Precision Target Discovery for Migraine: An Integrated GWAS-eQTL-PheWAS Pipeline
Abstract
1. Introduction
2. Results
2.1. SMR Main Analysis
2.2. Identification and Profiles of Screened Druggable Genes
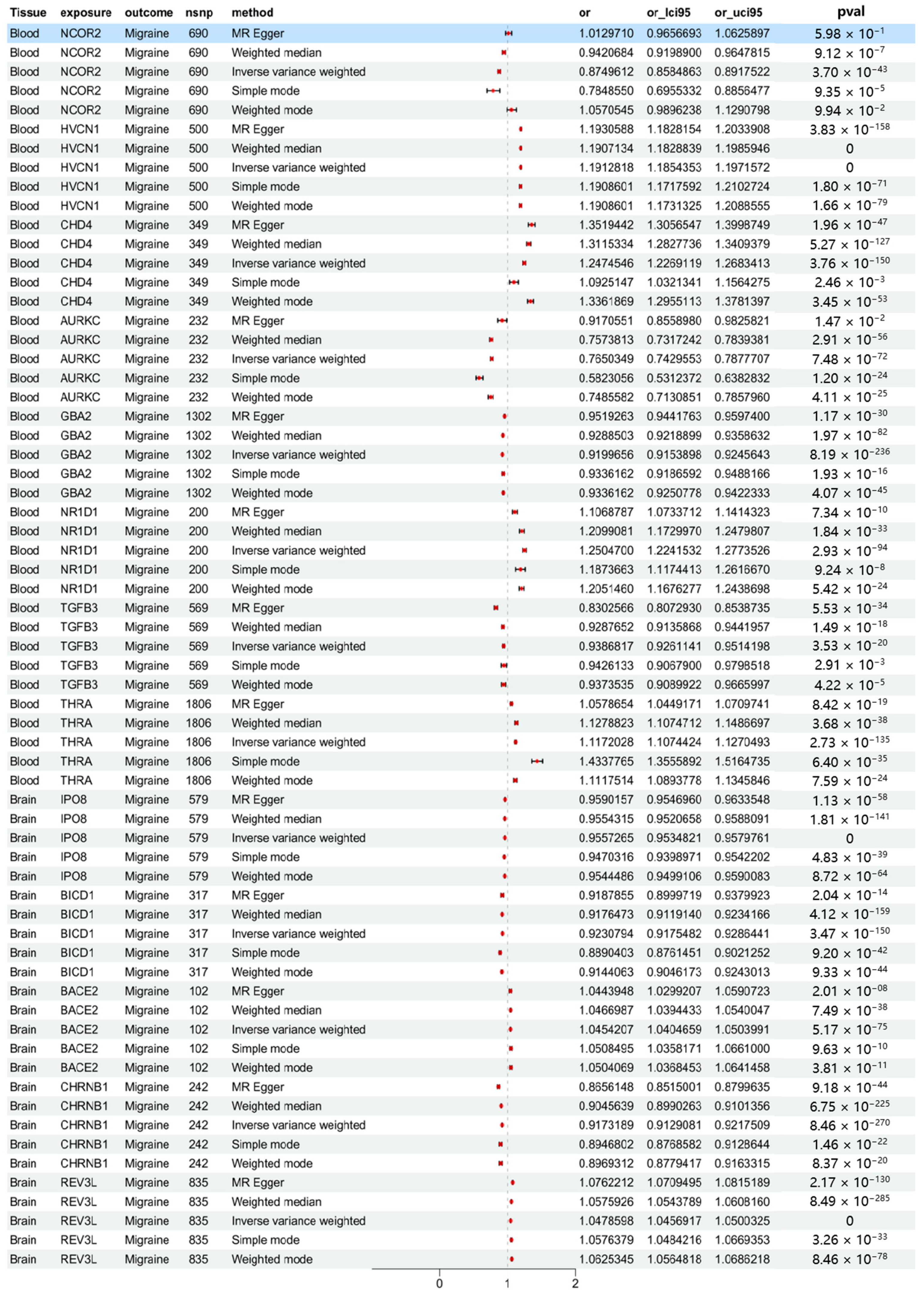
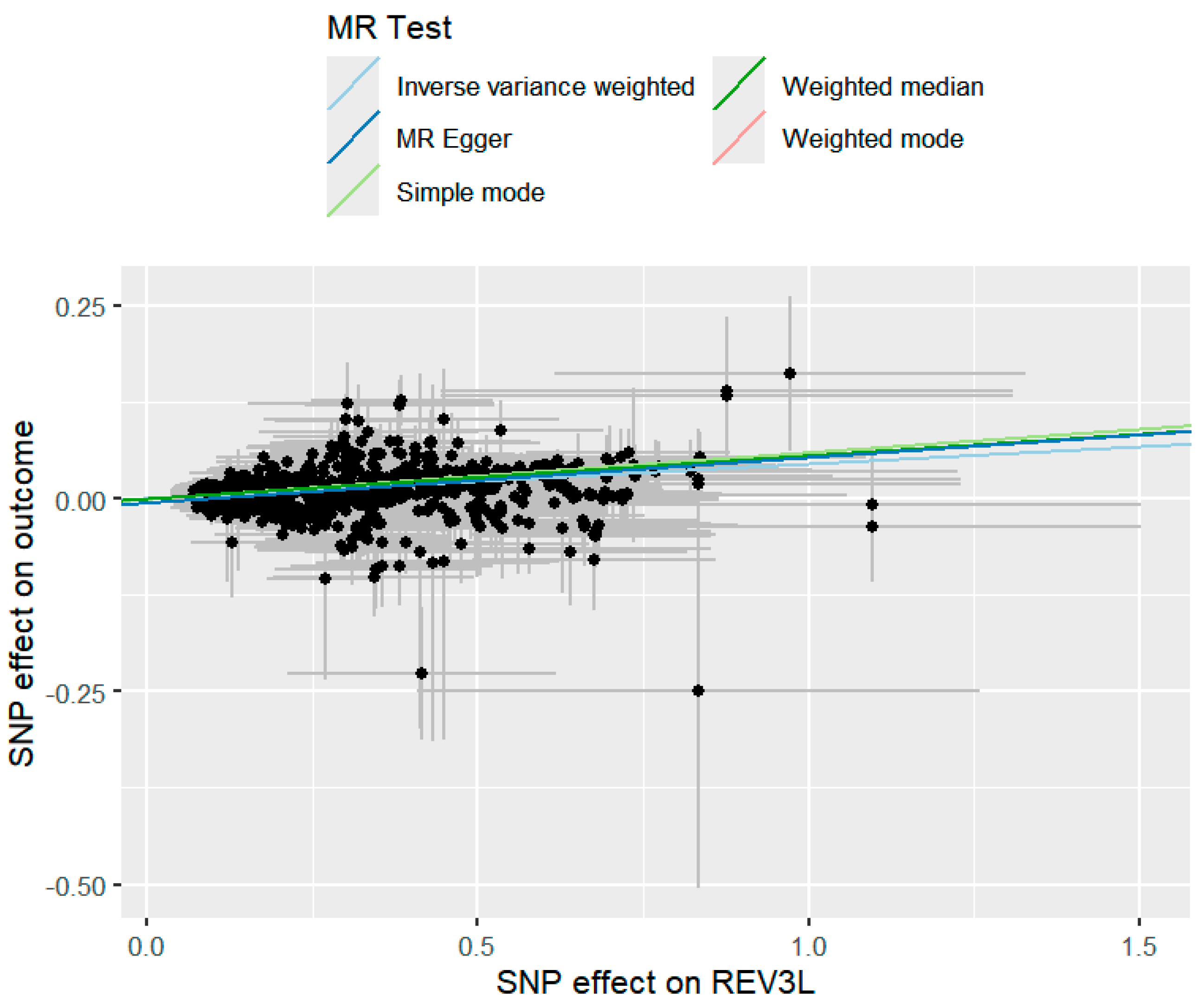
2.3. MR Analysis Results Based on Multiple Methods
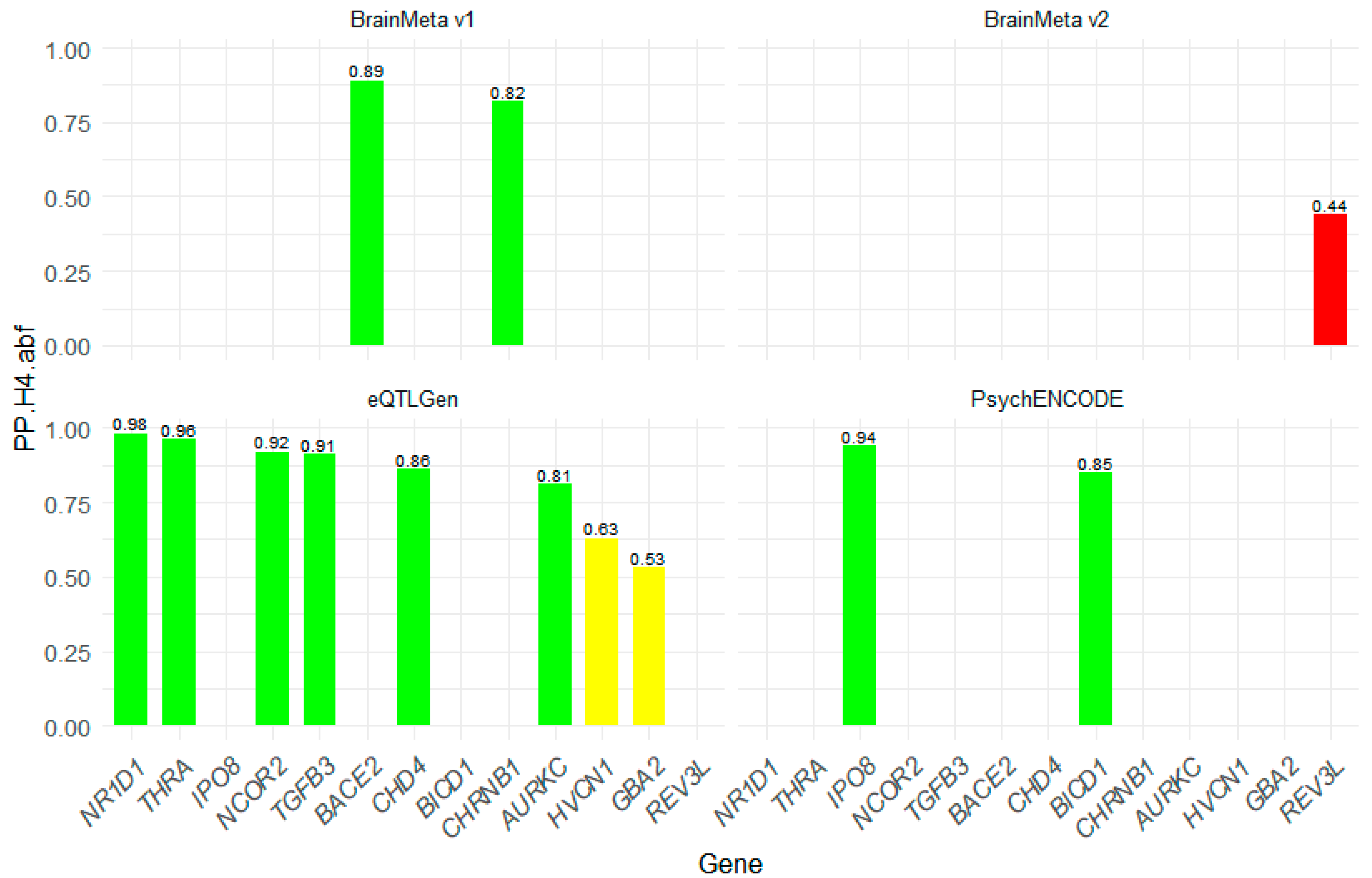
2.4. Colocalization Analysis of Druggable Genes
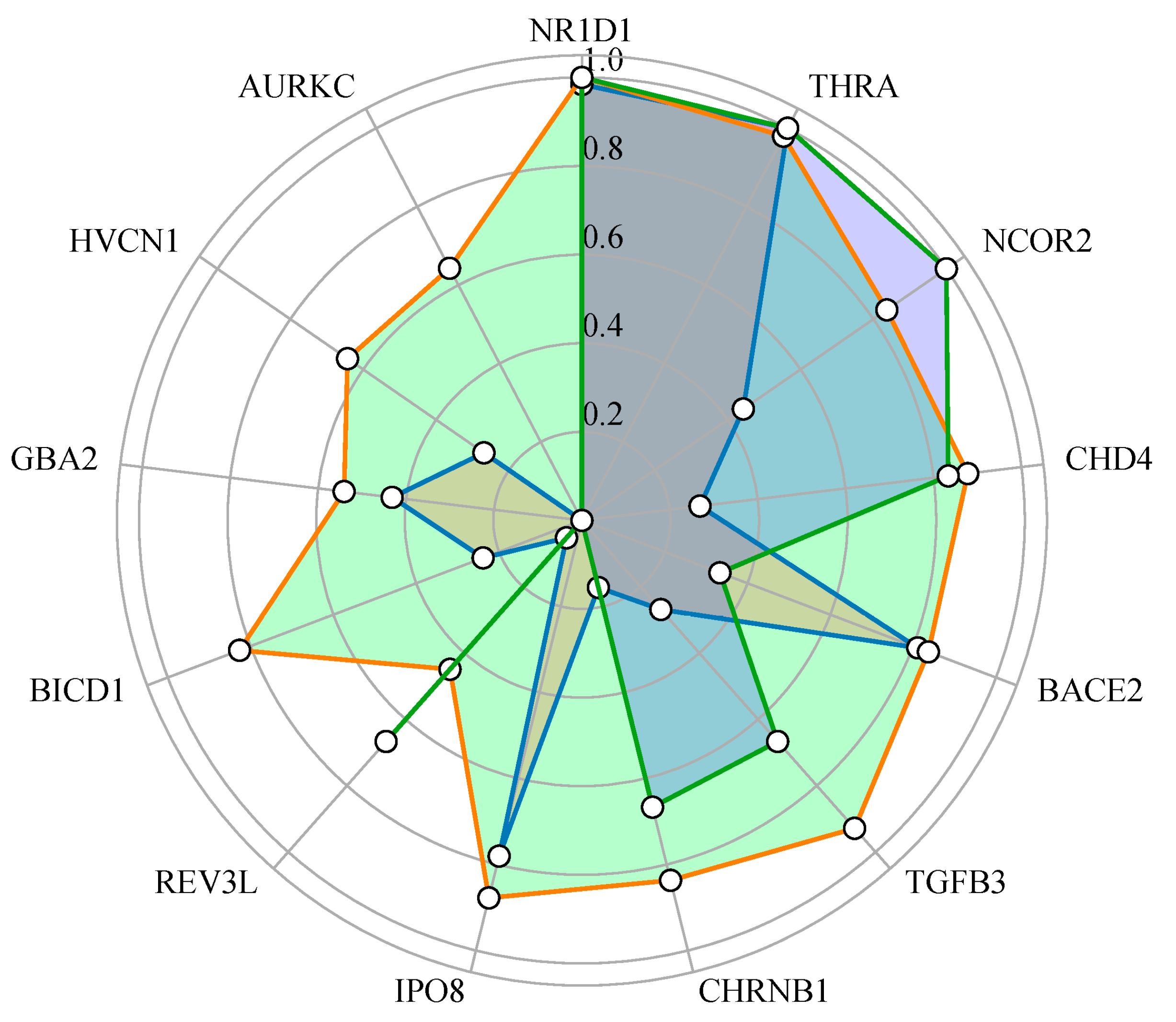
2.5. Exploration of the Druggability Potential of Genes
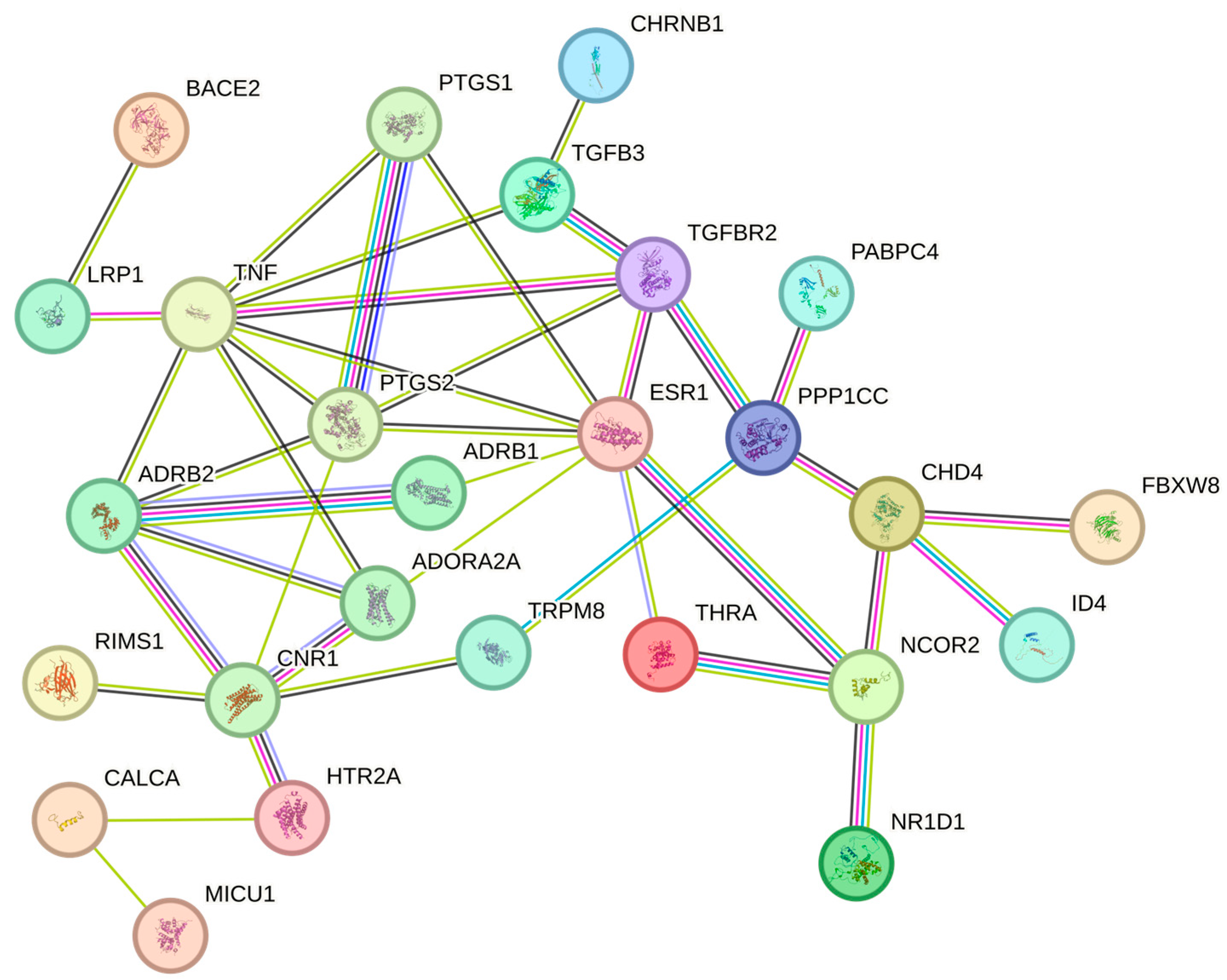
2.6. Enrichment Analysis
2.7. PheWAS Analysis
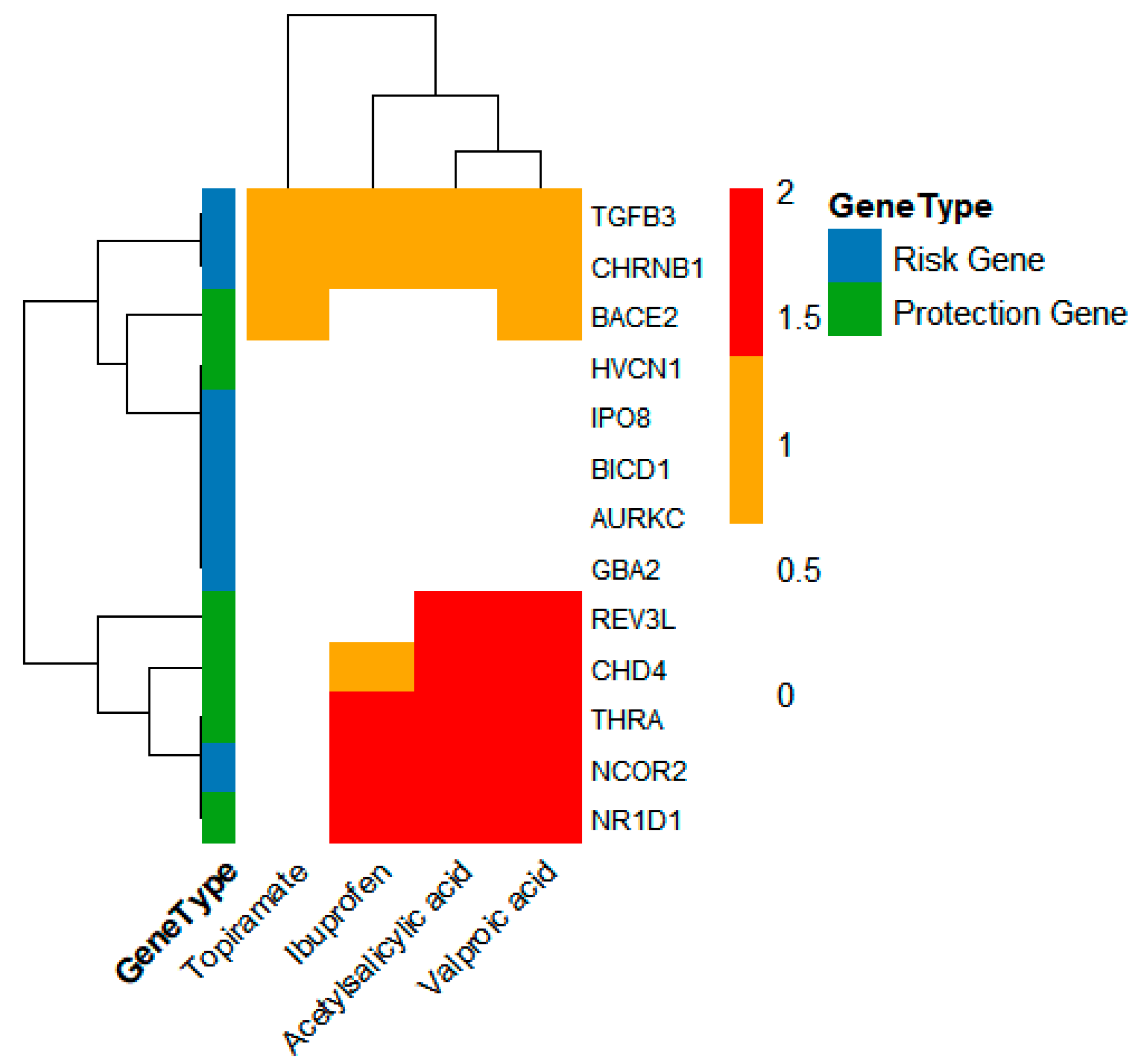
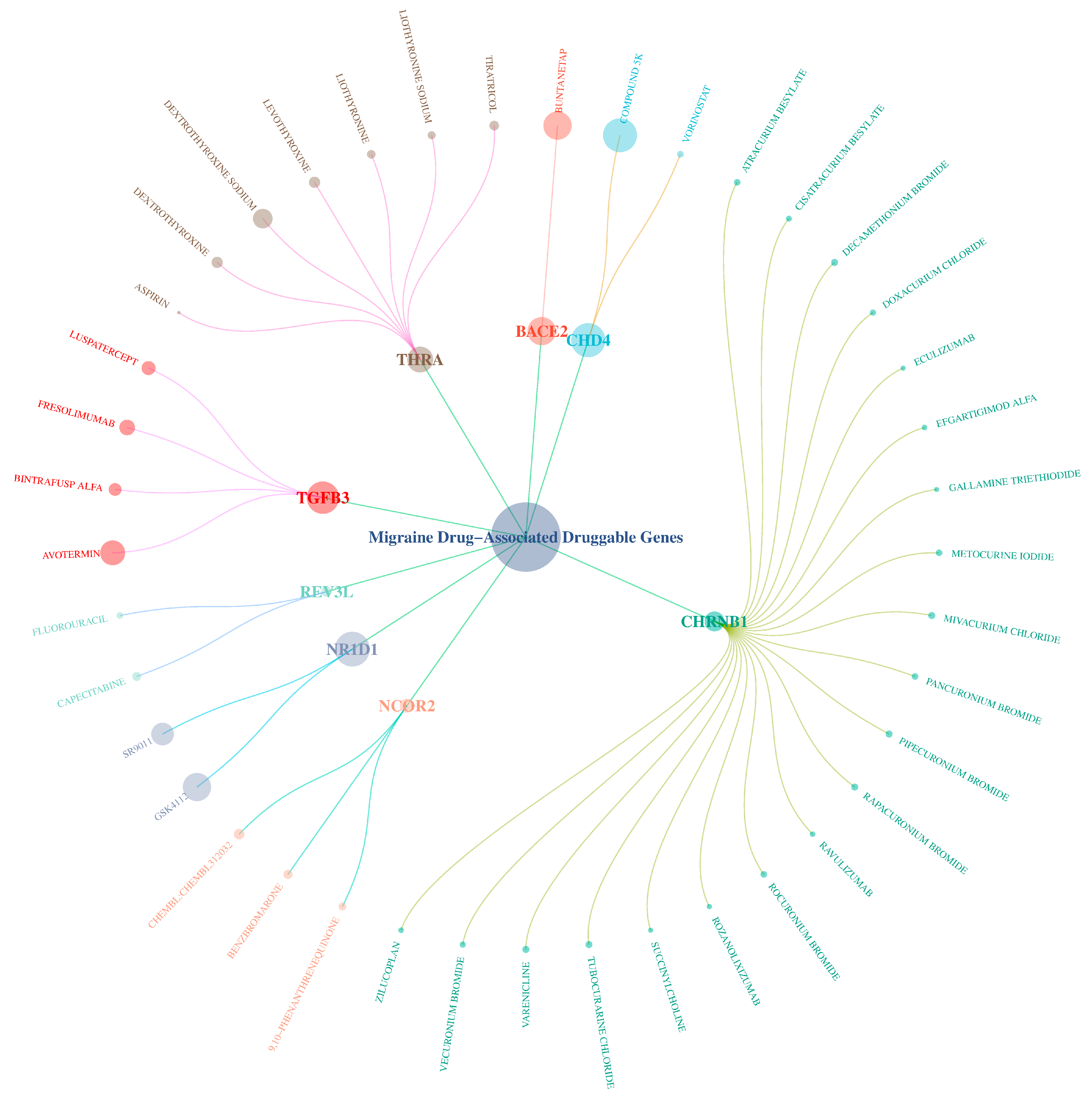
2.8. Potential Targeted Drug Prediction
3. Discussion
3.1. Causal Associations and Biological Mechanisms
3.2. Translational Implications: Drug Targets and Repurposing
3.3. Cross-Study Comparison and Verification
3.4. Limitations and Future Research Directions
- (1)
- Conduct cross-ancestry validation studies to explore the impact of gene-environment interaction on migraine;
- (2)
- Verify the functions of key genes (such as MICU1, UFL1, LY6G5C, PPP1CC) in the pathology of migraine through CRISPR/Cas9 or animal models;
- (3)
- Develop targeted regulatory strategies for genes such as NR1D1, THRA, NCOR2, CHD4, and evaluate their roles in neuroinflammation and synaptic plasticity;
- (4)
- Design multi-target combination therapies tailored to the different phases of migraine (e.g., acute vs. preventive), and integrate real-world adverse reaction data from monotherapies [55] to optimize treatment safety and efficacy.
4. Materials and Methods
4.1. Study Design
4.2. Screening of Genetic Tools for Target Gene Expression
4.3. Migraine GWAS Data
4.4. Data Analysis
4.5. Sensitivity Analysis of Key Target Genes
4.6. External Validation Analysis
4.7. Screening of Druggable Genes
4.8. Candidate Gene Selection and Functional Enrichment Analysis
4.9. PheWAS Analysis of Candidate Gene
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GWASs | Genome-wide association studies |
| eQTL | Expression quantitative trait loci |
| MR | Mendelian randomization |
| IV (IVs) | Instrumental variable (s) |
| SMR | Summary-based Mendelian randomization |
| HEIDI | Heterogeneity in dependent instruments |
| SNP(s) | Single nucleotide polymorphism(s) |
| PPI | Protein–protein interaction |
| PheWASs | Phenome-wide association studies |
| GO enrichment | Gene Ontology enrichment |
| LD | Linkage disequilibrium |
References
