Rapid G4 Ligand Screening Through Spectral Changes Using HT-SRCD with Minimal Material
Abstract
1. Introduction
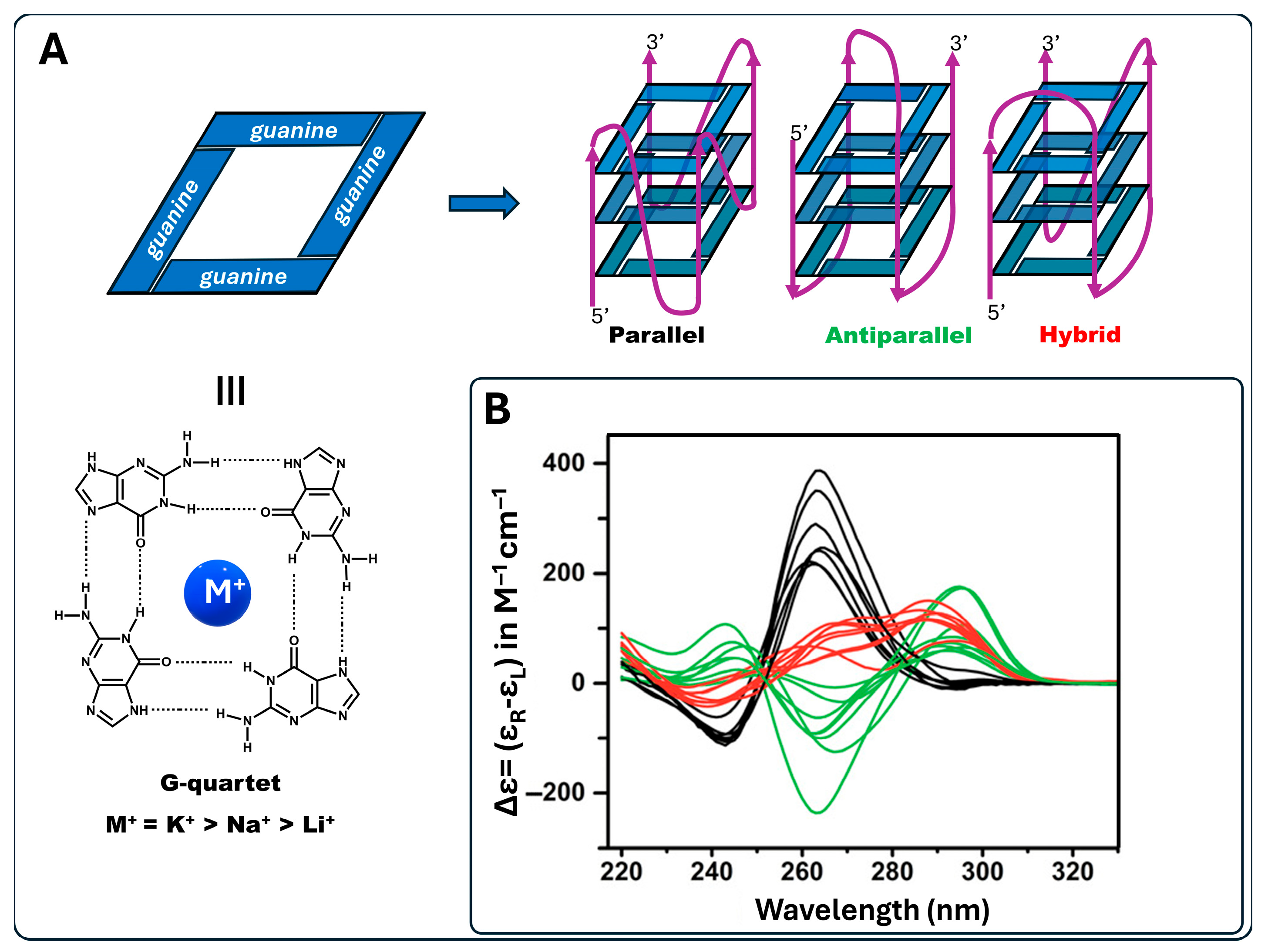
2. Results
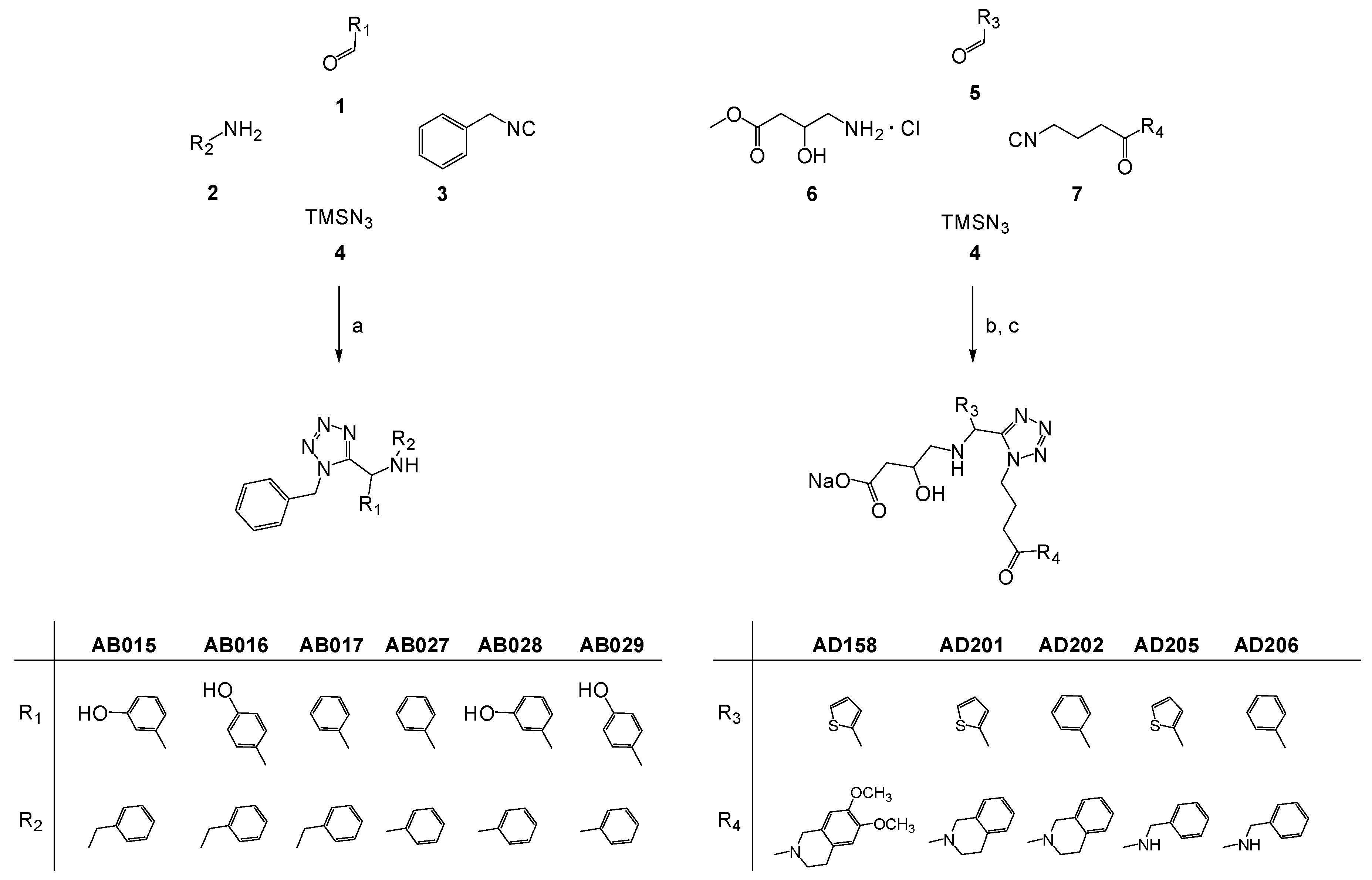
2.1. Ligand Design and Synthesis
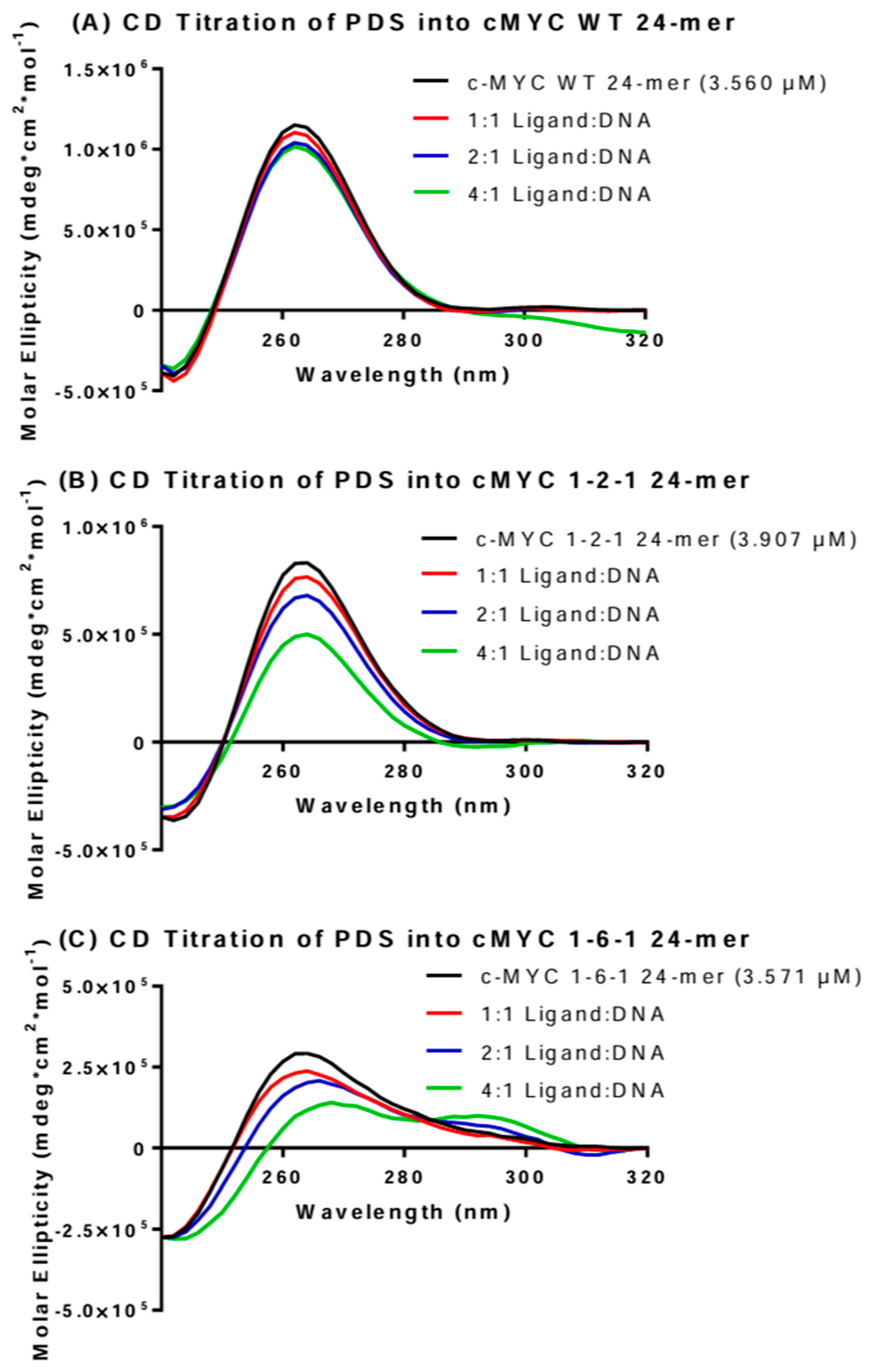
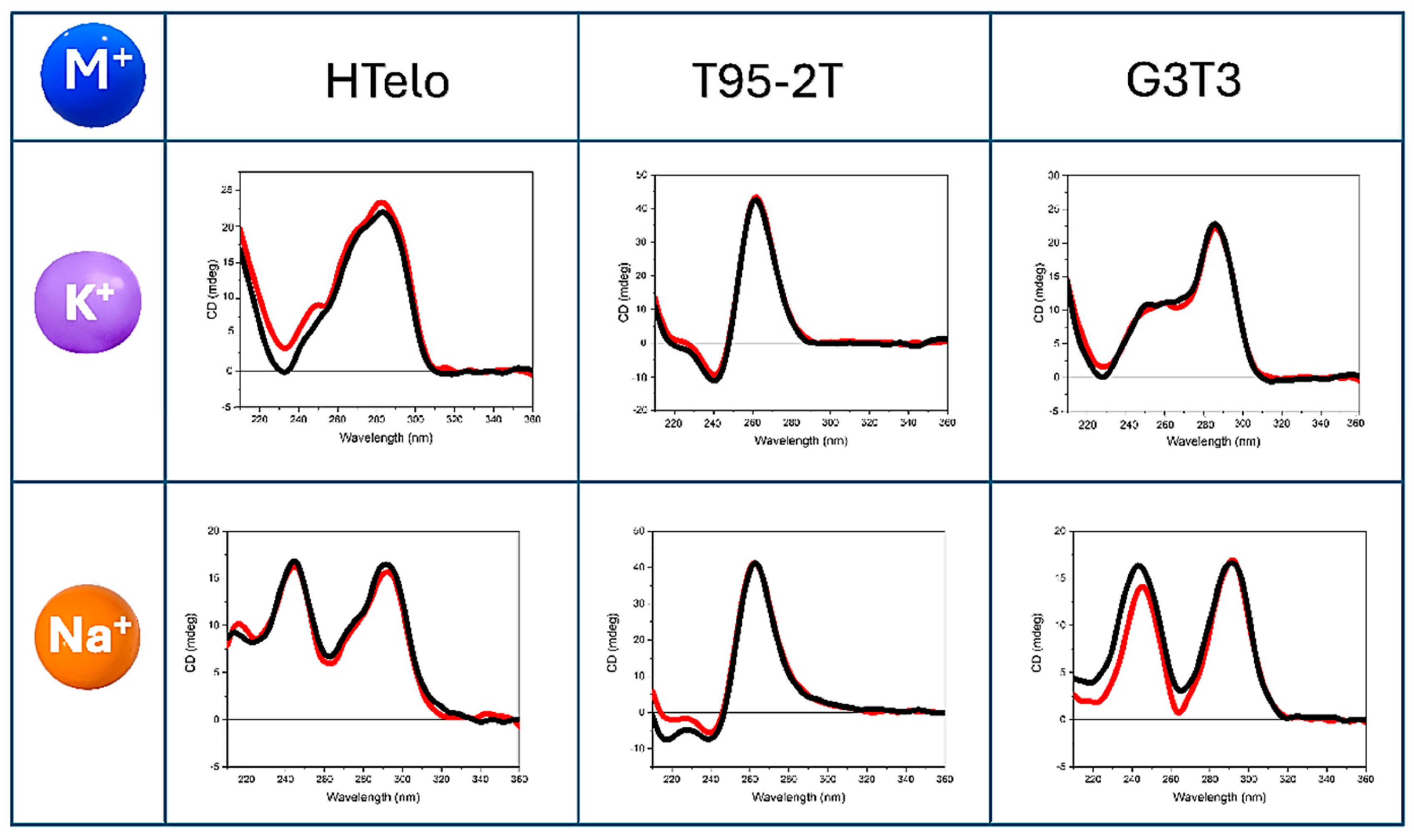
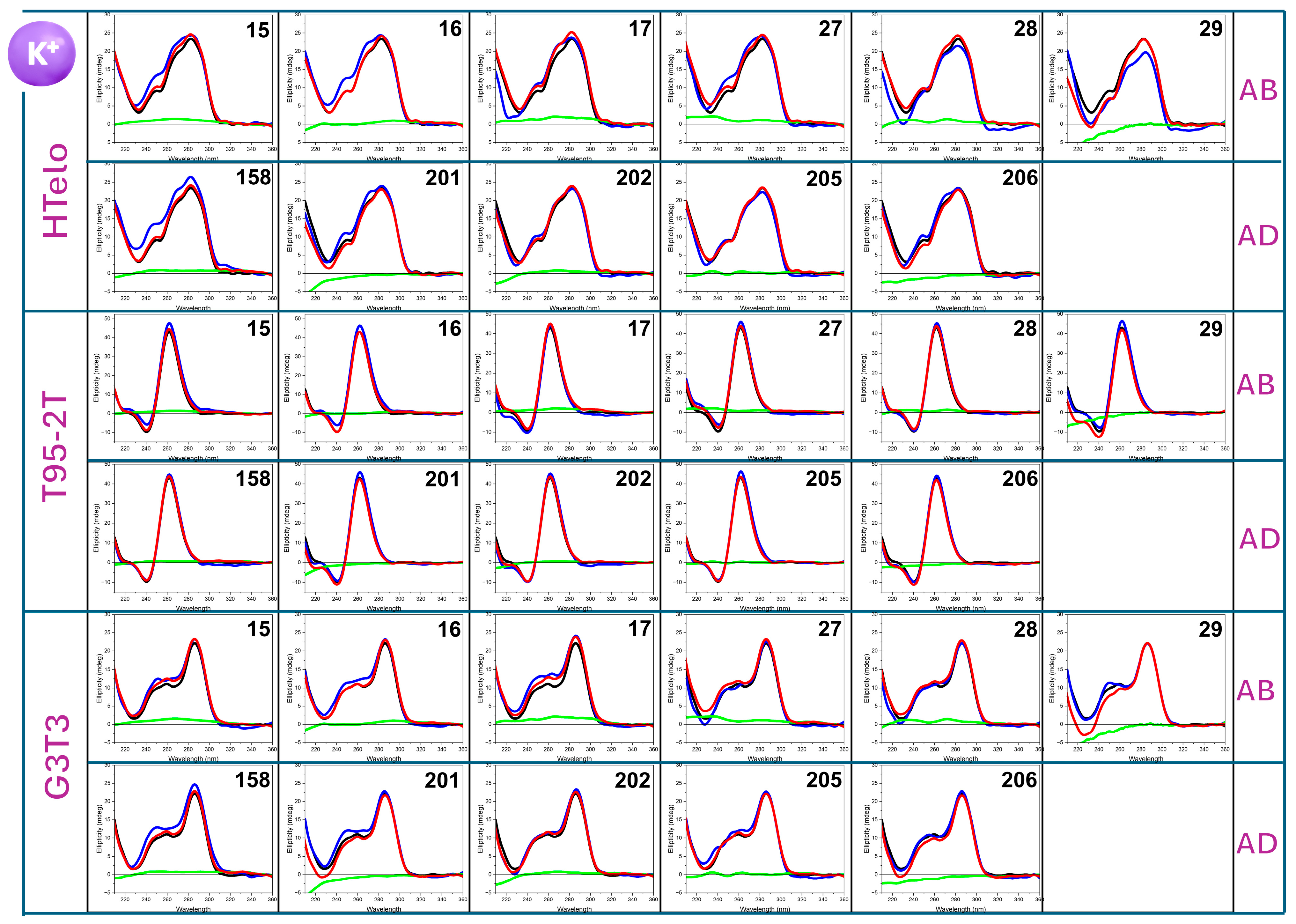
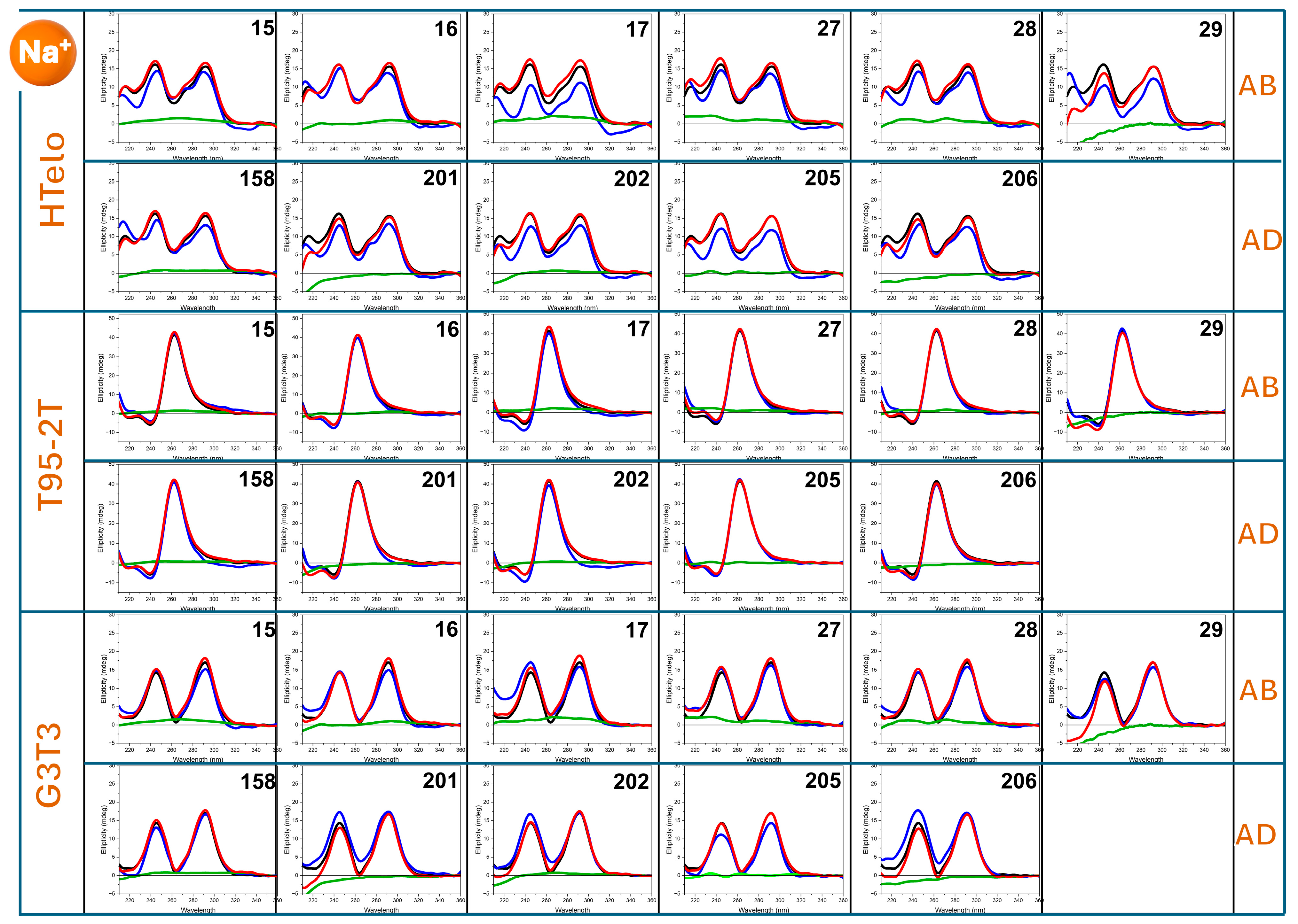
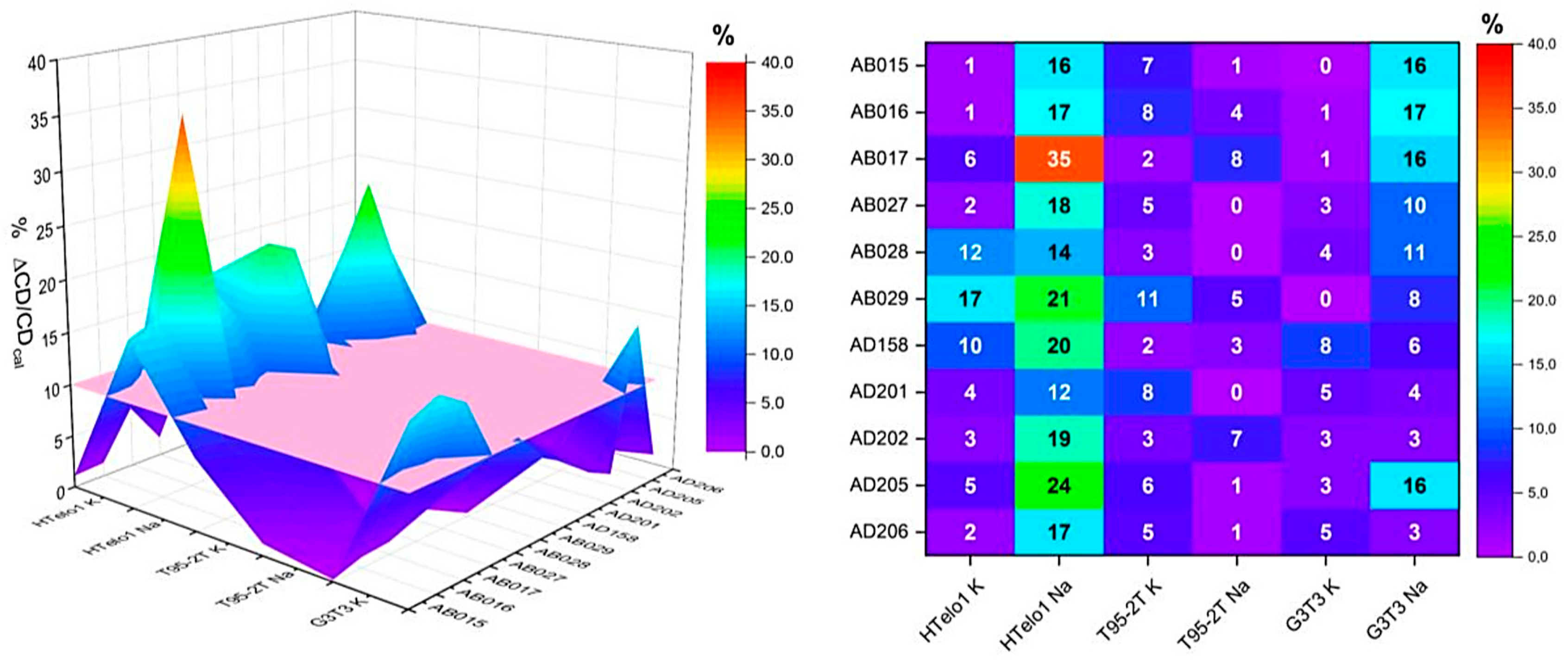
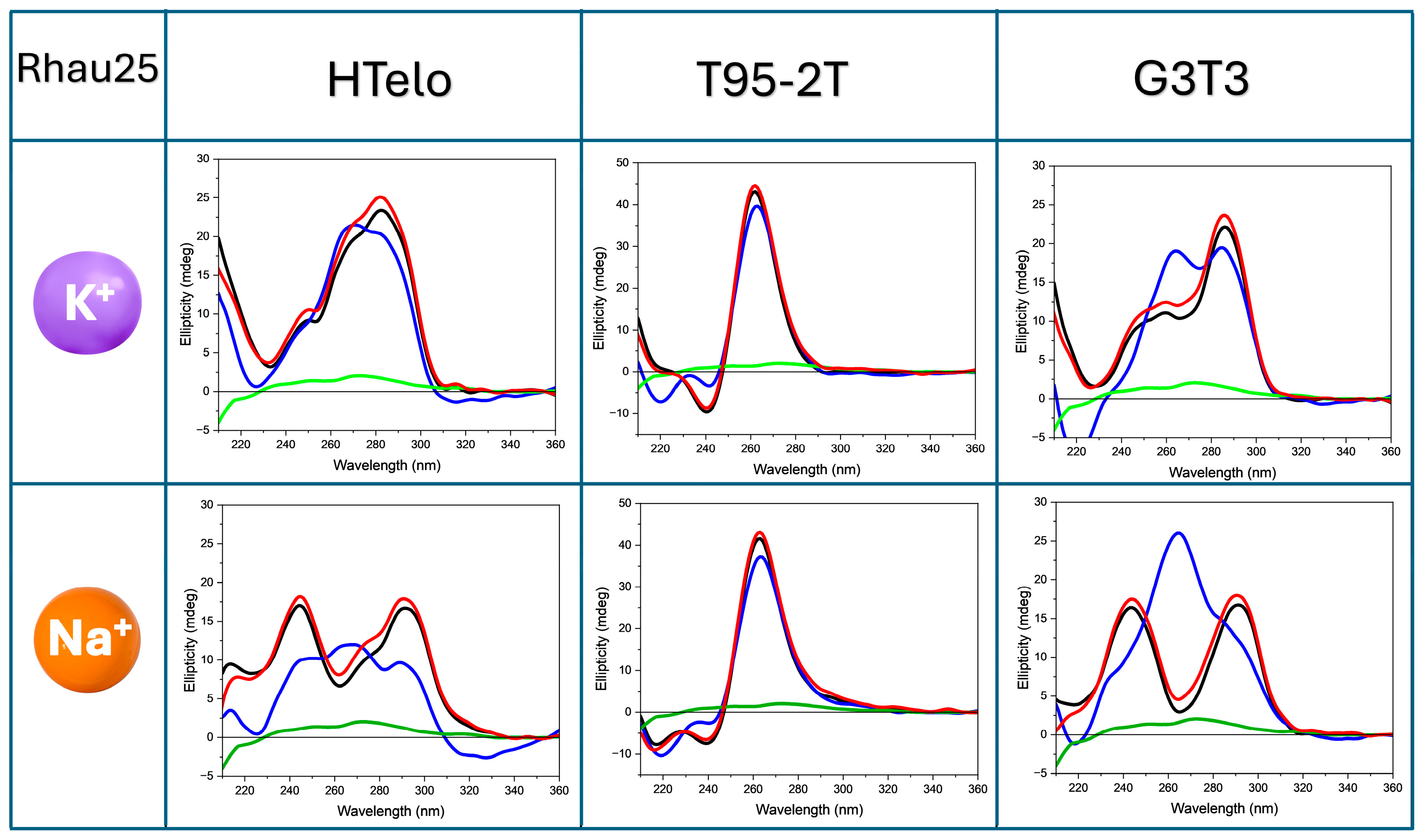
2.2. HT-SRCD Analysis of G4–Ligand Interactions
3. Materials and Methods
3.1. Ligand Synthesis
3.1.1. General MCR Procedure for the Synthesis of 1,5-Disubstituted α-Amino Tetrazoles Series AB
3.1.2. General MCR Procedure for the Synthesis of 1,5-Disubstituted α-Amino Tetrazoles Series AD
3.2. Peptide Synthesis
3.3. Preparation of G-Quadruplex Sequences
3.4. Preparation of Small-Molecule Ligands
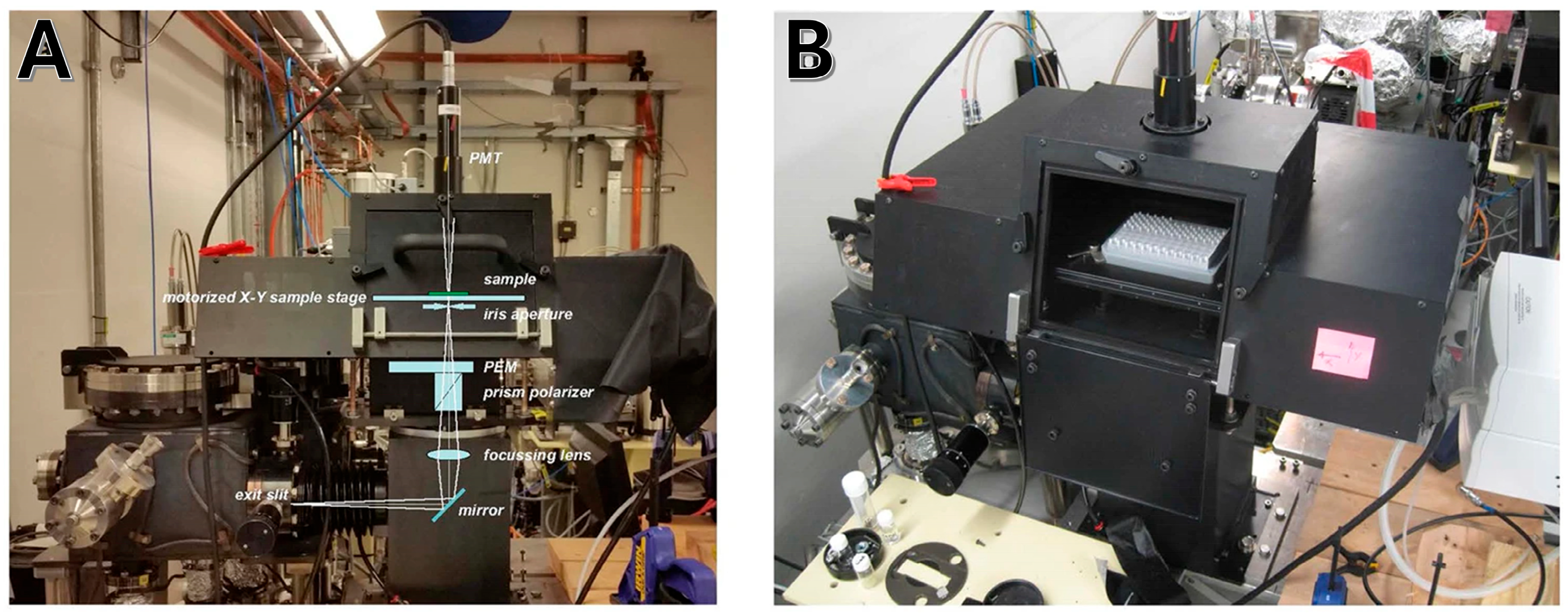
3.5. High-Throughput Synchrotron Radiation Circular Dichroism (HT-SRCD)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tateishi-Karimata, H.; Sugimoto, N. Chemical biology of non-canonical structures of nucleic acids for therapeutic applications. Chem. Commun. 2020, 56, 2379–2390. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef]
