Hydrolytic Enzymes in the Secretome of the Mushrooms P. eryngii and P. ostreatus: A Comparison Between the Two Species
Abstract
1. Introduction
2. Results and Discussion
2.1. Substrate Collection
2.2. Protein Content
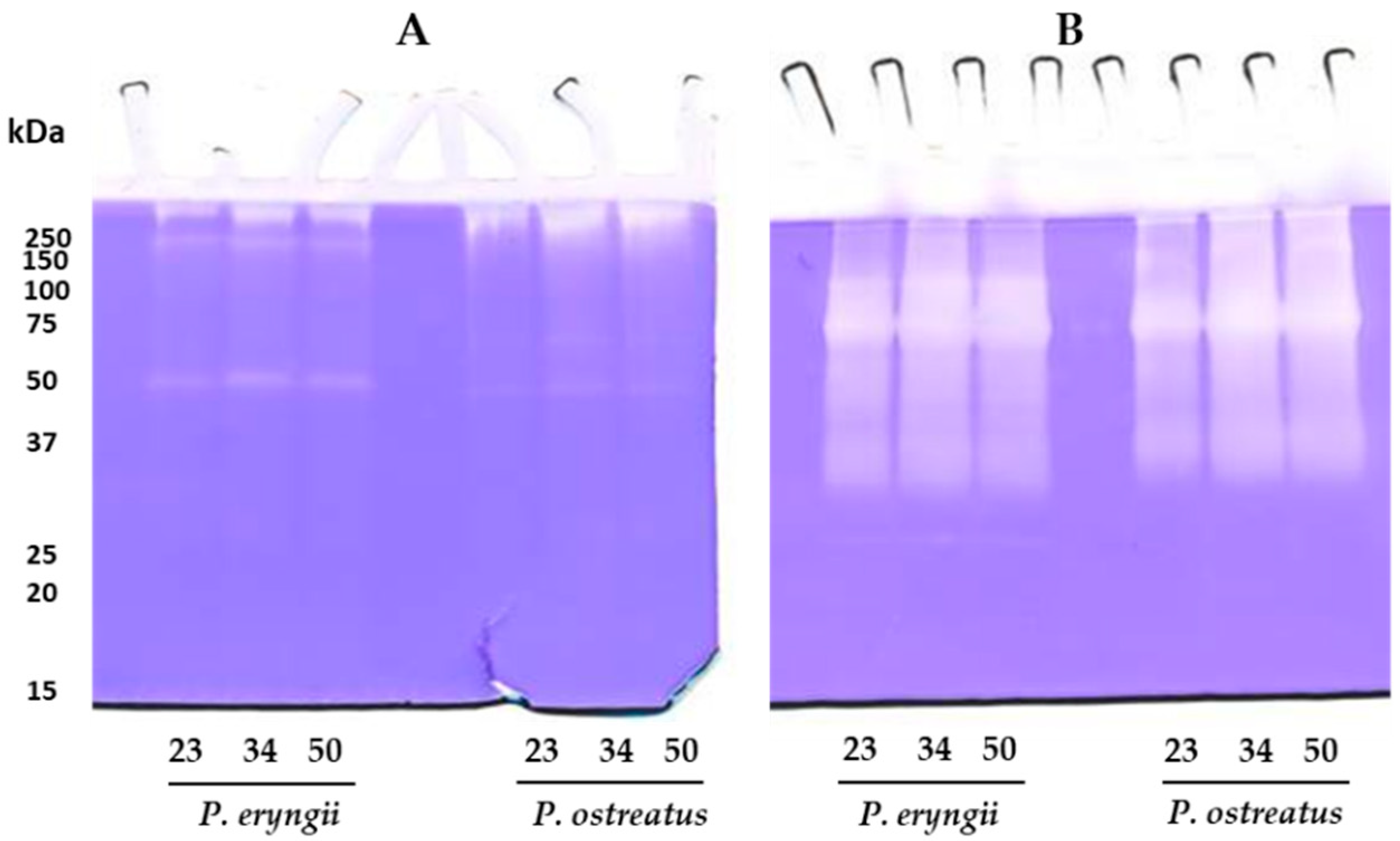
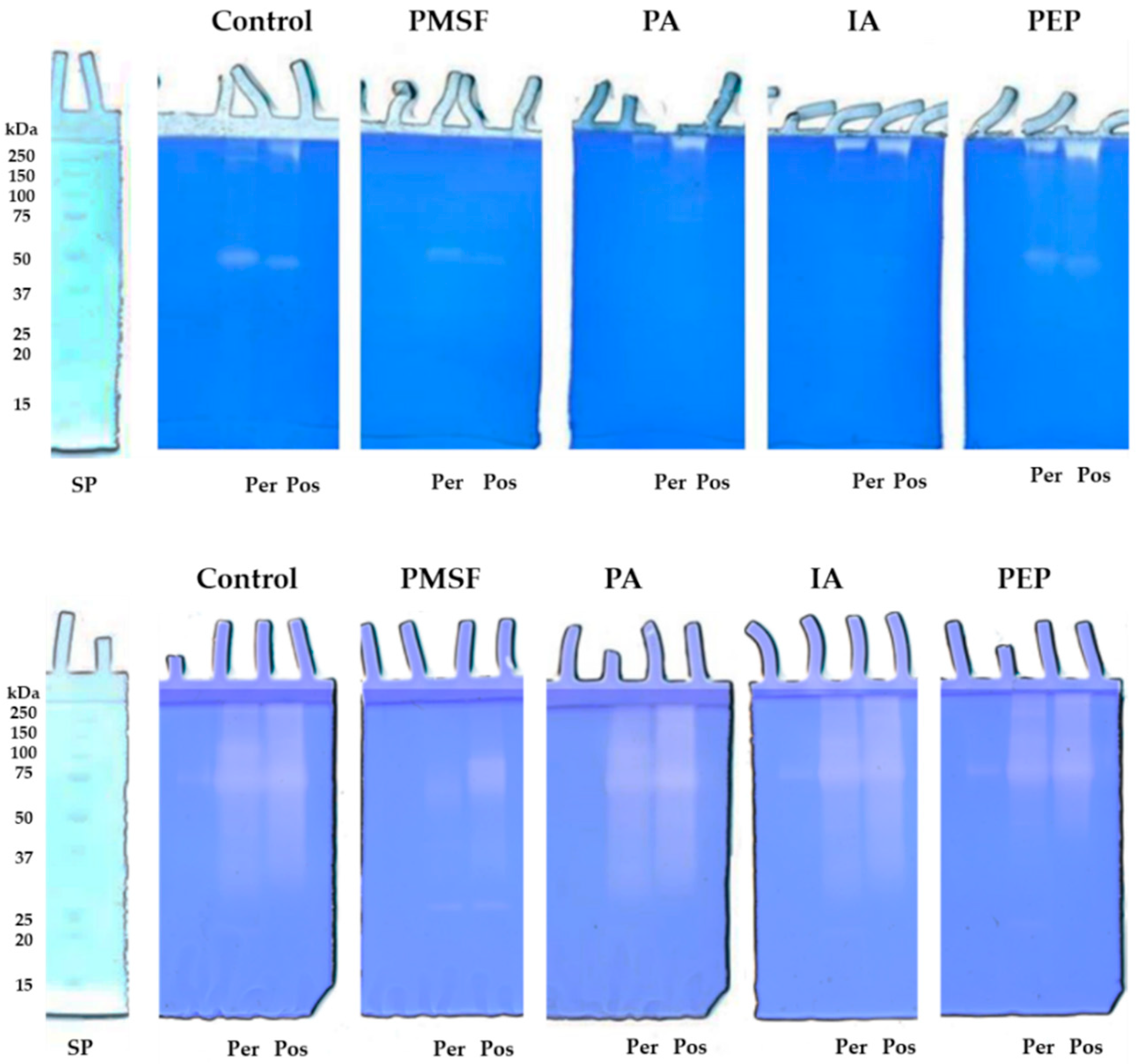
2.3. Total Proteolytic Activity Assessment and Zymographic Analysis
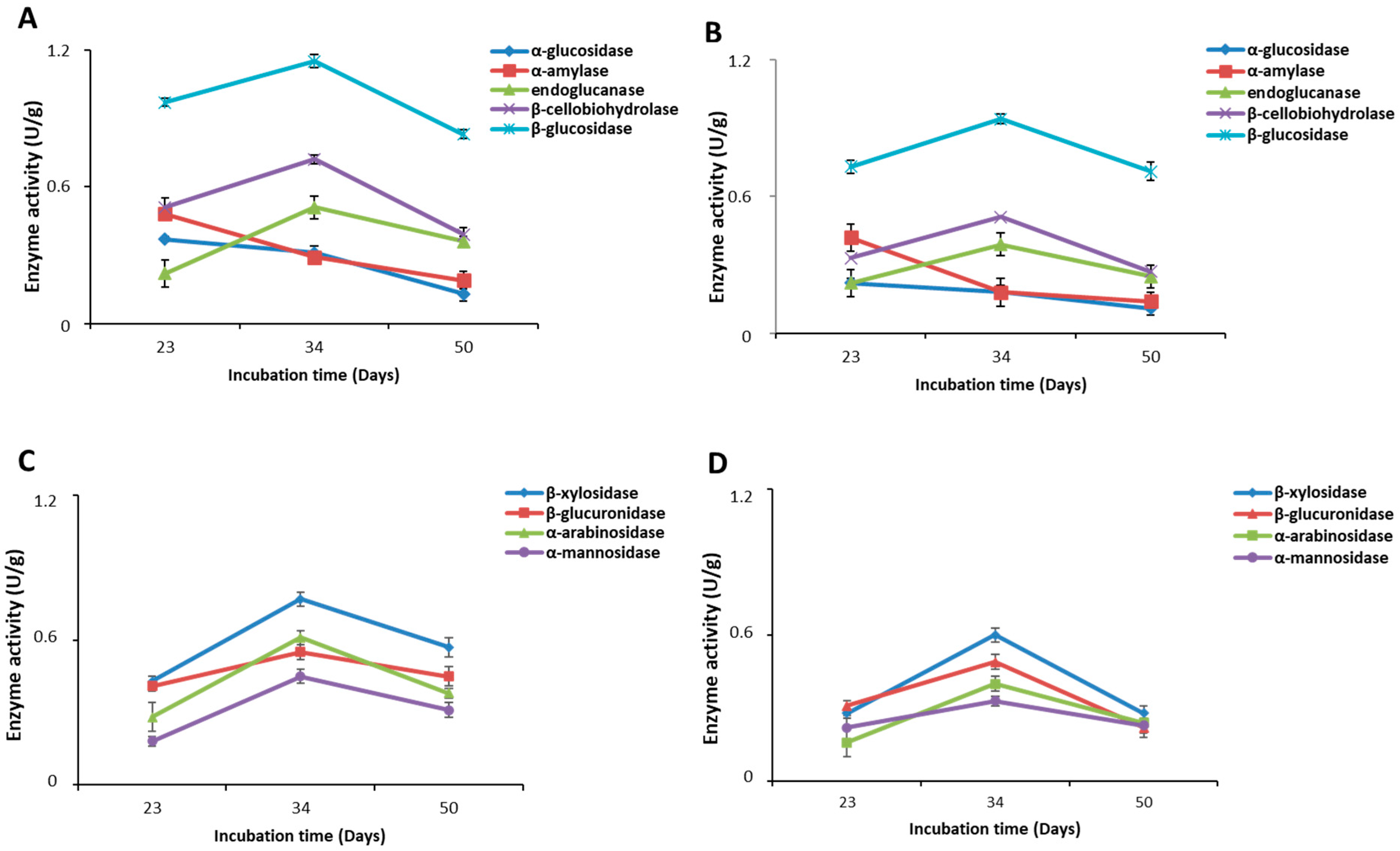
2.4. Carbohydrate-Hydrolyzing Enzymes
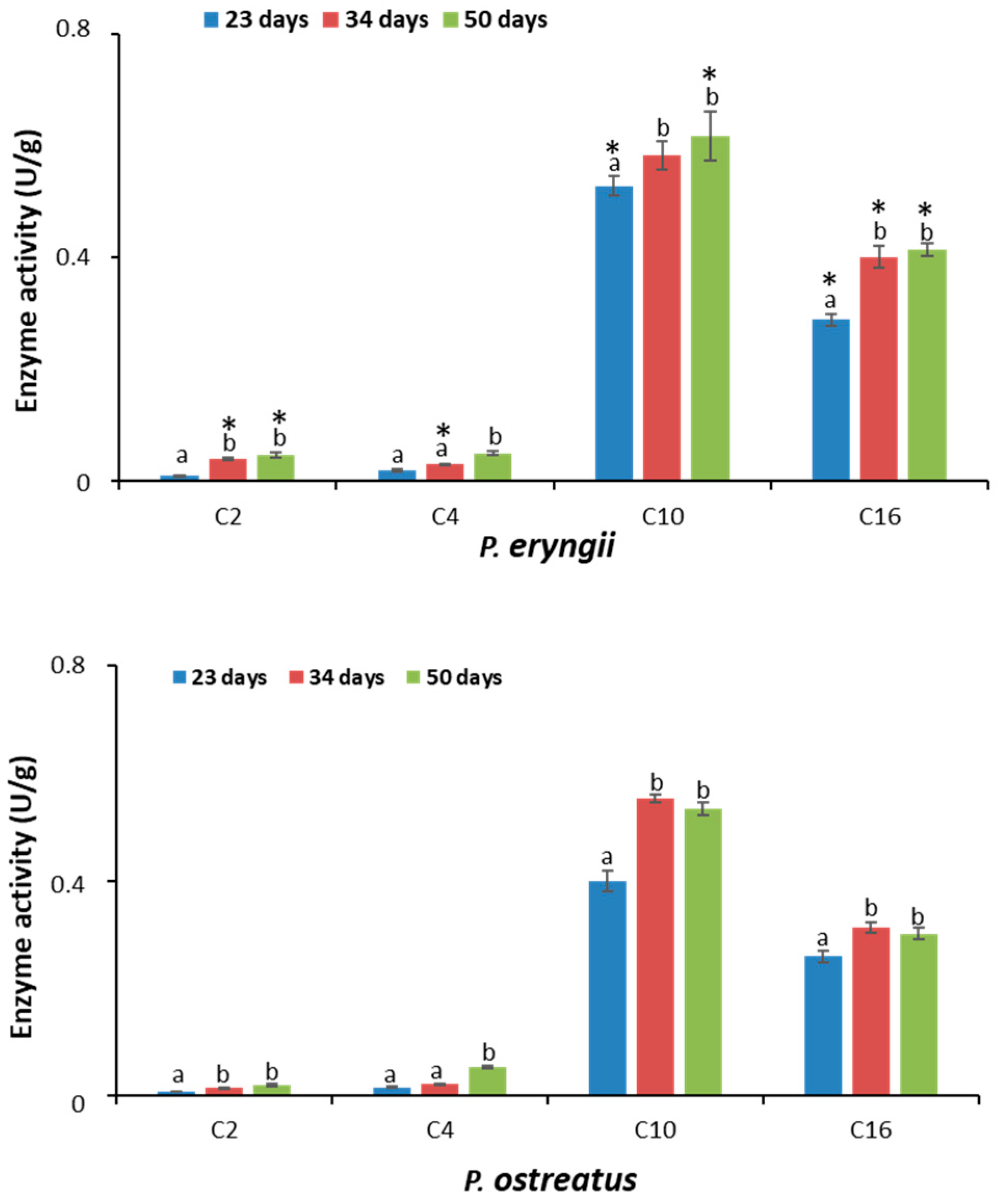
2.5. Lipolytic Activity
3. Materials and Methods
3.1. Mushroom Cultivation and Substrate Collection
3.2. Extraction of Extracellular Enzymes
3.3. Protein Content and Total Proteolytic Activity
3.4. Zymographic Analysis
3.5. Polysaccharide-Hydrolizing Activity
3.5.1. Cellulolytic and Amylolytic Activities
3.5.2. Hemicellulolytic Activity
3.6. Detection of Lipolytic Activity
4. Conclusions
5. Limitations of the Study
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ball, A.S.; Jackson, A.M. The recovery of lignocellulose-degrading enzymes from spent mushroom compost. Bioresour. Technol. 1995, 54, 311–314. [Google Scholar] [CrossRef]
- Phan, C.; Sabaratnam, V. Potential uses of spent mushroom substrate and its associated lignocellulosic enzymes. Appl. Microbiol. Biotechnol. 2012, 96, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Grujic, M.; Dojnov, B.; Potocnik, I.; Duduk, B.; Vujcic, Z. Spent mushroom compost as substrate for the production of industrially important hydrolytic enzymes by fungi Trichoderma spp. and Aspergillus niger in solid state fermentation. Int. Biodeterior. Biodegrad. 2015, 104, 290–298. [Google Scholar] [CrossRef]