- Stovner, L.J.; Hagen, K.; Linde, M.; Steiner, T.J. The global prevalence of headache: An update, with analysis of the influences of methodological factors on prevalence estimates. J. Headache Pain 2022, 23, 34. [Google Scholar] [CrossRef]
- Khan, J.; Asoom, L.I.A.; Sunni, A.A.; Rafique, N.; Latif, R.; Saif, S.A.; Almandil, N.B.; Almohazey, D.; AbdulAzeez, S.; Borgio, J.F. Genetics, pathophysiology, diagnosis, treatment, management, and prevention of migraine. Biomed. Pharmacother. 2021, 139, 111557. [Google Scholar] [CrossRef]
- Ha, H.; Gonzalez, A. Migraine headache prophylaxis. Am. Fam. Physician 2019, 99, 17–24. [Google Scholar]
- Urru, M.; Buonvicino, D.; Pistolesi, A.; Paccosi, S.; Chiarugi, A. Histone deacetylase inhibitors counteract CGRP signaling and pronociceptive sensitization in a rat model of medication overuse headache. J. Pain 2022, 23, 1874–1884. [Google Scholar] [CrossRef] [PubMed]
- Ducros, A. Genetics of migraine. Rev. Neurol. 2021, 177, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Lv, H.; Yang, D.; Zhang, L.; Li, X.; Ning, Y.; Tang, Y.; Wu, X.; Han, J. Drug-related side effects and adverse reactions in the treatment of migraine: A bibliometric and visual analysis. Front. Neurol. 2024, 15, 1342111. [Google Scholar] [CrossRef]
- Tasma, Z.; Garelja, M.L.; Jamaluddin, A.; Alexander, T.I.; Rees, T.A. Where are we now? Biased signalling of class B G protein-coupled receptor-targeted therapeutics. Pharmacol. Ther. 2025, 270, 108846. [Google Scholar] [CrossRef]
- Jia, Z.C.; Yang, X.; Wu, Y.K.; Li, M.; Das, D.; Chen, M.X.; Wu, J. The art of finding the right drug target: Emerging methods and strategies. Pharmacol. Rev. 2024, 76, 896–914. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, X.; Wang, Y.; Yu, Y.; Huang, N.; Li, G.; Li, X.; Wu, J.C.; Yang, S. Artificial intelligence in drug development. Nat. Med. 2025, 31, 45–59. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; Vries, J.D.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Irene, D.B.; Aster, V.E.H.; Michel, D.F.; Van den Maagdenberg, A.M.J.M.; Terwindt, G.M. Chapter 7—Genetics of migraine: Delineation of contemporary understanding of the genetic underpinning of migraine. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2023; Volume 198, pp. 85–103. [Google Scholar]