- Smargiasso, N.; Rosu, F.; Hsia, W.; Colson, P.; Baker, E.S.; Bowers, M.T.; De Pauw, E.; Gabelica, V. G-Quadruplex DNA Assemblies: Loop Length, Cation Identity, and Multimer Formation. J. Am. Chem. Soc. 2008, 130, 10208–10216. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef]
- Joachimi, A.; Benz, A.; Hartig, J.S. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorganic Med. Chem. 2009, 17, 6811–6815. [Google Scholar] [CrossRef]
- Lyu, J.; Shao, R.; Kwong Yung, P.Y.; Elsässer, S.J. Genome-wide mapping of G-quadruplex structures with CUT & Tag. Nucleic Acids Res. 2021, 50, e13. [Google Scholar]
- Yang, S.Y.; Chang, E.Y.C.; Lim, J.; Kwan, H.H.; Monchaud, D.; Yip, S.; Stirling Peter, C.; Wong, J.M.Y. G-quadruplexes mark alternative lengthening of telomeres. NAR Cancer 2021, 3, zcab031. [Google Scholar] [CrossRef]
- Jain, A.K.; Bhattacharya, S. Recent developments in the chemistry and biology of G-quadruplexes with reference to the DNA groove binders. Curr. Pharm. Des. 2012, 18, 1917–1933. [Google Scholar] [CrossRef]
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Domíniguez Moreno, H.; Schotta, G.; Richter, S.N. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 2021, 12, 3885. [Google Scholar] [CrossRef]
- Sato, K.; Knipscheer, P. G-quadruplex resolution: From molecular mechanisms to physiological relevance. DNA Repair 2023, 130, 103552. [Google Scholar] [CrossRef]
