev, K.I. Effect of Solvent on EPR Spectra of Cu(II) Complexes. Theor. Exp. Chem. 1971, 4, 296–300. [Google Scholar] [CrossRef]
- Kamada, K.; Namikawa, T.; Senatore, S.; Matthews, C.; Lenne, P.; Maury, O.; Andraud, C.; Ponce-Vargas, M.; Le Guennic, B.; Jacquemin, D.; et al. Boron Difluoride Curcuminoid Fluorophores with Enhanced Two-Photon Excited Fluorescence Emission and Versatile Living-Cell Imaging Properties. Chem.-Eur. J. 2016, 22, 5219–5232. [Google Scholar] [CrossRef]
- Ferreira, J.R.M.; Alves, M.; Sousa, B.; Vieira, S.I.; Silva, A.M.S.; Guieu, S.; Cunha, Â.; Nunes da Silva, R. Curcumin-Based Molecular Probes for Fluorescence Imaging of Fungi. Org. Biomol. Chem. 2023, 21, 1531–1536. [Google Scholar] [CrossRef]
- Bessho, T.; Constable, E.C.; Graetzel, M.; Hernandez Redondo, A.; Housecroft, C.E.; Kylberg, W.; Nazeeruddin, M.K.; Neuburger, M.; Schaffner, S. An Element of Surprise—Efficient Copper-Functionalized Dye-Sensitized Solar Cells. Chem. Commun. 2008, 32, 3717–3719. [Google Scholar] [CrossRef] [PubMed]
- Abbo, H.; Ashfaq, M.; Feizi-Dehnayebi, M.; Titinchi, S. Asymmetrical Curcumin Derivative: Synthesis, Structural Exploration, Hirshfeld Surface Analysis, and Computational Study. Struct. Chem. 2025, 36, 1855–1870. [Google Scholar] [CrossRef]
- Meza-Morales, W.; Alvarez-Ricardo, Y.; Obregón-Mendoza, M.A.; Arenaza-Corona, A.; Ramírez-Apan, M.T.; Toscano, R.A.; Poveda-Jaramillo, J.C.; Enríquez, R.G. Three New Coordination Geometries of Homoleptic Zn Complexes of Curcuminoids and Their High Antiproliferative Potential. RSC Adv. 2023, 13, 8577–8585. [Google Scholar] [CrossRef]
- Zhao, R.; Tan, T.; Sandström, C. NMR Studies on Puerarin and Its Interaction with Beta-Cyclodextrin. J. Biol. Phys. 2011, 37, 387–400. [Google Scholar] [CrossRef]
- Maheshwari, A.; Sharma, M.; Sharma, D. Complexation of Sodium Picosulphate with Beta Cyclodextrin: NMR Spectroscopic Study in Solution. J. Incl. Phenom. Macrocycl. Chem. 2013, 77, 337–342. [Google Scholar] [CrossRef]
