Exometabolome and Molecular Signatures Associated with HPV 16 in Cervical Cancer: Integrative Metabolomic and Transcriptomic Analysis for Biomarker Discovery
Abstract
1. Introduction
2. Results
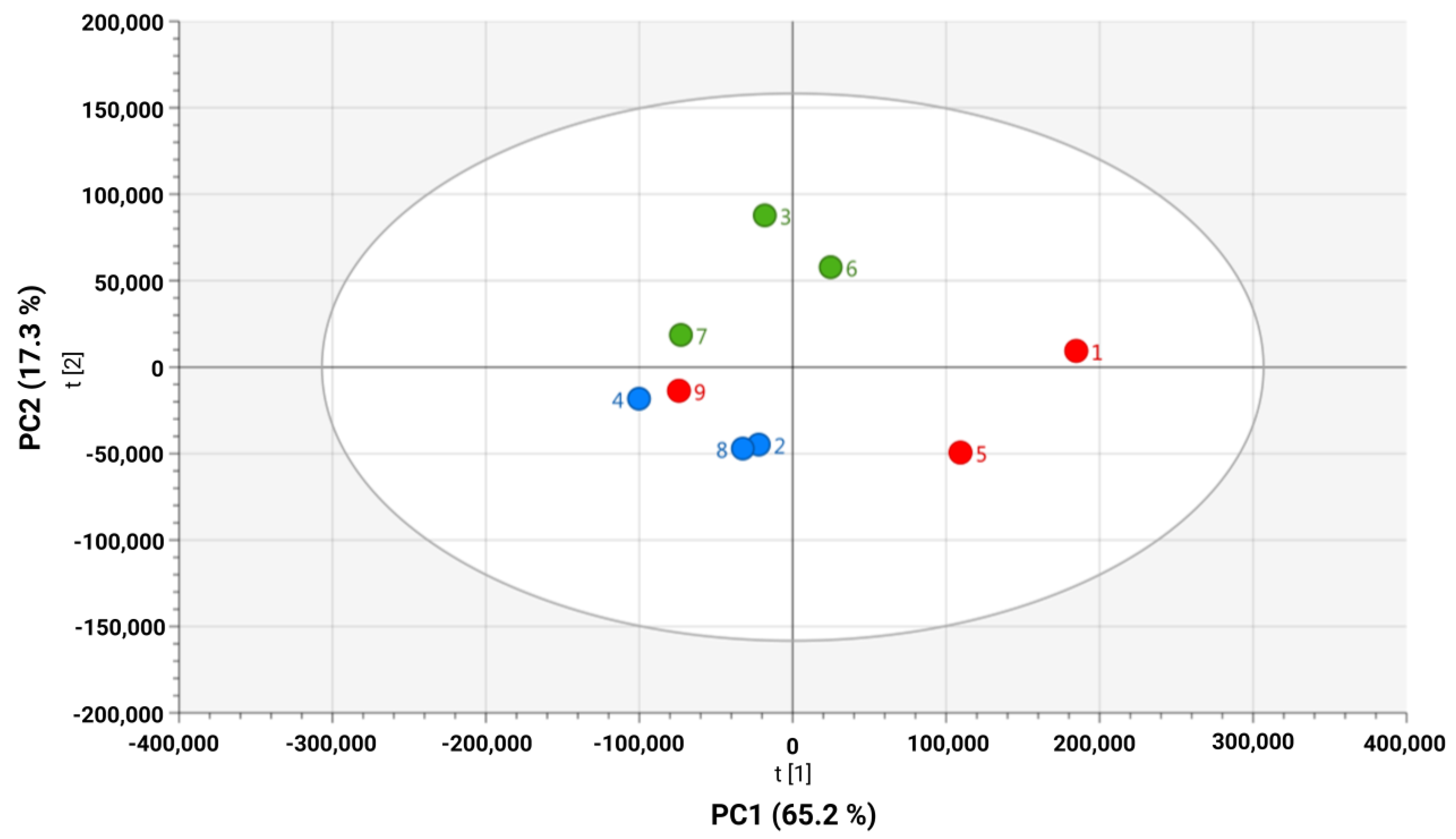
2.1. Unsupervised Analysis of Culture Media Reveals HPV 16–Driven Metabolic Discrimination in Cervical Cancer Cell Lines
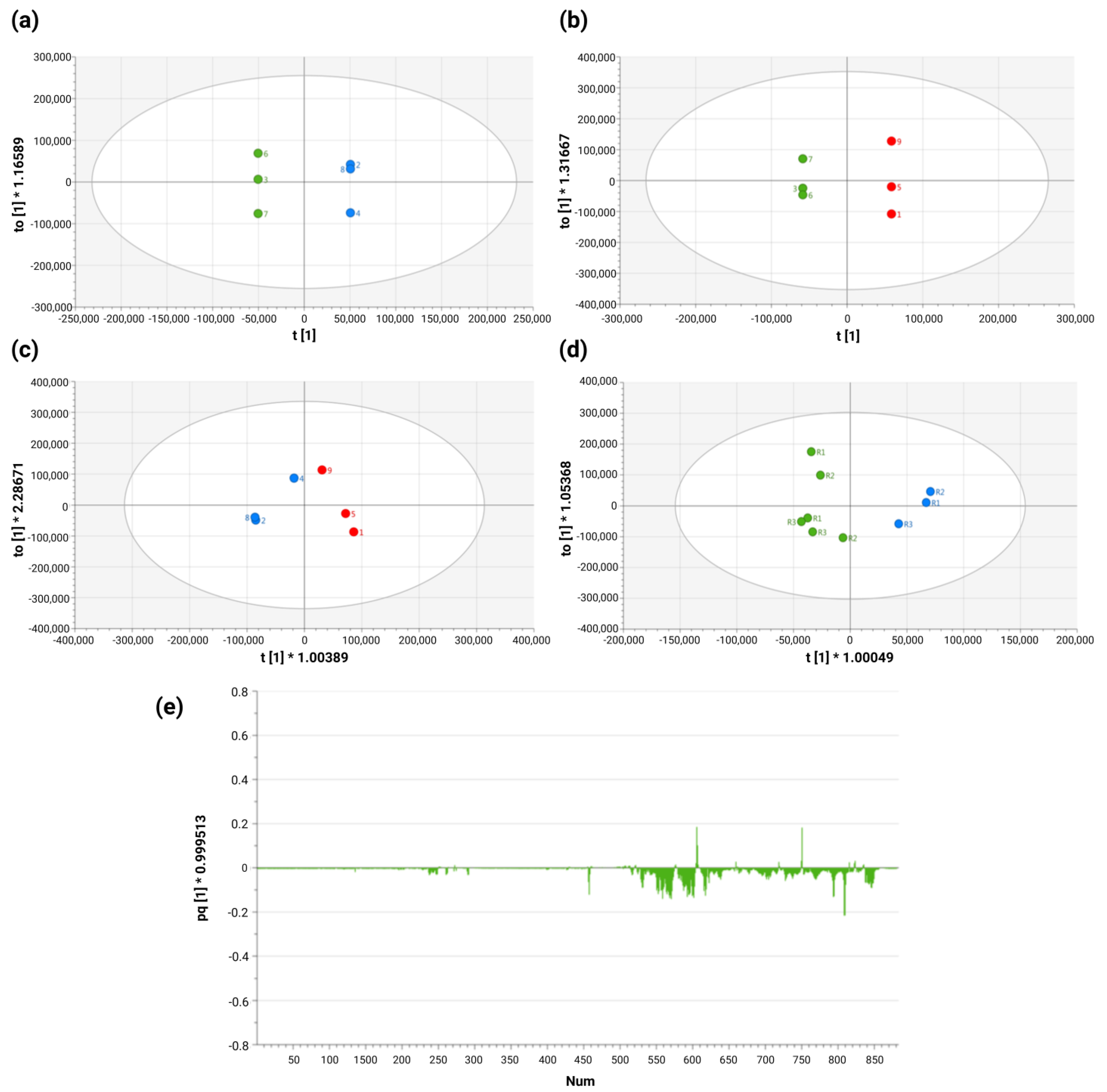
2.2. Supervised Analysis Reveals Significant Exometabolomic Differences Between HPV 16-Positive and -Negative Cervical Cancer Cell Lines
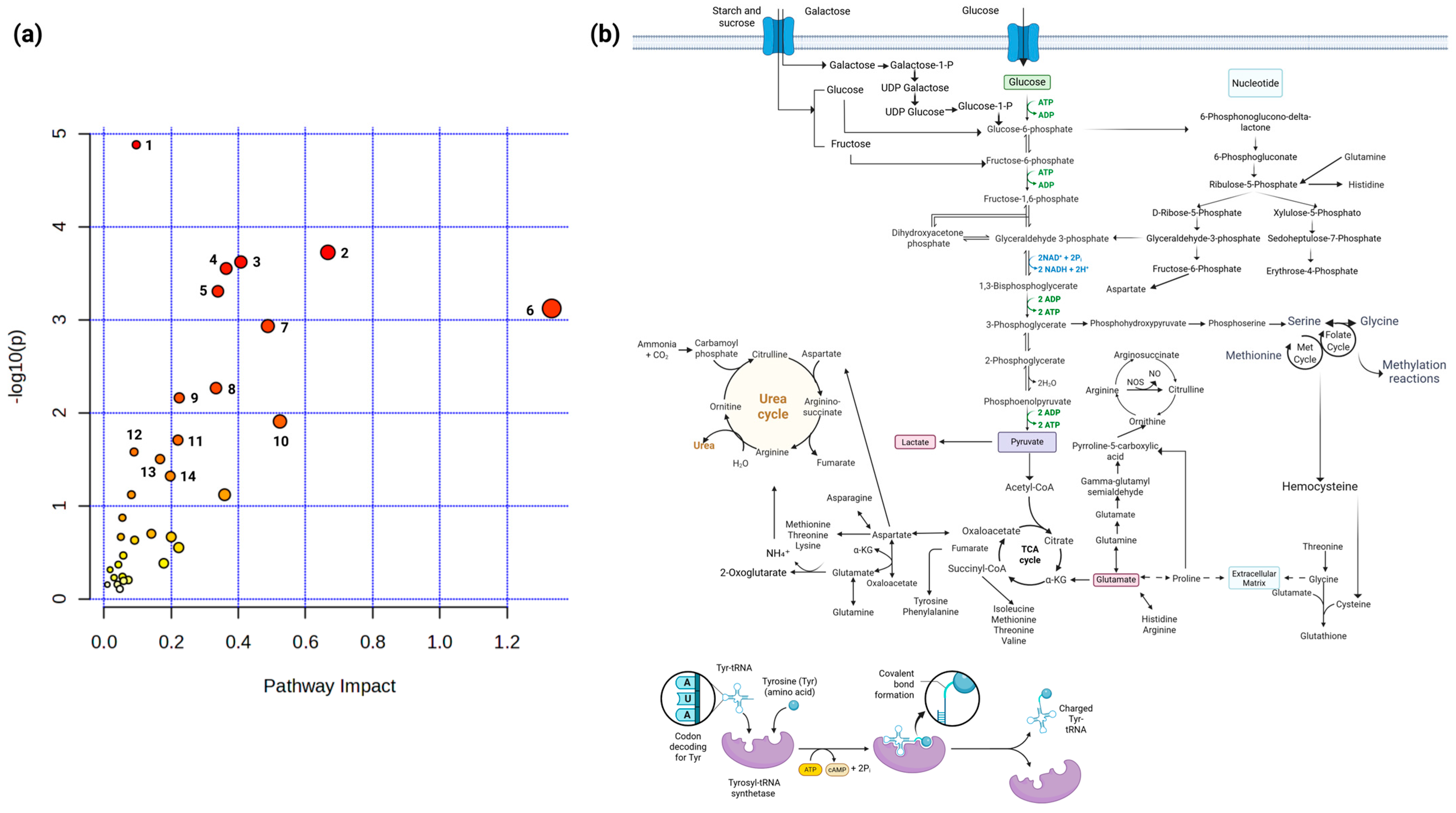
2.3. Specific Combinations of Metabolites Discriminate HPV 16-Negative and Positive Cells
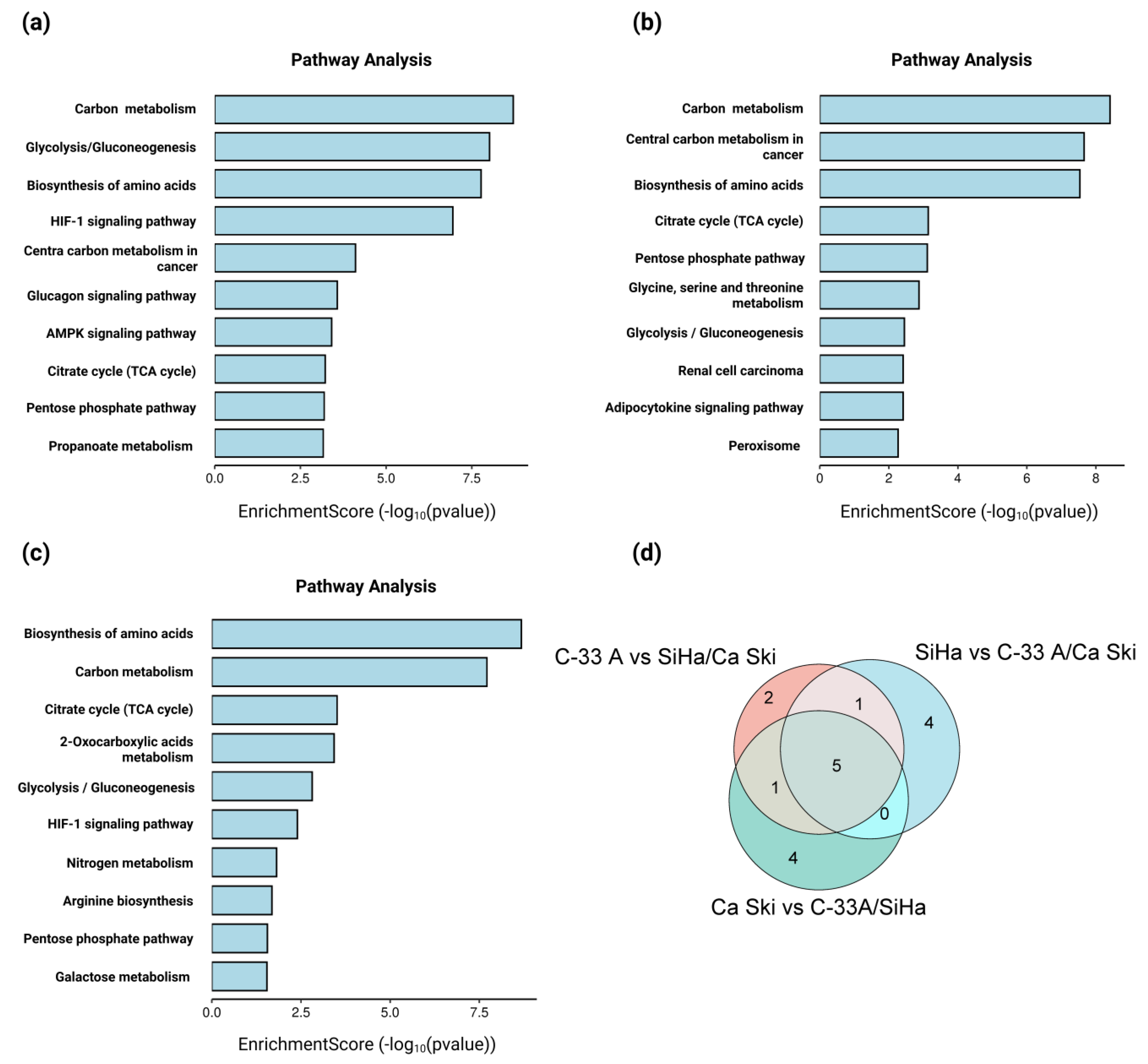
2.4. The Exometabolome Is Associated with the Expression of Genes Related to Metabolic Reprogramming in HPV 16-Positive and -Negative Cervical Cancer Cell Lines
3. Discussion
4. Materials and Methods
4.1. Workflow Diagram
4.2. Cell Culture
4.3. Sample Preparation and 1H-NMR Acquisition
4.4. Multivariate Analysis
4.5. Quantitative Metabolite Selection Analysis
4.6. Gene Expression Analysis
4.7. GO, KEGG and GSEA of Differentially Expressed Genes in C-33 A, SiHa, and Ca Ski Cells
4.8. Statistical Analysis of Gene Expression
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CC | Cervical cancer |
| HPV 16 | Human papillomavirus 16 |
| HPV-HR | High-risk HPVs |
| 1H-NMR | 1H nuclear magnetic resonance |
| VHL | von Hippel–Lindau |
| GEO | Gene Expression Omnibus |
| HIF-1α | Hypoxia-inducible factor-1α |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCA | Principal component analysis |
| GO | Gene Ontology |
| OPLS-DA | Orthogonal Partial Least Squares Discriminant Analysis |
| PGM | Phosphoglucomutase |
| PSAT1 | Phosphoserine Aminotransferase 1 |
| ALDOA, B | Aldolase A and B |
| HK2 | Hexokinase 2 |
| IDH3G | Isocitrate Dehydrogenase (NAD(+)) 3 Non-Catalytic Subunit Gamma |
| CA9 | Carbonic Anhydrase 9 |
| RPIA | Ribose 5-Phosphate Isomerase A |
| BSG | Basigin |
| HMG1 | High Mobility Group Box 1 |
| ASL | Argininosuccinate Lyase |
| PGK1 | Phosphoglycerate Kinase 1 |
| IDH3B | Isocitrate Dehydrogenase (NAD(+)) 3 Non-Catalytic Subunit Beta |
| ACSL3, 4 | Acyl-CoA Synthetase Long-Chain Family Member 3 and 4 |
| PADI2, 3, 4 | Peptidyl Arginine Deiminase 2, 3 and 4 |
| SUCLG2 | Succinate-CoA Ligase GDP-Forming Subunit Beta |