- Bolduc, F.; Garant, J.-M.; Allard, F.; Perreault, J.-P. Irregular G-quadruplexes Found in the Untranslated Regions of Human mRNAs Influence Translation. J. Biol. Chem. 2016, 291, 21751–21760. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.H.; Qian, S.H.; Wei, D.; Chen, Z.X. In vivo dynamics and regulation of DNA G-quadruplex structures in mammals. Cell Biosci. 2023, 13, 117. [Google Scholar] [CrossRef]
- Diaz Escarcega, R.; Marshall, P.; Tsvetkov, A.S. G-quadruplex DNA and RNA in cellular senescence. Front. Aging 2024, 5, 1491389. [Google Scholar] [CrossRef]
- Tian, T.; Chen, Y.-Q.; Wang, S.-R.; Zhou, X. G-Quadruplex: A Regulator of Gene Expression and Its Chemical Targeting. Chem 2018, 4, 1314–1344. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Neidle, S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. [Google Scholar] [CrossRef]
- Perrone, R.; Lavezzo, E.; Riello, E.; Manganelli, R.; Palù, G.; Toppo, S.; Provvedi, R.; Richter, S.N. Mapping and characterization of G-quadruplexes in Mycobacterium tuberculosis gene promoter regions. Sci. Rep. 2017, 7, 5743. [Google Scholar] [CrossRef]
- Evans, L.; Kotar, A.; Valentini, M.; Filloux, A.; Jamshidi, S.; Plavec, J.; Rahman, K.M.; Vilar, R. Identification and characterisation of G-quadruplex DNA-forming sequences in the Pseudomonas aeruginosa genome. RSC Chem. Biol. 2023, 4, 94–100. [Google Scholar] [CrossRef]