- Jiang, H.; Sun, H.; Zhang, S.; Hua, R.; Xu, Y.; Jin, S.; Gong, H.; Li, L. NMR Investigations of Inclusion Complexes between β-Cyclodextrin and Naphthalene/Anthraquinone Derivatives. J. Incl. Phenom. Macrocycl. Chem. 2007, 58, 133–138. [Google Scholar] [CrossRef]
- Saha, S.; Roy, A.; Roy, K.; Roy, M.N. Study to Explore the Mechanism to Form Inclusion Complexes of β-Cyclodextrin with Vitamin Molecules. Sci. Rep. 2016, 6, 35764. [Google Scholar] [CrossRef] [PubMed]
- Rachmawati, H.; Edityaningrum, C.A.; Mauludin, R. Molecular Inclusion Complex of Curcumin–β-Cyclodextrin Nanoparticle to Enhance Curcumin Skin Permeability from Hydrophilic Matrix Gel. AAPS PharmSciTech 2013, 14, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Sambasevam, K.; Mohamad, S.; Sarih, N.; Ismail, N. Synthesis and Characterization of the Inclusion Complex of β-Cyclodextrin and Azomethine. Int. J. Mol. Sci. 2013, 14, 3671–3682. [Google Scholar] [CrossRef]
- Yuan, H.; Peng, Q.; Yongwei, W.; Dongsheng, Z.; Mingwan, Z.; Rui, L.; Nan, J. Host-Guest Interaction of β-Cyclodextrin with Isomeric Ursolicacid and Oleanolic Acid: Physicochemical Characterization Andmolecular Modeling Study. J. Biomed. Res. 2017, 31, 395. [Google Scholar] [CrossRef]
- Alizadeh, N.; Poorbagher, N. Host-Guest Inclusion Complexes of Sulfabenzamide with β- and Methyl-β-Cyclodextrins: Characterization, Antioxidant Activity and DFT Calculation. J. Mol. Struct. 2022, 1260, 132809. [Google Scholar] [CrossRef]
- Nguyen, A.-T.; Pham, M.Q.; Nguyen, P.-H.; To, D.C.; Dang, N.Q.; Nguyen, T.-H.; Nguyen, H.-T.; Nguyen, T.-D.; Thi Pham, K.-H.; Tran, M.-H. Identification of Natural Curcumins as Potential Dual Inhibitors of PTP1B and α-Glucosidase through Experimental and Computational Study. Kuwait J. Sci. 2025, 52, 100312. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Hirata, R.; Yasui, H.; Hattori, M.; Sakurai, H. Inhibitory Effect of CuSO4 on α-Glucosidase Activity in ddY Mice. Metallomics 2010, 2, 67–73. [Google Scholar] [CrossRef]
- Malik, N.P.; Ashiq, U.; Jamal, R.A.; Gul, S.; Lateef, M. Design, Synthesis, In Vitro and In Silico Alpha Glucosidase and Lipoxygenase Inhibition Studies of Copper(II) Oxamide Complexes. ChemistrySelect 2024, 9, e202400398. [Google Scholar] [CrossRef]