| PGM1 | Phosphoglucomutase 1 |
| IDH1, 2 and 5 | Isocitrate Dehydrogenase 1, 2 and 5 |
| PKM | Pyruvate Kinase M1/2 |
| HIF1A | Hypoxia Inducible Factor 1 Subunit Alpha |
| TKT | Transketolase |
| SLC2A1 | Solute Carrier Family 2 Member 1 |
| GLUD1 | Glutamate Dehydrogenase 1 |
| PHGDH | Phosphoglycerate Dehydrogenase |
| SHMT1 | Serine Hydroxymethyltransferase 1 |
| SHM2 | Serine Hydroxymethyltransferase 2 |
| PDH | Pyruvate Dehydrogenase |
| PSPH | Phosphoserine Phosphatase |
| ASS | Argininosuccinate Synthase |
| FUM1 | Fumarate hydratase 1 |
| IDHG | Isocitrate Dehydrogenase Gamma |
| GLS | Glutaminase |
| HMG1 | High Mobility Group 1 |
| PFKFB2 | 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 2 |
| ACSM2B | Acyl-CoA Synthetase Medium-Chain Family Member 2B |
| PADI1 | Peptidyl Arginine Deiminase 1 |
| PFKM | Phosphofructokinase, Muscle |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase |
| ACC1 | Acetyl-CoA carboxylase 1 |
| SDH2 | Succinate Dehydrogenase 2 or B |
| ICDH | Isocitrate Dehydrogenase |
| ACLB2 | ATP-citrate synthase beta chain protein 2 |
| ACCY | Pyruvate oxidase/decarboxylase |
| ACSS3 | Acyl-CoA Synthetase Short Chain Family Member 3 |
| PGD6 | phosphogluconate dehydrogenase 6 |
| LDHAL6B | Lactate Dehydrogenase A-Like 6B |
| AACS | Acetoacetyl-CoA Synthetase |
| CPT1A | Carnitine Palmitoyltransferase 1A |
| ENO1 | Enolase 1 |
| MDH1 | Malate Dehydrogenase 1 |
| FASN | Fatty Acid Synthase |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef]
- de Villiers, E.-M.; Fauquet, C.; Broker, T.R.; Bernard, H.-U.; zur Hausen, H. Classification of papillomaviruses (HPV). Virology 2004, 324, 17–27. [Google Scholar] [CrossRef]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The biology and life-cycle of human papillomaviruses. Vaccine 2012, 30, F55–F70. [Google Scholar] [CrossRef] [PubMed]
- Harden, M.E.; Munger, K. Human papillomavirus molecular biology. Mutat. Res.-Rev. Mutat. Res. 2017, 772, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Estêvão, D.; Costa, N.R.; da Costa, R.M.G.; Medeiros, R. Hallmarks of HPV carcinogenesis: The role of E6, E7 and E5 oncoproteins in cellular malignancy. Biochim. Biophys. Acta-Gene Regul. Mech. 2019, 1862, 153–162. [Google Scholar] [CrossRef]
- Muñoz, N. New perspective for cervical cancer prevention based on human papilloma virus. Biomed. Rev. Inst. Nac. Salud 2007, 26, 471–474. [Google Scholar]