- Hautakangas, H.; Winsvold, B.S.; Ruotsalainen, S.E.; Pirinen, M. Genome-wide analysis of 102,084 migraine cases identifies 123 risk loci and subtype-specific risk alleles. Nat. Genet. 2022, 54, 152–160. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, L.; Ji, J.; Yan, R.; Guo, P.; Gong, W.; Xue, F.; Zhou, X.; Yuan, Z. Efficient mendelian randomization analysis with self-adaptive determination of sample structure and multiple pleiotropic effects. Am. J. Hum. Genet. 2025, 112, 1962–1978. [Google Scholar] [CrossRef]
- Rasooly, D.; Patel, C.J. Conducting a reproducible mendelian randomization analysis using the R analytic statistical environment. Curr. Protoc. Hum. Genet. 2019, 101, e82. [Google Scholar] [CrossRef]
- Burgess, S.; Mason, A.M.; Grant, A.J.; Slob, E.A.W.; Gkatzionis, A.; Zuber, V.; Patel, A.; Tian, H.; Liu, C.; Haynes, W.G.; et al. Using genetic association data to guide drug discovery and development: Review of methods and applications. Am. J. Hum. Genet. 2023, 110, 195–214. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, T.; Luo, J.; Jiang, Z.; Chen, W.; Chen, H.; Qi, T.; Yang, J. SMR-portal: An online platform for integrative analysis of GWAS and xQTL data to identify complex trait genes. Nat. Methods 2025, 22, 220–222. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, X.; Zhang, S.; Qi, C.; Zhang, Z.; Ma, R.; Xiang, L.; Chen, L.; Zhu, Y.; Tang, C.; et al. Oxidative stress gene expression, DNA methylation, and gut microbiota interaction trigger crohn’s disease: A multi-omics mendelian randomization study. BMC Med. 2023, 21, 179. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Q.; Ni, H.; Ni, J.; Yang, S.; Ji, J. Causal factors for migraine in mendelian randomization studies: A systematic review and meta-analysis. Front. Neurol. 2025, 16, 1660995. [Google Scholar] [CrossRef]
- Yang, S.; Guo, J.; Kong, Z.; Deng, M.; Da, J.; Lin, X.; Peng, S.; Fu, J.; Luo, T.; Ma, J.; et al. Causal effects of gut microbiota on sepsis and sepsis-related death: Insights from genome-wide mendelian randomization, single-cell RNA, bulk RNA sequencing, and network pharmacology. J. Transl. Med. 2024, 22, 10. [Google Scholar] [CrossRef]
- Lin, P.W.; Lin, Z.R.; Wang, W.W.; Guo, A.S.; Chen, Y.X. Identification of immune-inflammation targets for intracranial aneurysms: A multiomics and epigenome-wide study integrating summary-data-based mendelian randomization, single-cell-type expression analysis, and DNA methylation regulation. Int. J. Surg. 2025, 111, 346–359. [Google Scholar] [CrossRef]
- Hardiman, G. An introduction to systems analytics and integration of big omics data. Genes 2020, 11, 245. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Small, D.S.; Thompson, S.G. A review of instrumental variable estimators for mendelian randomization. Stat. Methods Med. Res. 2017, 26, 2333–2355. [Google Scholar] [CrossRef]