- Peng, X.; Liu, K.; Hu, X.; Gong, D.; Zhang, G. Hesperetin-Cu(II) Complex as Potential α-Amylase and α-Glucosidase Inhibitor: Inhibition Mechanism and Molecular Docking. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 290, 122301. [Google Scholar] [CrossRef]
- Mestrelab Research S.L. MestReNova®, Version 2020 v14.2.0; Mestrelab Research S.L.: Santiago de Compostela, Spain, 2020.
- Sheldrick, G.M. A Short History of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From Visualization to Analysis, Design and Prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A Program for Hirshfeld Surface Analysis, Visualization and Quantitative Analysis of Molecular Crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- Laali, K.K.; Rathman, B.M.; Bunge, S.D.; Qi, X.; Borosky, G.L. Fluoro-Curcuminoids and Curcuminoid-BF2 Adducts: Synthesis, X-Ray Structures, Bioassay, and Computational/Docking Study. J. Fluor. Chem. 2016, 191, 29–41. [Google Scholar] [CrossRef]
- Abonia, R.; Laali, K.K.; Raja Somu, D.; Bunge, S.D.; Wang, E.C. A Flexible Strategy for Modular Synthesis of Curcuminoid-BF2/Curcuminoid Pairs and Their Comparative Antiproliferative Activity in Human Cancer Cell Lines. ChemMedChem 2020, 15, 354–362. [Google Scholar] [CrossRef]
- Zhou, T.; Zhang, S.; Liu, S.; Cong, H.; Xuan, L. Daphnodorin Dimers from Edgeworthia Chrysantha with α-Glucosidase Inhibitory Activity. Phytochem. Lett. 2010, 3, 242–247. [Google Scholar] [CrossRef]
- Ye, X.-P.; Song, C.-Q.; Yuan, P.; Mao, R.-G. α-Glucosidase and α-Amylase Inhibitory Activity of Common Constituents from Traditional Chinese Medicine Used for Diabetes Mellitus. Chin. J. Nat. Med. 2010, 8, 349–352. [Google Scholar] [CrossRef]
- Abarca, R.L.; Rodríguez, F.J.; Guarda, A.; Galotto, M.J.; Bruna, J.E. Characterization of Beta-Cyclodextrin Inclusion Complexes Containing an Essential Oil Component. Food Chem. 2016, 196, 968–975. [Google Scholar] [CrossRef]