- Lizano, M.; Berumen, J.; García-Carrancá, A. HPV-related Carcinogenesis: Basic Concepts Viral Types and Variants. Arch. Med. Res. 2009, 40, 428–434. [Google Scholar] [CrossRef]
- Hay, N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 2016, 16, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Soga, T. Cancer metabolism: Key players in metabolic reprogramming. Cancer Sci. 2013, 104, 275–281. [Google Scholar] [CrossRef]
- Pinheiro, C.; Garcia, E.A.; Morais-Santos, F.; Scapulatempo-Neto, C.; Mafra, A.; Steenbergen, R.D.M.; Boccardo, E.; Villa, L.L.; Baltazar, F.; Longatto-Filho, A. Lactate transporters and vascular factors in HPV-induced squamous cell carcinoma of the uterine cervix. BMC Cancer 2014, 14, 751. [Google Scholar] [CrossRef]
- Abudula, A.; Rouzi, N.; Xu, L.; Yang, Y.; Hasimu, A. Tissue-based metabolomics reveals potential biomarkers for cervical carcinoma and HPV infection. Bosn. J. Basic Med. Sci. 2020, 5, 78–87. [Google Scholar] [CrossRef]
- Li, B.; Sui, L. Metabolic reprogramming in cervical cancer and metabolomics perspectives. Nutr. Metab. 2021, 18, 93. [Google Scholar] [CrossRef]
- Arizmendi-Izazaga, A.; Navarro-Tito, N.; Jiménez-Wences, H.; Evaristo-Priego, A.; Priego-Hernández, V.D.; Dircio-Maldonado, R.; Zacapala-Gómez, A.E.; Mendoza-Catalán, M.Á.; Illades-Aguiar, B.; De Nova Ocampo, M.A.; et al. Bioinformatics Analysis Reveals E6 and E7 of HPV 16 Regulate Metabolic Reprogramming in Cervical Cancer, Head and Neck Cancer, and Colorectal Cancer Through the PHD2-VHL-CUL2-ELOC-HIF-1α Axis. Curr. Issues Mol. Biol. 2024, 46, 6199–6222. [Google Scholar] [CrossRef]
- Martínez-Ramírez, I.; Carrillo-García, A.; Contreras-Paredes, A.; Ortiz-Sánchez, E.; Cruz-Gregorio, A.; Lizano, M. Regulation of cellular metabolism by high-risk human papillomaviruses. Int. J. Mol. Sci. 2018, 19, 1839. [Google Scholar] [CrossRef]
- Arizmendi-Izazaga, A.; Navarro-Tito, N.; Jiménez-Wences, H.; Mendoza-Catalán, M.A.; Martínez-Carrillo, D.N.; Zacapala-Gómez, A.E.; Olea-Flores, M.; Dircio-Maldonado, R.; Torres-Rojas, F.I.; Soto-Flores, D.G.; et al. Metabolic reprogramming in cancer: Role of hpv 16 variants. Pathogens 2021, 10, 347. [Google Scholar] [CrossRef]