- Carvalho, J.; Lopes-Nunes, J.; Campello, M.P.C.; Paulo, A.; Milici, J.; Meyers, C.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Human Papillomavirus G-Rich Regions as Potential Antiviral Drug Targets. Nucleic Acid Ther. 2021, 31, 68–81. [Google Scholar] [CrossRef]
- Perrone, R.; Nadai, M.; Frasson, I.; Poe, J.A.; Butovskaya, E.; Smithgall, T.E.; Palumbo, M.; Palù, G.; Richter, S.N. A Dynamic G-Quadruplex Region Regulates the HIV-1 Long Terminal Repeat Promoter. J. Med. Chem. 2013, 56, 6521–6530. [Google Scholar] [CrossRef] [PubMed]
- Harpster, C.; Boyle, E.; Musier-Forsyth, K.; Kankia, B. HIV-1 genomic RNA U3 region forms a stable quadruplex-hairpin structure. Biophys. Chem. 2021, 272, 106567. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.A.; Nguyen, T.T.T.; Dang, D.T. Specific binding of G-quadruplex in SARS-CoV-2 RNA by RHAU peptide. Curr. Res. Struct. Biol. 2024, 7, 100126. [Google Scholar] [CrossRef]
- Luo, D.; Liu, C.; Muturi, E.; Hu, Z.; Hong, W.; Gao, C.; Li, J.; Wei, H.; Wei, D.; Yang, H. Inhibition of SARS-CoV-2 replication in cells by G-quadruplex ligands. Med. Chem. Res. 2024, 33, 2169–2179. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem. Int. Ed. Engl. 2018, 57, 7171–7175. [Google Scholar] [CrossRef]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Imagawa, Y.; Nagano, K.; Maeda, R.; Nagahama, N.; Torii, T.; Kinoshita, N.; Takamiya, N.; Kawauchi, K.; Tatesishi-Karimata, H.; et al. Simple and fast screening for structure-selective G-quadruplex ligands. Chem. Commun. 2023, 59, 4891–4894. [Google Scholar] [CrossRef]
- Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Circular dichroism and guanine quadruplexes. Methods 2012, 57, 64–75. [Google Scholar] [CrossRef]
- Hussain, R.; Jávorfi, T.; Rudd, T.R.; Siligardi, G. High-throughput SRCD using multi-well plates and its applications. Sci. Rep. 2016, 6, 38028. [Google Scholar] [CrossRef]
- Siligardi, G.; Hussain, R. CD Spectroscopy: An Essential Tool for Quality Control of Protein Folding. In Structural Proteomics. Methods in Molecular Biology; Owens, R., Ed.; Humana Press: New York, NY, USA, 2015; Volume 1261. [Google Scholar]
- Hussain, R.; Jávorfi, T.; Hughes, C.S.; Siligardi, G. Circular Dichroism and Synchrotron Radiation Circular Dichroism Applications to Biomaterials. In Radiation in Bioanalysis; Pereira, A., Tavares, P., Limão-Vieira, P., Eds.; Springer: Cham, Switzerland, 2019; Volume 8. [Google Scholar]
- Hussain, R.; Siligardi, G. Characterisation of Conformational and Ligand Binding Properties of Membrane Proteins Using Synchrotron Radiation Circular Dichroism (SRCD). In The Next Generation in Membrane Protein Structure Determination. Advances in Experimental Medicine and Biology; Moraes, I., Ed.; Springer: Cham, Switzerland, 2016; Volume 922. [Google Scholar]