- Ariff, I.N.M.; Bahrin, E.K.; Ramli, N.; Abd-Aziz, S. Direct use of spent mushroom substrate from Pleurotus pulmonarius as a readily delignified feedstock for cellulase production. Waste Biomass Valorization 2019, 10, 839–850. [Google Scholar] [CrossRef]
- Branà, M.T.; Sergio, L.; Haidukowski, M.; Logrieco, A.F.; Altomare, C. Degradation of Aflatoxin B1 by a Sustainable Enzymatic Extract from Spent Mushroom Substrate of Pleurotus eryngii. Toxins 2020, 12, 49. [Google Scholar] [CrossRef]
- Duran, K.; Magnin, J.; America, A.H.P.; Peng, M.; Hilgers, R.; de Vries, R.P.; Baars, J.J.P.; van Berkel, W.J.H.; Kuyper, T.W.; Kabel, M.A. The secretome of Agaricus bisporus: Temporal dynamics of plant polysaccharides and lignin degradation. iScience 2023, 26, 107087. [Google Scholar] [CrossRef]
- Kumla, J.; Suwannarach, N.; Sujarit, K.; Penkhrue, W.; Kakumyan, P.; Jatuwong, K.; Vadthanarat, S.; Lumyong, S. Cultivation of mushrooms and their lignocellulolytic enzyme pro duction through the utilization of agro-industrial waste. Molecules 2020, 25, 2811. [Google Scholar] [CrossRef]
- Martín, C.; Zervakis, G.I.; Xiong, S.; Koutrotsiosc, G.; Strætkvern, K.O. Spent substrate from mushroom cultivation: Exploitation potential toward various applications and value-added products. Bioengineered 2023, 14, 2252138. [Google Scholar] [CrossRef]