- Karekar, A.K.; Dandekar, S.P. Cancer metabolomics: A tool of clinical utility for early diagnosis of gynaecological cancers. Indian J. Med. Res. 2021, 154, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Bailey, N.J.C.; Johnson, H.E. Measuring the metabolome: Current analytical technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, L.; Huang, X.; Peng, Q.; Zhang, S.; Tang, J.; Wang, J.; Gui, D.; Zeng, F. High-throughput metabolomics identifies new biomarkers for cervical cancer. Discov. Oncol. 2024, 15, 9. [Google Scholar] [CrossRef] [PubMed]
- Scotto, L.; Gopeshwar, N.; Nandula, S.V.; Arias-Pulido, H.; Subramaniyam, S.; Schneider, A.; Kaufmann, A.M.; Wright, J.D.; Pothuri, B.; Mansukhani, M.; et al. Identification of Copy Number Gain and Overexpressed Genes on Chromosome Arm 20q by an Integrative Genomic Approach in Cervical Cancer: Potential Role in Progression. Genes Chromosomes Cancer 2008, 47, 755–765. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; Macdonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E.I. Killing-Off of Tumor Cells In Vitro. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Kell, D.B.; Brown, M.; Davey, H.M.; Dunn, W.B.; Spasic, I.; Oliver, S.G. Metabolic footprinting and systems biology: The medium is the message. Nat. Rev. Microbiol. 2005, 3, 557–565. [Google Scholar] [CrossRef]
- Xu, H.; Liu, M.; Song, Y.; Liu, L.; Xu, F.; Chen, J.; Zhan, H.; Zhang, Y.; Chen, Y.; Lu, M.; et al. Metabolomics variation profiling of vaginal discharge identifies potential targets for cervical cancer early warning. Acta Biochim. Biophys. Sin. 2022, 54, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, Q.; Wang, T.; Liang, H.; Wang, Y.; Duan, Y.; Song, M.; Wang, Y. Exploring metabolomics biomarkers for evaluating the effectiveness of concurrent radiochemotherapy for cervical cancers. Transl. Cancer Res. 2020, 9, 2734–2747. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Xu, M.; Chen, J.; Chen, T.; Wang, Q.; Zhang, R.; Qiu, J. Plasma-Based Metabolomics Profiling of High-Risk Human Papillomavirus and their Emerging Roles in the Progression of Cervical Cancer. BioMed Res. Int. 2022, 2022, 6207701. [Google Scholar] [CrossRef]
- Vuckovic, D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2012, 403, 1523–1548. [Google Scholar] [CrossRef]