- Kwan, T.O.C.; Kolek, S.A.; Danson, A.E.; Reis, R.I.; Camacho, I.S.; Shaw Stewart, P.D.; Moraes, I. Measuring Protein Aggregation and Stability Using High-Throughput Biophysical Approaches. Front. Mol. Biosci. 2022, 16, 890862. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Townsend, D.J.; Mala, B.; Hughes, E.; Hussain, R.; Siligardi, G.; Fullwood, N.J.; Middleton, D.A. Circular Dichroism Spectroscopy Identifies the β-Adrenoceptor Agonist Salbutamol As a Direct Inhibitor of Tau Filament Formation in Vitro. ACS Chem. Neurosci. 2020, 11, 2104–2116. [Google Scholar] [CrossRef]
- Honisch, C.; Ragazzi, E.; Hussain, R.; Brazier, J.; Siligardi, G.; Ruzza, P. Interaction of a Short Peptide with G-Quadruplex-Forming Sequences: An SRCD and CD Study. Pharmaceutics 2021, 13, 1104. [Google Scholar] [CrossRef]
- Marcaccini, S.; Torroba, T. The use of the Ugi four-component condensation. Nat. Protoc. 2007, 2, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Ulgheri, F.; Spanu, P.; Deligia, F.; Loriga, G.; Fuggetta, M.P.; de Haan, I.; Chandgudge, A.; Groves, M.; Domling, A. Design, synthesis and biological evaluation of 1,5-disubstituted α-amino tetrazole derivatives as non-covalent inflammasome-caspase-1 complex inhibitors with potential application against immune and inflammatory disorders. Eur. J. Med. Chem. 2022, 229, 114002. [Google Scholar] [CrossRef] [PubMed]