- Horn, S.J.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 2012, 5, 45. [Google Scholar] [CrossRef]
- Singh, A.D.; Abdullah, N.; Vikineswary, S. Optimization of extraction of bulk enzymes from spent mushroom compost. J. Chem. Technol. Biotechnol. 2003, 78, 743–752. [Google Scholar] [CrossRef]
- Ko, H.G.; Park, S.H.; Kim, S.H.; Kim, S.H.; Park, H.G.; Park, W.M. Detection and recovery of hydrolytic enzymes from spent compost of four mushroom species. Folia Microbiol. 2005, 50, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Lee, Y.H.; Kang, H.W. Efficient recovery of lignocellulolytic enzymes of spent mushroom compost from oyster mushrooms, Pleurotus spp., and potential use in dye decolorization. Mycobiology 2013, 41, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Kim, J.K.; Lee, Y.H. Production of lignocellulolytic enzymes from spent mushroom compost of Pleurotus eryngii. Korean J. Mycol. 2012, 40, 152–158. [Google Scholar] [CrossRef]
- Cohen, R.; Persky, L.; Hadar, Y. Biotechnological applications and potential of wood-degrading mushrooms of the genus Pleurotus. Appl. Microbiol. Biotechnol. 2002, 58, 582–594. [Google Scholar] [CrossRef]
- Inácio, F.D.; Ferreira, R.O.; de Araujo, C.A.; Brugnari, T.; Castoldi, R.; Peralta, R.M.; de Souza, C.G. Proteases of Wood Rot Fungi with Emphasis on the Genus Pleurotus. Biomed. Res. Int. 2015, 2015, 290161. [Google Scholar] [CrossRef]