- Priego-Hernández, V.D.; Arizmendi-Izazaga, A.; Soto-Flores, D.G.; Santiago-Ramón, N.; Feria-Valadez, M.D.; Navarro-Tito, N.; Jiménez-Wences, H.; Martínez-Carrillo, D.N.; Salmerón-Bárcenas, E.G.; Leyva-Vázquez, M.A.; et al. Expression of HIF-1α and Genes Involved in Glucose Metabolism Is Increased in Cervical Cancer and HPV-16-Positive Cell Lines. Pathogens 2023, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Echelman, D.; Feldman, S. Management of cervical precancers: A global perspective, Hematol. Oncol. Clin. N. Am. 2012, 26, 31–44. [Google Scholar] [CrossRef]
- Kim, M.A.; Oh, J.K.; Kim, B.W.; Chay, D.; Park, D.C.; Kim, S.M.; Kang, E.S.; Kim, J.H.; Cho, C.H.; Shin, H.R.; et al. Prevalence and seroprevalence of low-risk human papillomavirus in Korean women. J. Korean Med. Sci. 2012, 27, 922–928. [Google Scholar] [CrossRef]
- Fontenelle, L.J.; Henderson, J.F. Sources of nitrogen as rate-limiting factors for purine biosynthesis de novo tn ehrlich ascites tumor cells. Biochim. Biophys. Acta 1969, 77, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Gaglio, D.; Soldati, C.; Vanoni, M.; Alberghina, L.; Chiaradonna, F. Glutamine Deprivation Induces Abortive S-Phase Rescued by Deoxyribonucleotides in K-Ras Transformed Fibroblasts. PLoS ONE 2009, 4, e4715. [Google Scholar] [CrossRef]
- Ramesh, D.; Vijayakumar, B.G.; Kannan, T. Therapeutic potential of uracil and its derivatives in countering pathogenic and physiological disorders. Eur. J. Med. Chem. 2020, 207, 112801. [Google Scholar] [CrossRef] [PubMed]
- Hilvo, M.; De Santiago, I.; Gopalacharyulu, P.; Schmitt, W.D.; Budczies, J.; Kuhberg, M. Accumulated metabolites of hydroxybutyric acid serve as diagnostic and prognostic biomarkers of ovarian high-grade serous carcinomas. Cancer Res. 2015, 4, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Vettukattil, R.; Eline, T.; Ann, V.; Kærn, J.; Davidson, B.; Bathen, T.F. Proton magnetic resonance metabolomic characterization of ovarian serous carcinoma effusions: Chemotherapy-related effects and comparison with malignant mesothelioma and breast carcinoma. Hum. Pathol. 2013, 9, 1859–1866. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Katada, S.; Imhof, A.; Sassone-corsi, P. Essay Connecting Threads: Epigenetics and Metabolism. Cell 2012, 148, 24–28. [Google Scholar] [CrossRef]
- Kalyanaraman, B. Teaching the basics of cancer metabolism: Developing antitumor strategies by exploiting the differences between normal and cancer cell metabolism. Redox Biol. 2017, 12, 833–842. [Google Scholar] [CrossRef]