- Burgess, S.; Bowden, J.; Fall, T.; Ingelsson, E.; Thompson, S.G. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology 2017, 28, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Ni, H.; Yang, Q.; Ni, J.; Ji, J.; Yang, S.; Peng, F. Evaluating the bidirectional causal effects of alzheimer’s disease across multiple conditions: A systematic review and meta-analysis of mendelian randomization studies. Int. J. Mol. Sci. 2025, 26, 3589. [Google Scholar] [CrossRef]
- Bowden, J.; Holmes, M.V. Meta-analysis and mendelian randomization: A review. Res. Synth. Methods 2019, 10, 486–496. [Google Scholar] [CrossRef]
- Porcu, E.; Rüeger, S.; Lepik, K.; Santoni, F.A.; Reymond, A.; Kutalik, Z. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat. Commun. 2019, 10, 3300. [Google Scholar] [CrossRef]
- Athanasios, A.; Charalampos, V.; Vasileios, T.; Ashraf, G.M. Protein-protein interaction (PPI) network: Recent advances in drug discovery. Curr. Drug Metab. 2017, 18, 5–10. [Google Scholar] [CrossRef]
- Bastarache, L.; Denny, J.C.; Roden, D.M. Phenome-wide association studies. JAMA 2022, 327, 75–76. [Google Scholar] [CrossRef]
- The GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Gleason, K.J.; Yang, F.; Pierce, B.L.; He, X.; Chen, L.S. Primo: Integration of multiple GWAS and omics QTL summary statistics for elucidation of molecular mechanisms of trait-associated SNPs and detection of pleiotropy in complex traits. Genome Biol. 2020, 21, 236. [Google Scholar] [CrossRef] [PubMed]
- Su, W.M.; Gu, X.J.; Dou, M.; Duan, Q.Q.; Jiang, Z.; Yin, K.F.; Cai, W.C.; Cao, B.; Wang, Y.; Chen, Y.P. Systematic druggable genome-wide mendelian randomisation identifies therapeutic targets for alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 2023, 94, 954–961. [Google Scholar] [CrossRef]
- Yin, K.F.; Chen, T.; Gu, X.J.; Su, W.M.; Jiang, Z.; Lu, S.J.; Cao, B.; Chi, L.Y.; Gao, X.; Chen, Y.P. Systematic druggable genome-wide mendelian randomization identifies therapeutic targets for sarcopenia. J. Cachexia Sarcopenia Muscle 2024, 15, 1324–1334. [Google Scholar] [CrossRef]
- Bonanno, L.; Lo Buono, V.; De Salvo, S.; Ruvolo, C.; Torre, V.; Bramanti, P.; Marino, S.; Corallo, F. Brain morphologic abnormalities in migraine patients: An observational study. J. Headache Pain 2020, 21, 39. [Google Scholar] [CrossRef] [PubMed]