- Cha, W.S.; Park, S.S.; Kim, S.J.; Choi, D. Biochemical and enzymatic properties of a fibrinolytic enzyme from Pleurotus eryngii cultivated under solid-state conditions using corn cob. Bioresour. Technol. 2010, 101, 6475–6481. [Google Scholar] [CrossRef]
- Cui, L.; Liu, Q.H.; Wang, H.X.; Ng, T.B. An alkaline protease from fresh fruiting bodies of the edible mushroom Pleurotus citrinopileatus. Appl. Microbiol. Biot. 2007, 75, 81–85. [Google Scholar] [CrossRef]
- Palmieri, G.; Giardina, P.; Bianco, C.; Fontanella, B.; Sannia, G. Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 2000, 66, 920–924. [Google Scholar] [CrossRef]
- Eisele, N.; Linke, D.; Nimtz, M.; Berger, R.G. Heterologous expression, refolding and characterization of a salt activated subtilase from Pleurotus ostreatus. Process Biochem. 2011, 46, 1840–1846. [Google Scholar] [CrossRef]
- Pawar, K.S.; Singh, P.N.; Singh, S.K. Fungal alkaline proteases and their potential applications in different industries. Front. Microbiol. 2023, 14, 1138401. [Google Scholar] [CrossRef]
- Bano, S.; Umar Dahot, M.; Naqvi, S.H.A. Optimization of culture conditions for the production of protease by Pleurotus eryngii. Pak. J. Biotechnol. 2016, 13, 193–198. [Google Scholar]
- Contato, A.G.; Inácio, F.D.; Bueno, P.S.A.; Nolli, M.M.; Janeiro, V.; Peralta, R.M.; de Souza, C.G.M. Pleurotus pulmonarius: A protease-producing white rot fungus in lignocellulosic residues. Int. Microbiol. 2023, 26, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Verma, V.; Dubey, V.K.; Srivastava, A.; Garg, S.K.; Singh, V.P.; Arora, P.K. Industrial applications of fungal lipases: A review. Front. Microbiol. 2023, 14, 1142536. [Google Scholar] [CrossRef] [PubMed]