- Jang, M.; Rhee, J.E.; Jang, D.H.; Kim, S.S. Gene Expression Profiles are Altered in Human Papillomavirus-16 E6 D25E-Expressing Cell Lines. Virol. J. 2011, 8, 6–11. [Google Scholar] [CrossRef]
- Ortiz-Pedraza, Y.; Muñoz-Bello, J.O.; Ramos-Chávez, L.A.; Martínez-Ramírez, I.; Olmedo-Nieva, L.; López-Saavedra, A.; Manzo-Merino, J.; la Cruz, V.P.-D.; Lizano, M. HPV16 E6 and E7 Oncoproteins Stimulate the Glutamine Pathway Maintaining Cell Proliferation in a SNAT1-Dependent Fashion. Viruses 2023, 15, 324. [Google Scholar] [CrossRef]
- Cuninghame, S.; Jackson, R.; Lees, S.J.; Zehbe, I. Two common variants of human papillomavirus type 16 E6 differentially deregulate sugar metabolism and hypoxia signalling in permissive human keratinocytes. J. Gen. Virol. 2017, 98, 2310–2319. [Google Scholar] [CrossRef]
- Mulukutla, B.C.; Yongky, A.; Le, T.; Mashek, D.G.; Hu, W.S. Regulation of Glucose Metabolism—A Perspective From Cell Bioprocessing. Trends Biotechnol. 2016, 34, 638–651. [Google Scholar] [CrossRef]
- Guo, Y.; Meng, X.; Ma, J.; Zheng, Y.; Wang, Q.; Wang, Y.; Shang, H. Human papillomavirus 16 E6 contributes HIF-1α induced warburg effect by attenuating the VHL-HIF-1α interaction. Int. J. Mol. Sci. 2014, 15, 7974–7986. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Huang, J.; Sui, M. Targeting arginine metabolism pathway to treat arginine-dependent cancers. Cancer Lett. 2015, 364, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jarrold, J.; Davies, C.C. PRMTs and Arginine Methylation: Cancer’s Best-Kept Secret? Trends Mol. Med. 2019, 25, 993–1009. [Google Scholar] [CrossRef]
- Lizano, M.; Carrillo-García, A.; de la Cruz-Hernández, E.; Castro-Muñoz, L.J.A. Contreras-Paredes, Promising predictive molecular biomarkers for cervical cancer (Review). Int. J. Mol. Med. 2024, 53, 50. [Google Scholar] [CrossRef]
- Puchades-Carrasco, L.; Jantus-Lewintre, E.; Pérez-Rambla, C.; García-García, F.; Lucas, R.; Calabuig, S.; Blasco, A.; Dopazo, J.; Camps, C.; Pineda-Lucena, A. Serum metabolomic profiling facilitates the non-invasive identification of metabolic biomarkers associated with the onset and progression of non-small cell lung cancer. Oncotarget 2016, 7, 12904. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Foxall, P.J.D.; Spraul, M.; Farrant, R.D.; Lindon, J.C. 750 MHz 1H and 1H-13C NMR Spectroscopy of Human Blood Plasma. Anal. Chem. 1995, 67, 793–811. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Zhou, M.; Hampton, C.Y.; Benigno, B.B.; Walker, L.D.E.; Gray, A.; McDonald, J.F.; Fernández, F.M. Ovarian cancer detection from metabolomic liquid chromatography/mass spectrometry data by support vector machines. BMC Bioinform. 2009, 10, 259. [Google Scholar] [CrossRef]