- Reid, X.J.; Zhong, Y.; Mackay, J.P. How does CHD4 slide nucleosomes? Biochem. Soc. Trans. 2024, 52, 1995–2008. [Google Scholar] [CrossRef]
- Weiss, K.; Lachlan, K. CHD4 neurodevelopmental disorder. In Genereviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Liu, X.; Huang, S.; Liu, C.; Liu, X.; Shen, Y.; Cui, Z. PPP1CC is associated with astrocyte and microglia proliferation after traumatic spinal cord injury in rats. Pathol. Res. Pract. 2017, 213, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Hagman, J.R.; Arends, T.; Laborda, C.; Knapp, J.R.; Harmacek, L.; O’Connor, B.P. Chromodomain helicase DNA-binding 4 (CHD4) regulates early B cell identity and V(D)J recombination. Immunol. Rev. 2022, 305, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Goodman, J.V.; Yamada, T.; Yang, Y.; Kong, L.; Wu, D.Y.; Zhao, G.; Gabel, H.W.; Bonni, A. The chromatin remodeling enzyme Chd4 regulates genome architecture in the mouse brain. Nat. Commun. 2020, 11, 3419. [Google Scholar] [CrossRef]
- Foley, K.; McKee, C.; Nairn, A.C.; Xia, H. Regulation of synaptic transmission and plasticity by protein phosphatase 1. J. Neurosci. 2021, 41, 3040–3050. [Google Scholar] [CrossRef]
- Barker, H.M.; Craig, S.P.; Spurr, N.K.; Cohen, P.T. Sequence of human protein serine/threonine phosphatase 1 gamma and localization of the gene (PPP1CC) encoding it to chromosome bands 12q24.1–q24.2. Biochim. Biophys. Acta 1993, 1178, 228–233. [Google Scholar] [CrossRef]
- Ogunlaja, O.; Karsan, N.; Goadsby, P. Emerging drugs for the prevention of migraine. Expert Opin. Emerg. Drugs 2021, 26, 271–280. [Google Scholar] [CrossRef]
- Messoud, A.; Jan, H.; Håkan, A.; Debbie, L.H.; Yadira, F.M.; Thien, P.D.; Roberto, D.I.; David, W.D. Pharmacotherapies for migraine and translating evidence from bench to bedside. Mayo Clin. Proc. 2024, 99, 285–299. [Google Scholar] [CrossRef]
- Zhang, C.; He, Y.; Liu, L. Identifying therapeutic target genes for migraine by systematic druggable genome-wide mendelian randomization. J. Headache Pain 2024, 25, 100. [Google Scholar] [CrossRef]
- Deng, Z.; Fan, T.; Xiao, C.; Tian, H.; Zheng, Y.; Li, C.; He, J. TGF-β signaling in health, disease, and therapeutics. Signal Transduct. Target. Ther. 2024, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Tianhe, S.; Zhiyu, H.; Wei-Ching, L.; Jianping, Y.; Wei, Y.L.; Jia, W.; Jean-Michel, V.; Jeff, L.; Patrick, C.; Surinder, J.; et al. TGFβ2 and TGFβ3 isoforms drive fibrotic disease pathogenesis. Sci. Transl. Med. 2021, 13, eabe0407. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Zhao, L.; Mei, Y.; Qiu, D.; Li, X.; Zhang, P.; Zhang, M.; Cao, J.; Wang, Y. Proteome-wide mendelian randomization identified potential drug targets for migraine. J. Headache Pain 2024, 25, 148. [Google Scholar] [CrossRef]