- Kim, J.-S. Study of Flavonoid/Hydroxypropyl-β-Cyclodextrin Inclusion Complexes by UV-Vis, FT-IR, DSC, and X-Ray Diffraction Analysis. Prev. Nutr. Food Sci. 2020, 25, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhou, T.-P.; Zou, W.; Wang, Y.; Wang, K.; Yang, Y.; Liu, C.; Tu, Z.; Liu, Q.; Yuan, Y. Formation of β-Cyclodextrin Inclusion Complexes with a Series of Structurally Related Parabens: Preparation, Physicochemical Characterization and Antifungal Properties. Carbohydr. Polym. Technol. Appl. 2025, 11, 100924. [Google Scholar] [CrossRef]
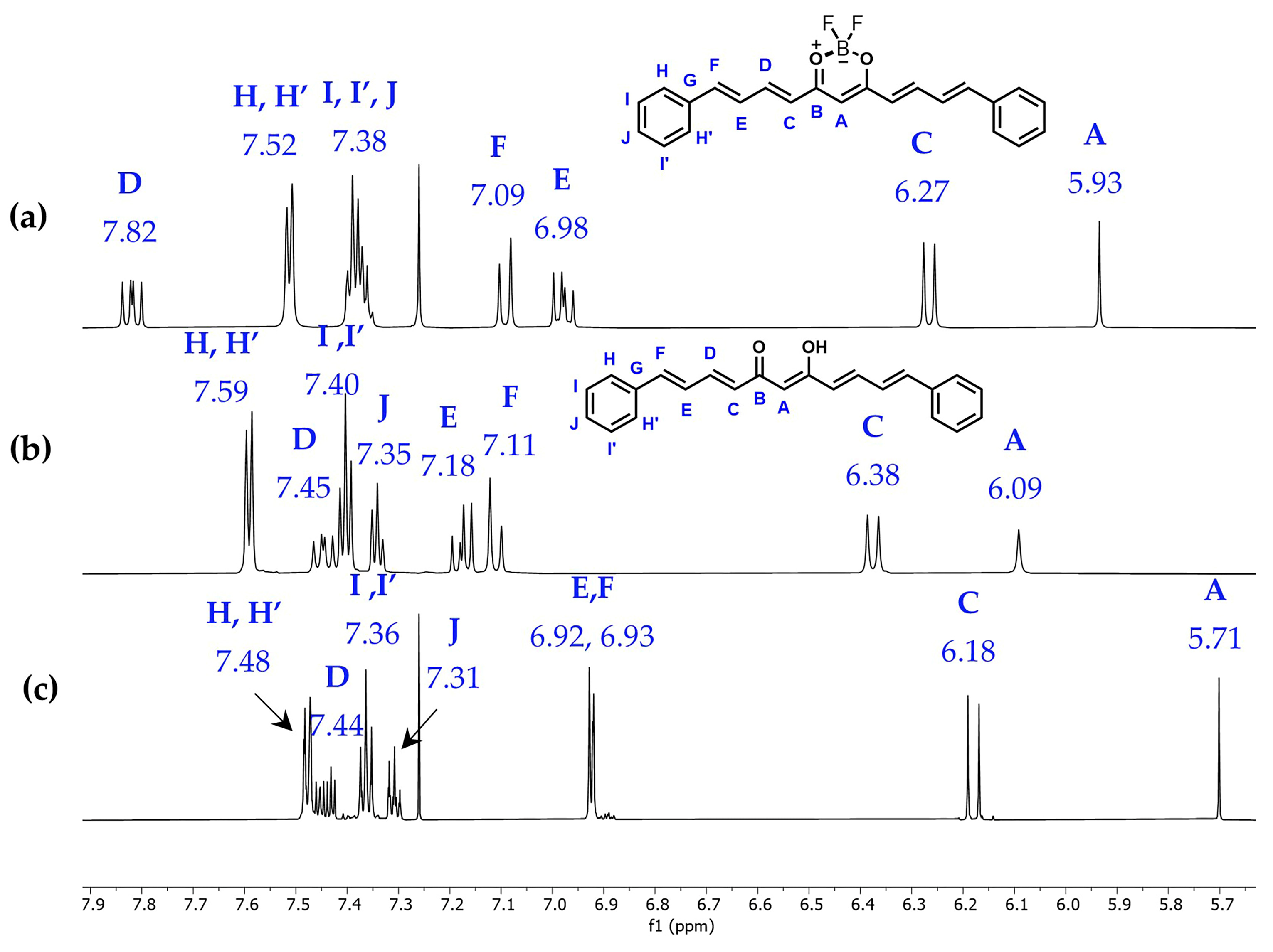


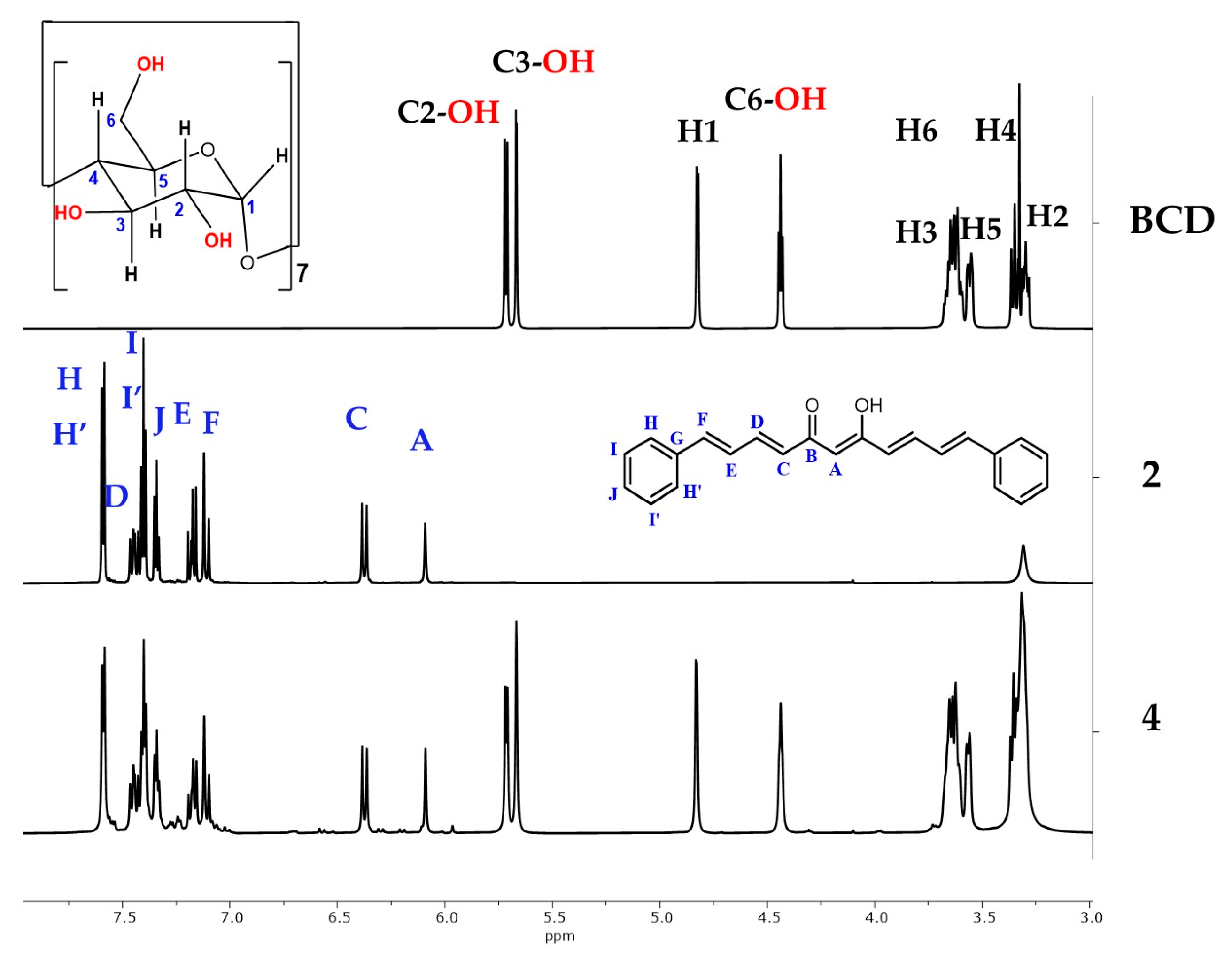
| Compound 1 (CDCl3) | Compound 2 (DMSO-d6) | Compound 2 (CDCl3) | ||||
|---|---|---|---|---|---|---|
| 13C (δ, ppm) | 1H (δ, ppm) | 13C (δ, ppm) | 1H (δ, ppm) | 13C (δ, ppm) | 1H (δ, ppm) | |
| A | 102.22 | 5.93 | 101.34 | 6.09 (s) | 101.87 | 5.71 (s) |
| B | 179.59 | 182.88 | 183.24 | |||
| C | 124.22 | 6.27 | 127.74 | 6.38 (d) | 127.87 | 6.18 (d) |
| D | 147.66 | 7.82 | 141.00 | 7.45 (dd) | 140.88 | 7.44 (m) |
| E | 126.64 | 6.98 | 127.36 | 7.18 (dd) | 127.22 | 6.92 (d) * |
| F | 145.13 | 7.09 | 140.34 | 7.11 (d) | 140.38 | 6.93 (d) * |
| G | 135.8 | 136.10 | 136.44 | |||
| H, H’ | 128.01 | 7.52 | 127.25 | 7.59 (m) *** | 127.22 | 7.48 (m) |
| I, I’ | 129.14 | 7.38 | 128.87 | 7.40 (m) ** | 128.97 | 7.36 (m) |
| J | 130.27 | 7.38 | 129.06 | 7.34 (m) ** | 129.15 | 7.31 (m) |
| -OH | 16.06 (br, s) | 15.85 (br, s) | ||||
| Compound | Absorption λ (nm) | ε (M−1·cm−1) [log ε] | Emission λ (nm) |
|---|---|---|---|
| 1 | 487 | 75,372.75 [4.9] | 567.01 |
| 2 | 434 | 63,305.95 [4.8] | 511.04 |
| 3 | 435 | 120,156.79 [5.1] | 512.98 |
| Compound | 1 | 2 | 3 |
|---|---|---|---|
| CCDC (deposition number) | 2,483,477 | 2,483,476 | 2,483,475 |
| Empirical formula | C23H19BF2O2 | C27H28O3 | C46H38CuO4 |
| Formula weight | 376.19 | 400.49 | 718.30 |
| Crystal system | Triclinic | Orthorhombic | Monoclinic |
| Space group | P-1 | Cmc21 | C2/c |
| Unit cell dimensions | |||
| a (Å) | a = 9.1069 (6) | a = 28.5258 (9) | a = 41.1114 (11) |
| b (Å) | b = 10.0955 (7) | b = 9.7389 (3) | b = 5.56190 (10) |