- Krugener, S.; Zelena, K.; Zorn, H.; Nimtz, M.; Berger, R.G. Heterologous expression of an extracellular lipase from the basidiomycete Pleurotus sapidus. J. Mol. Catal. B Enzym. 2009, 57, 16–21. [Google Scholar] [CrossRef]
- Zelena, K.; Krugener, S.; Lunkenbein, S.; Zorn, H.; Berger, R.G. Functional expression of the lipase gene Lip2 of Pleurotus sapidus in Escherichia coli. Biotechnol. Lett. 2009, 31, 395–401. [Google Scholar] [CrossRef]
- Piscitelli, A.; Tarallo, V.; Guarino, L.; Sannia, G.; Birolo, L.; Pezzella, C. New lipases by mining of Pleurotus ostreatus genome. PLoS ONE 2017, 12, e0185377. [Google Scholar] [CrossRef]
- Dedousi, M.; Melanouri, E.M.; Diamantopoulou, P. Carposome productivity of Pleurotus ostreatus and Pleurotus eryngii growing on agro-industrial residues enriched with nitrogen, calcium salts and oils. Carbon Resour. Convers. 2023, 6, 150–165. [Google Scholar] [CrossRef]
- Nakagame, S.; Minagawa, H.; Motegi, N. Purification and Characterization of Class III Lipase from a White-Rot Fungus Pleurotus ostreatus. Appl. Biochem. Biotechnol. 2023, 195, 1085–1095. [Google Scholar] [CrossRef]
- Xie, C.; Luo, W.; Li, Z.; Yan, L.; Zhu, Z.; Wang, J.; Hu, Z.; Peng, Y. Secretome analysis of Pleurotus eryngii reveals enzymatic composition for ramie stalk degradation. Electrophoresis 2016, 37, 310–320. [Google Scholar] [CrossRef]