- Cuny, Z.G. Characterization of Pyridostatin and its Interactions with c-MYC G-Quadruplexes. Bachelor Thesis, Mississippi State University, Starkville, MS, USA, 2017. [Google Scholar]
- Islam, B.; Stadlbauer, P.; Krepl, M.; Havrila, M.; Haider, S.; Sponer, J. Structural Dynamics of Lateral and Diagonal Loops of Human Telomeric G-Quadruplexes in Extended MD Simulations. J. Chem. Theory Comput. 2018, 14, 5011–5026. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.W.; Amrane, S.; Bouaziz, S.; Xu, W.; Mu, Y.; Patel, D.J.; Luu, K.N.; Phan, A.T. Structure of the Human Telomere in K+ Solution: A Stable Basket-Type G-Quadruplex with Only Two G-Tetrad Layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. [Google Scholar] [CrossRef]
- Villani, G. Quantum Mechanical Investigation of the G-Quadruplex Systems of Human Telomere. ACS Omega 2018, 3, 9934–9944. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, C.-X.; Di Felice, R.; Sponer, J.; Islam, B.; Stadlbauer, P.; Ding, Y.; Mao, L.; Mao, Z.-W.; Qin, P.Z. Conformations of Human Telomeric G-Quadruplex Studied Using a Nucleotide-Independent Nitroxide Label. Biochemistry 2016, 55, 360–372. [Google Scholar] [CrossRef]
- Hussain, R.; Benning, K.; Javorfi, T.; Longo, E.; Rudd, T.R.; Pulford, B.; Siligardi, G. CDApps: Integrated software for experimental planning and data processing at beamline B23, Diamond Light Source. J. Synchrotron Radiat. 2015, 22, 465–468. [Google Scholar] [CrossRef]