- Wang, L.; Tang, Y.; Liu, S.; Mao, S.; Ling, Y.; Liu, D.; He, X.; Wang, X. Metabonomic Profiling of Serum and Urine by 1H NMR-Based Spectroscopy Discriminates Patients with Chronic Obstructive Pulmonary Disease and Healthy Individuals. PLoS ONE 2013, 8, e65675. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; et al. NCBI GEO: Archive for gene expression and epig enomics data sets: 23-year update. Nucleic Acids Res. 2024, 52, D138–D144. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]


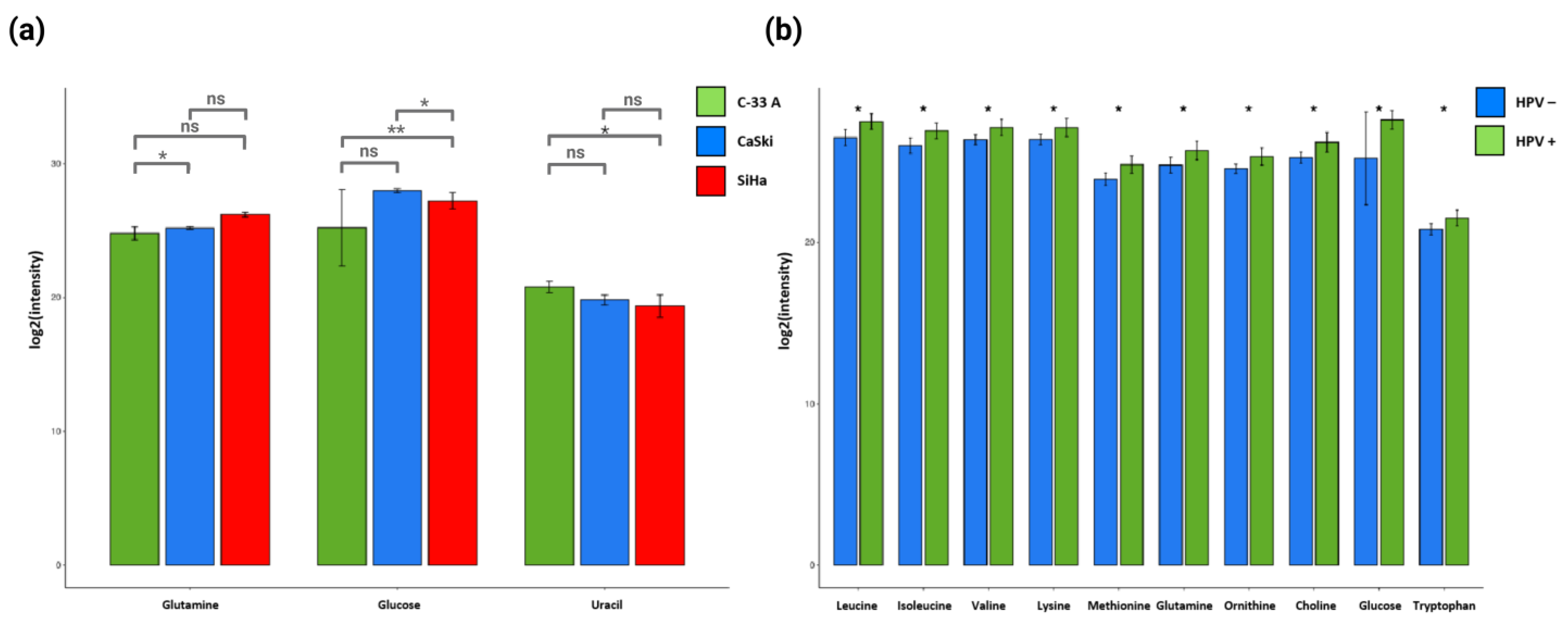


| Metabolite | Integration Area | T-Test SiHa vs. CaSki | T-Test C-33 A vs. SiHa | T-Test C-33 A vs. CaSki | T-Test HPV− vs. HPV+ |
|---|---|---|---|---|---|
| Leucine | 0.9442–0.9676 | 0.2407 | 0.0679 | 0.1018 | 0.0204 * |
| Isoleucine | 0.9889–1.0137 | 0.2556 | 0.0772 | 0.1214 | 0.0269 * |
| Valine | 1.0141–1.0419 | 0.2849 | 0.1150 | 0.1468 | 0.0313 * |
| 3-hydroxybutyrate | 1.1986–1.2222 | 0.3157 | 0.3516 | 0.8938 | 0.5077 |
| 3-hydroxyisovalerate | 1.2505–1.2595 | 0.1539 | 0.8354 | 0.1721 | 0.3751 |
| Alanine | 1.4511–1.4935 | 0.3036 | 0.1459 | 0.2276 | 0.0724 |
| Lysine | 1.6981–1.7369 | 0.2092 | 0.1081 | 0.2252 | 0.0458 * |
| Acetic acid | 1.8904–1.9226 | 0.2193 | 0.6337 | 0.1897 | 0.3280 |
| Methionine | 2.1293–2.1333 | 0.1189 | 0.0584 | 0.1362 | 0.0296 * |
| Glutamate | 2.3255–2.3534 | 0.2576 | 0.2503 | 0.9440 | 0.2456 |
| Pyruvate | 2.3586–2.3681 | 0.2742 | 0.2559 | 0.7578 | 0.2140 |
| Glutamine | 2.4254–2.4571 | 0.0153 * | 0.0039 ** | 0.2779 | 0.0358 * |
| Citrate | 2.6594–2.6741 | 0.3286 | 0.3681 | 0.8566 | 0.4766 |
| 2-oxoglutarate | 2.9752–3.0056 | 0.2067 | 0.2635 | 0.4724 | 0.3942 |
| Ornithine | 3.0491–3.0755 | 0.9719 | 0.2148 | 0.0946 | 0.0217* |
| Choline | 3.1807–3.1953 | 0.1825 | 0.1000 | 0.1130 | 0.0364 * |
| Glycine | 3.546–3.5583 | 0.3019 | 0.1619 | 0.2208 | 0.0536 |
| Lactate | 4.0728–4.133 | 0.2313 | 0.3054 | 0.3710 | 0.4316 |
| Pyroglutamate | 4.142–4.187 | 0.2502 | 0.2899 | 0.7799 | 0.4000 |
| Threonine | 4.2218–4.2753 | 0.1645 | 0.1909 | 0.7717 | 0.3239 |