- Pena, A.; Babiker, R.; Chaduli, D.; Lipzen, A.; Wang, M.; Chovatia, M.; Rencoret, J.; Marques, G.; Sanchez-Ruiz, M.I.; Kijpornyongpan, T.; et al. A multiomic approach to understand how Pleurotus eryngii transforms non-woody lignocellulosic material. J. Fungi 2021, 7, 426. [Google Scholar] [CrossRef]
- Petraglia, T.; Latronico, T.; Liuzzi, G.M.; Fanigliulo, A.; Crescenzi, A.; Rossano, R. Edible Mushrooms as Source of Fibrin(ogen)olytic Enzymes: Comparison between Four Cultivated Species. Molecules 2022, 27, 8145. [Google Scholar] [CrossRef] [PubMed]
- Sabotic, J.; Trcek, T.; Popovic, T.; Brzin, J. Basidiomycetes harbour a hidden treasure of proteolytic diversity. J. Biotechnol. 2007, 128, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Fueyo, E.; Ruiz-Duenas, F.J.; Lopez-Lucendo, M.F.; Perez-Boada, M.; Rencoret, J.; Gutiérrez, A.; Pisabarro, A.G.; Ramírez, L.; Martínez, A.T. A secretomic view of woody and nonwoody lignocellulose degradation by Pleurotus ostreatus. Biotechnol. Biofuels 2016, 9, 49. [Google Scholar] [CrossRef]
- Verger, R. Interfacial activation of lipases: Facts and artefacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Larocca, M.; Rossano, R.; Santamaria, M.; Riccio, P. Analysis of pineapple [Ananas comosus (L.) Merr.] fruit proteinases by 2-D zymography and direct identification of the major zymographic spots by mass spectrometry. Food Chem. 2010, 123, 1334–1342. [Google Scholar] [CrossRef]