| Strand | Sequence |
|---|---|
| HTelo1 | 5′-TAGGGTTAGGGTTAGGGTTAGGG-3′ |
| T95-2T | 5′-TTGGGTGGGTGGGTGGGT-3′ |
| G3T3 | 5′-GGGTTTGGGTTTGGGTTTGGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotondo, M.; Honisch, C.; Spanu, P.; Ulgheri, F.; Loriga, G.; Beccu, A.; Hussain, R.; Biondi, B.; Ruzza, P.; Siligardi, G. Rapid G4 Ligand Screening Through Spectral Changes Using HT-SRCD with Minimal Material. Molecules 2025, 30, 3322. https://doi.org/10.3390/molecules30163322
Rotondo M, Honisch C, Spanu P, Ulgheri F, Loriga G, Beccu A, Hussain R, Biondi B, Ruzza P, Siligardi G. Rapid G4 Ligand Screening Through Spectral Changes Using HT-SRCD with Minimal Material. Molecules. 2025; 30(16):3322. https://doi.org/10.3390/molecules30163322
Chicago/Turabian StyleRotondo, Martina, Claudia Honisch, Pietro Spanu, Fausta Ulgheri, Giovanni Loriga, Andrea Beccu, Rohanah Hussain, Barbara Biondi, Paolo Ruzza, and Giuliano Siligardi. 2025. "Rapid G4 Ligand Screening Through Spectral Changes Using HT-SRCD with Minimal Material" Molecules 30, no. 16: 3322. https://doi.org/10.3390/molecules30163322
APA StyleRotondo, M., Honisch, C., Spanu, P., Ulgheri, F., Loriga, G., Beccu, A., Hussain, R., Biondi, B., Ruzza, P., & Siligardi, G. (2025). Rapid G4 Ligand Screening Through Spectral Changes Using HT-SRCD with Minimal Material. Molecules, 30(16), 3322. https://doi.org/10.3390/molecules30163322