| c (Å) | c = 10.7715 (7) | c = 7.6957 (2) | c = 16.1765 (4) |
| α (°) | α = 74.270 (2) | α = 90 | α = 90 |
| β (°) | β = 85.586 (2) | β = 90 | β = 100.2810 (10) |
| γ (°) | γ = 85.457 (2) | γ = 90 | γ = 90 |
| Volume (Å3) | 948.65 (11) | 2137.94 (11) | 3639.49 (15) |
| Z | 2 | 4 | 4 |
| Density (calculate) (mg/m3) | 1.317 | 1.244 | 1.311 |
| Absorption coefficient (mm−1) | 0.095 | 0.080 | 0.644 |
| F (000) | 392 | 856 | 1500 |
| Crystal size (mm3) | 0.291 × 0.239 × 0.178 | 0.400 × 0.280 × 0.200 | 0.346 × 0.106 × 0.089 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavera-Hernández, R.; Sánchez-Obregón, R.; Obregón-Mendoza, M.A.; Nieto-Camacho, A.; Ramírez-Apan, M.T.; Pérez-González, L.L.; Enríquez, R.G. Synthesis of First Copper Metal Complex of C=C Extended Curcuminoid Analogue: Structure, β-Cyclodextrin Association, and Biological Properties. Molecules 2025, 30, 3943. https://doi.org/10.3390/molecules30193943
Tavera-Hernández R, Sánchez-Obregón R, Obregón-Mendoza MA, Nieto-Camacho A, Ramírez-Apan MT, Pérez-González LL, Enríquez RG. Synthesis of First Copper Metal Complex of C=C Extended Curcuminoid Analogue: Structure, β-Cyclodextrin Association, and Biological Properties. Molecules. 2025; 30(19):3943. https://doi.org/10.3390/molecules30193943
Chicago/Turabian StyleTavera-Hernández, Rosario, Rubén Sánchez-Obregón, Marco A. Obregón-Mendoza, Antonio Nieto-Camacho, María Teresa Ramírez-Apan, Leidys L. Pérez-González, and Raúl G. Enríquez. 2025. "Synthesis of First Copper Metal Complex of C=C Extended Curcuminoid Analogue: Structure, β-Cyclodextrin Association, and Biological Properties" Molecules 30, no. 19: 3943. https://doi.org/10.3390/molecules30193943
APA StyleTavera-Hernández, R., Sánchez-Obregón, R., Obregón-Mendoza, M. A., Nieto-Camacho, A., Ramírez-Apan, M. T., Pérez-González, L. L., & Enríquez, R. G. (2025). Synthesis of First Copper Metal Complex of C=C Extended Curcuminoid Analogue: Structure, β-Cyclodextrin Association, and Biological Properties. Molecules, 30(19), 3943. https://doi.org/10.3390/molecules30193943