- Nuottamo, M.E.; Häppölä, P.; Artto, V.; Hautakangas, H.; Pirinen, M.; Hiekkalinna, T.; Ellonen, P.; Lepistö, M.; Hämäläinen, E. NCOR2 is a novel candidate gene for migraine-epilepsy phenotype. Cephalalgia 2022, 42, 631–644. [Google Scholar] [CrossRef]
- Sun, X.; Chen, B.; Qi, Y.; Wei, M.; Chen, W.; Wu, X.; Wang, Q.; Li, J.; Lei, X.; Luo, J. Multi-omics mendelian randomization integrating GWAS, eQTL and pQTL data revealed GSTM4 as a potential drug target for migraine. J. Headache Pain 2024, 25, 117. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Han, F.; Liu, H. Challenges of big data analysis. Natl. Sci. Rev. 2014, 1, 293–314. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Zhao, Q.; Lawlor, D.A.; Sheehan, N.A.; Thompson, J.; Smith, G.D. Improving the accuracy of two-sample summary-data mendelian randomization: Moving beyond the NOME assumption. Leuk. Res. 2018, 48, 728–742. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, W.; Zhang, F.; Wei, J.; Wang, S.; Min, K.; Chen, Y.; Yang, H.; Lv, X. Exploring the bidirectional causal associations between pain and circulating inflammatory proteins: A mendelian randomization study. Clin. Exp. Pharmacol. Physiol. 2024, 51, e13905. [Google Scholar] [CrossRef]
- Greco, M.F.D.; Minelli, C.; Sheehan, N.A.; Thompson, J.R. Detecting pleiotropy in mendelian randomisation studies with summary data and a continuous outcome. Stat. Med. 2015, 34, 2926–2940. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Liu, X.; Sun, H.; Chen, Y.; Fu, C.; Xu, X.; Yang, S.; Tang, Y. A novel approach for the screening analysis of anticancer compounds from traditional chinese medicine by a G-quadruplex functionalized magnetic system. Anal. Methods 2020, 12, 528–534. [Google Scholar] [CrossRef]
- Hu, Q.; Hu, H.; Hu, M.; Zhang, J.; Gou, L.; Shi, S.; Zhou, J.; Zhou, N.; Huang, Z. Use of failure mode and effect analysis to reduce patient safety risks in purchasing prescription drugs from online pharmacies in China. Front. Med. 2022, 9, 913214. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.A. The benjamini-hochberg method in the case of discrete test statistics. Int. J. Biostat. 2007, 3, 11. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.F.; Chalumeau, M.; Cohen, R.; Korevaar, D.A.; Khoshnood, B.; Bossuyt, P.M.M. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J. Clin. Epidemiol. 2015, 68, 299–306. [Google Scholar] [CrossRef]