| Glucose | 5.2005–5.2506 | 0.1387 | 0.2339 | 0.0288 * | 0.0498 * |
| Uracil | 5.7844–5.8088 | 0.4696 | 0.0462 * | 0.0773 | 0.0637 |
| Fumarate | 6.5–6.5142 | 0.3342 | 0.2578 | 0.4848 | 0.1814 |
| Tyrosine | 6.8701–6.9202 | 0.2801 | 0.1660 | 0.2971 | 0.0771 |
| Histidine | 7.0391–7.1008 | 0.3013 | 0.1875 | 0.3953 | 0.1051 |
| Phenylalanine | 7.3529–7.443 | 0.2571 | 0.1543 | 0.2425 | 0.0647 |
| Tryptophan | 7.7141–7.741 | 0.3551 | 0.1391 | 0.1629 | 0.0390 * |
| Formate | 8.4346–8.4565 | 0.4352 | 0.2927 | 0.3567 | 0.1388 |
| Nicotinate | 8.589–8.6114 | 0.5253 | 0.5427 | 0.1556 | 0.2038 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arizmendi-Izazaga, A.; Navarro-Tito, N.; Campos-Viguri, G.E.; Jiménez-Wences, H.; Acevedo-Quiroz, M.E.; Salmerón-Bárcenas, E.G.; Illades-Aguiar, B.; Leyva-Vázquez, M.A.; Ortiz-Ortiz, J. Exometabolome and Molecular Signatures Associated with HPV 16 in Cervical Cancer: Integrative Metabolomic and Transcriptomic Analysis for Biomarker Discovery. Molecules 2025, 30, 3909. https://doi.org/10.3390/molecules30193909
Arizmendi-Izazaga A, Navarro-Tito N, Campos-Viguri GE, Jiménez-Wences H, Acevedo-Quiroz ME, Salmerón-Bárcenas EG, Illades-Aguiar B, Leyva-Vázquez MA, Ortiz-Ortiz J. Exometabolome and Molecular Signatures Associated with HPV 16 in Cervical Cancer: Integrative Metabolomic and Transcriptomic Analysis for Biomarker Discovery. Molecules. 2025; 30(19):3909. https://doi.org/10.3390/molecules30193909
Chicago/Turabian StyleArizmendi-Izazaga, Adán, Napoleón Navarro-Tito, Gabriela Elizabeth Campos-Viguri, Hilda Jiménez-Wences, Macdiel Emilio Acevedo-Quiroz, Eric Genaro Salmerón-Bárcenas, Berenice Illades-Aguiar, Marco Antonio Leyva-Vázquez, and Julio Ortiz-Ortiz. 2025. "Exometabolome and Molecular Signatures Associated with HPV 16 in Cervical Cancer: Integrative Metabolomic and Transcriptomic Analysis for Biomarker Discovery" Molecules 30, no. 19: 3909. https://doi.org/10.3390/molecules30193909
APA StyleArizmendi-Izazaga, A., Navarro-Tito, N., Campos-Viguri, G. E., Jiménez-Wences, H., Acevedo-Quiroz, M. E., Salmerón-Bárcenas, E. G., Illades-Aguiar, B., Leyva-Vázquez, M. A., & Ortiz-Ortiz, J. (2025). Exometabolome and Molecular Signatures Associated with HPV 16 in Cervical Cancer: Integrative Metabolomic and Transcriptomic Analysis for Biomarker Discovery. Molecules, 30(19), 3909. https://doi.org/10.3390/molecules30193909