- Qi, T.; Wu, Y.; Zeng, J.; Zhang, F.; Xue, A.; Jiang, L.; Zhu, Z.; Kemper, K.; Yengo, L.; Zheng, Z.; et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 2018, 9, 2282. [Google Scholar] [CrossRef]
- GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef]
- Fromer, M.; Roussos, P.; Sieberts, S.K.; Johnson, J.S.; Kavanagh, D.H.; Perumal, T.M.; Ruderfer, D.M.; Oh, E.C.; Topol, A.; Shah, H.R.; et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016, 19, 1442–1453. [Google Scholar] [CrossRef]
- Ng, B.; White, C.C.; Klein, H.-U.; Sieberts, S.K.; McCabe, C.; Patrick, E.; Xu, J.; Yu, L.; Gaiteri, C.; Bennett, D.A.; et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 2017, 20, 1418–1426. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Wu, Y.; Fang, H.; Zhang, F.; Liu, S.; Zeng, J.; Yang, J. Genetic control of RNA splicing and its distinct role in complex trait variation. Nat. Genet. 2022, 54, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Finan, C.; Gaulton, A.; Kruger, F.A.; Lumbers, R.T.; Shah, T.; Engmann, J.; Galver, L.; Kelley, R.; Karlsson, A.; Santos, R.; et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 2017, 9, eaag1166. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shih, A.; Freudenberg-Hua, Y.; Fury, W.; Yang, Y. Beyond standard pipeline and p < 0.05 in pathway enrichment analyses. Comput. Biol. Chem. 2021, 92, 107455. [Google Scholar] [CrossRef]

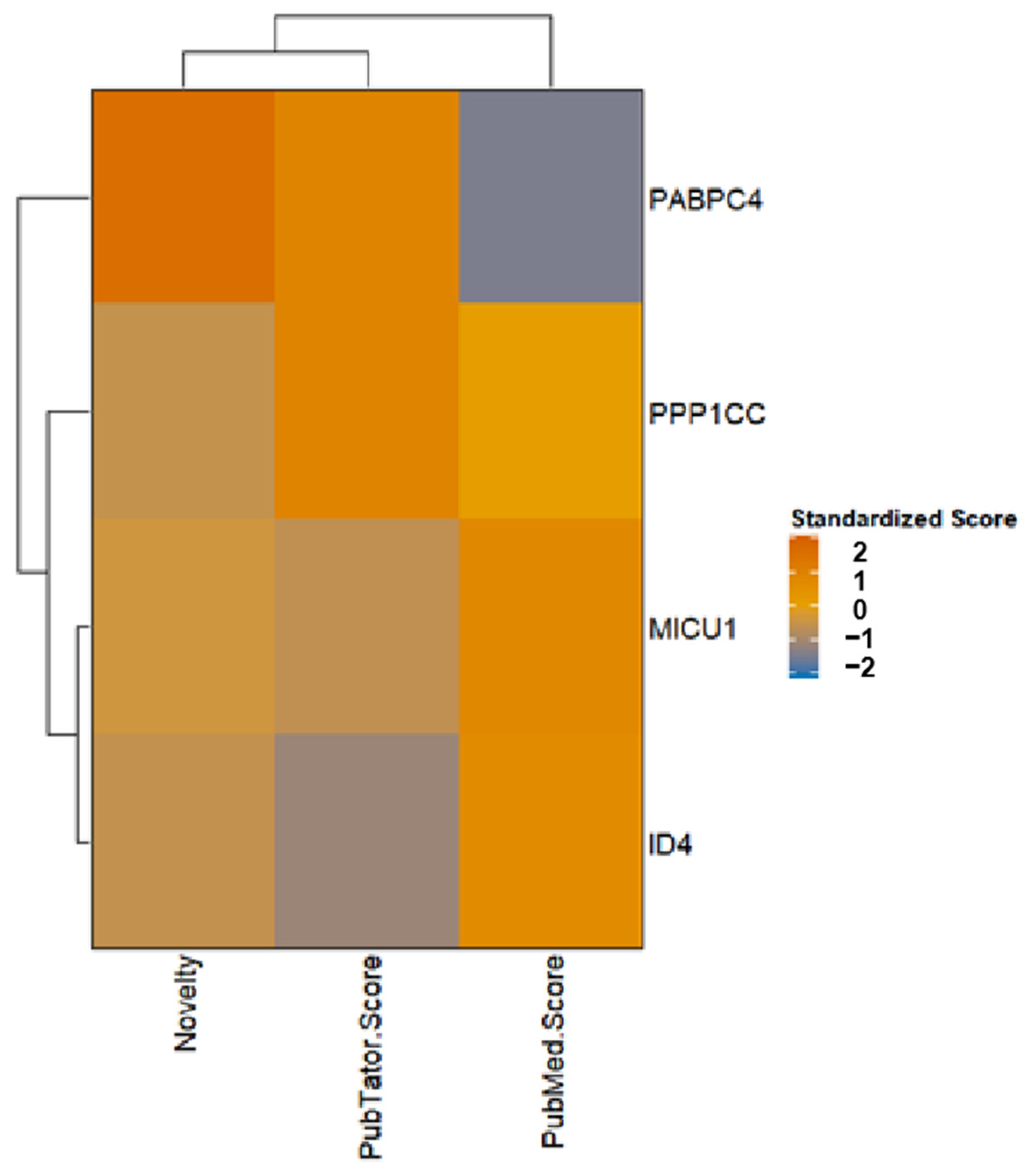
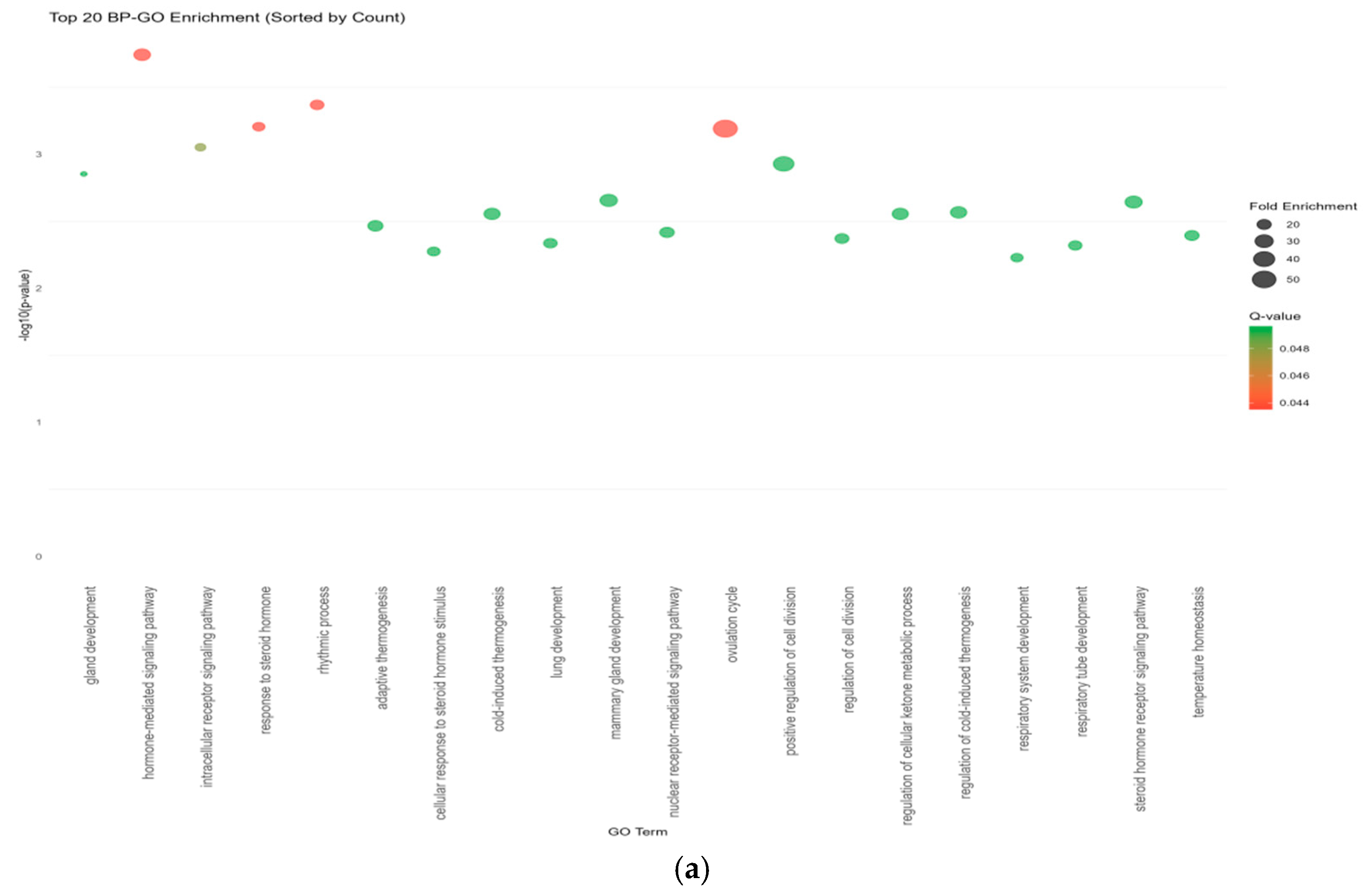
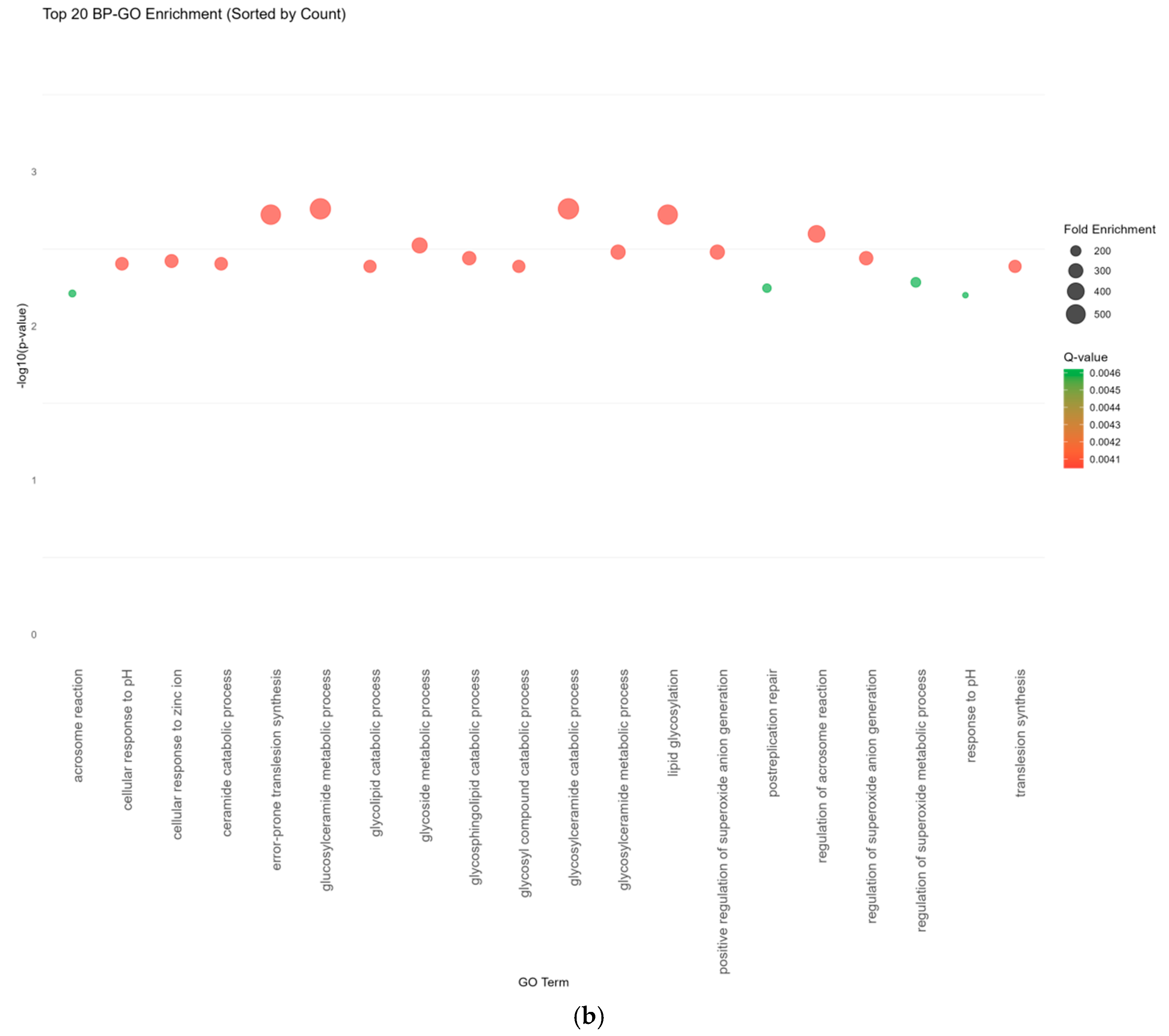
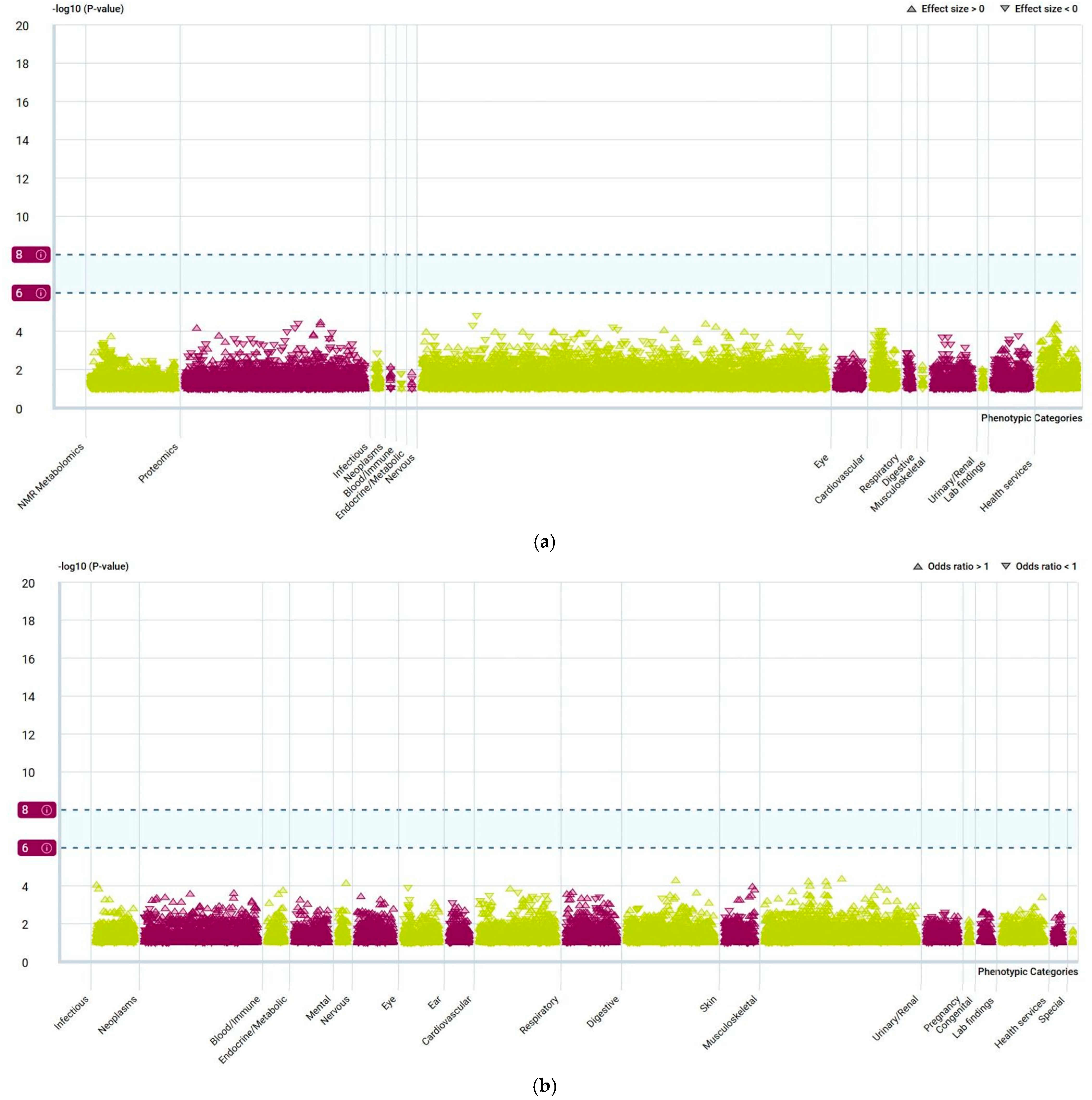

| Source | Tissue | Gene |
|---|---|---|
| eQTLGen | Whole Blood | TJP2, B9D2, CCDC97, SNORA63, THRA, NR1D1, LY6G5C, RNFT2, PABPC4, ZC3H6, HRK, PPP1CC, SHISA5, UBE2D3-AS1, DNAJB12, RALGAPA1, CISD2, FAM8A1, PRR13, SFXN2, NCOR2, KLF10, GBA2, RGP1, PAM16, TGFB3, HVCN1, CHD4, ZMYND12, CCDC18-AS1, AURKC, RCCD1, RPS16 (novel transcript), RP11-252K23.2 (novel transcript), CTC-444N24.8 (novel transcript) |
| PsychENCODE | Prefrontal Cortex | KIAA0040, CFDP1, SLC9B1, CARF, ID4, CISD2, HMGXB3, RABAC1, RIMS1, UBE2D3-AS1, POLR1B, INPP5B, TREX1, TMA7, IPO8, SHISA5, EAPP, PACSIN3, SNX6, BICD1, RALGAPA1, CCDC18-AS1, ZMYND12 |
| GTEx V8 | Cerebellar Hemisphere | UFL1, LY6G5C, POC5, RCCD1, EXOSC5 |
| Cerebellum | UFL1, LY6G5C, RP11-753C18.8 | |
| Whole Blood | UFL1, LY6G5C, FBXW8, LY6G5B, CTD-2509G16.2, RALGAPA1, PABPC4, HRK, ATP5SL |
| Tissue | Gene | Source | Chr | Top SNP | b_SMR | se_SMR | p_SMR | p_HEIDI | nsnp_HEIDI | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| Blood | AURKC | eQTLGen | 19 | rs2074858 | −0.309 | 0.082 | 1.68 × 10−4 | 0.277 | 20 | 0.049 |
| Blood | CHD4 | eQTLGen | 12 | rs7969177 | 0.281 | 0.073 | 1.17 × 10−4 | 0.149 | 20 | 0.037 |
| Blood | GBA2 | eQTLGen | 9 | rs10814274 | −0.068 | 0.017 | 7.54 × 10−5 | 0.173 | 20 | 0.030 |
| Blood | HVCN1 | eQTLGen | 12 | rs113511140 | 0.168 | 0.044 | 1.17 × 10−4 | 0.306 | 20 | 0.037 |
| Blood | NCOR2 | eQTLGen | 12 | rs1271309 | −0.312 | 0.078 | 6.86 × 10−5 | 0.312 | 20 | 0.029 |
| Blood | NR1D1 | eQTLGen | 17 | rs883871 | 0.177 | 0.038 | 4.05 × 10−6 | 0.556 | 20 | 0.005 |
| Blood | TGFB3 | eQTLGen | 14 | rs146047341 | −0.122 | 0.032 | 1.15 × 10−4 | 0.251 | 20 | 0.037 |
| Blood | THRA | eQTLGen | 17 | rs883871 | 0.121 | 0.026 | 3.20 × 10−6 | 0.595 | 20 | 0.004 |
| Brain—Prefrontal Cortex | BICD1 | PsychENCODE | 12 | rs11051973 | −0.087 | 0.023 | 1.13 × 10−4 | 0.213 | 20 | 0.038 |
| Brain—Prefrontal Cortex | IPO8 | PsychENCODE | 12 | rs61923728 | −0.045 | 0.011 | 1.91 × 10−5 | 0.717 | 20 | 0.014 |
| Brain—Hippocampus | BACE2 | BrainMeta v1 | 21 | rs914187 | 0.051 | 0.013 | 7.08 × 10−5 | 0.887 | 20 | 0.013 |
| Brain—Cerebellum | CHRNB1 | BrainMeta v1 | 17 | rs60488855 | −0.101 | 0.026 | 8.94 × 10−5 | 0.053 | 20 | 0.042 |
| Brain—Cerebellum | REV3L | BrainMeta v2 | 6 | rs12214097 | 0.052 | 0.015 | 3.53 × 10−4 | 0.683 | 20 | 0.046 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Liu, Q.; Zhu, H.; Zhou, X.; Li, X.; Hu, M.; Peng, F.; Ji, J.; Yang, S. Precision Target Discovery for Migraine: An Integrated GWAS-eQTL-PheWAS Pipeline. Molecules 2025, 30, 3921. https://doi.org/10.3390/molecules30193921
Liu X, Liu Q, Zhu H, Zhou X, Li X, Hu M, Peng F, Ji J, Yang S. Precision Target Discovery for Migraine: An Integrated GWAS-eQTL-PheWAS Pipeline. Molecules. 2025; 30(19):3921. https://doi.org/10.3390/molecules30193921
Chicago/Turabian StyleLiu, Xianting, Qingming Liu, Haoning Zhu, Xiao Zhou, Xinyao Li, Ming Hu, Fu Peng, Jianguang Ji, and Shu Yang. 2025. "Precision Target Discovery for Migraine: An Integrated GWAS-eQTL-PheWAS Pipeline" Molecules 30, no. 19: 3921. https://doi.org/10.3390/molecules30193921
APA StyleLiu, X., Liu, Q., Zhu, H., Zhou, X., Li, X., Hu, M., Peng, F., Ji, J., & Yang, S. (2025). Precision Target Discovery for Migraine: An Integrated GWAS-eQTL-PheWAS Pipeline. Molecules, 30(19), 3921. https://doi.org/10.3390/molecules30193921