- Latronico, T.; Petraglia, T.; Sileo, C.; Bilancia, D.; Rossano, R.; Liuzzi, G.M. Inhibition of MMP-2 and MMP-9 by Dietary Antioxidants in THP-1 Macrophages and Sera from Patients with Breast Cancer. Molecules 2024, 29, 1718. [Google Scholar] [CrossRef]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- McCleary, B.V.; Draga, A. Measurement of β-glucan in mushrooms and mycelial products. J. AOAC Int. 2016, 99, 2. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]

| Proteins (mg prot/g) | Total Proteolytic Activity (U/g) | |||||
|---|---|---|---|---|---|---|
| Day 23 | Day 34 | Day 50 | Day 23 | Day 34 | Day 50 | |
| P. eryngii | * 1.33 ± 0.11 a | * 2.74 ± 0.16 b | * 1.97 ± 0.15 c | * 589.63 ± 12.85 a | * 790.33 ± 9.37 b | * 773.93 ± 19.34 b |
| P. ostreatus | 0.94 ± 0.06 a | 1.79 ± 0.14 b | 1.26 ± 0.11 c | 512.93 ± 12.27 a | 594.95 ± 13.06 b | 603.52 ± 8.11 b |
| P. eryngii (U/g) | P. ostreatus (U/g) | |||||
|---|---|---|---|---|---|---|
| Enzymes | Day 23 | Day 34 | Day 50 | Day 23 | Day 34 | Day 50 |
| α-glucosidase | * 0.37 ± 0.01 a | * 0.31 ± 0.03 b | 0.13 ± 0.03 c | 0.25 ± 0.02 a | 0.18 ± 0.03 b | 0.11 ± 0.01 c |
| α-amylase | 0.48 ± 0.02 a | * 0.29 ± 0.01 b | 0.19 ± 0.04 c | 0.42 ± 0.06 a | 0.21 ± 0.03 b | 0.14 ± 0.02 c |
| endoglucanase | 0.22 ± 0.06 a | * 0.51 ± 0.05 b | * 0.36 ± 0.02 c | 0.22 ± 0.06 a | 0.39 ± 0.05 b | 0.25 ± 0.05 a |
| β-cellobiohydrolase | * 0.51 ± 0.04 a | * 0.72 ± 0.02 b | * 0.39 ± 0.03 c | 0.33 ± 0.04 a | 0.51 ± 0.02 b | 0.27 ± 0.03 a |
| β-glucosidase | * 0.97 ± 0.02 a | * 1.15 ± 0.03 b | * 0.83 ± 0.02 c | 0.73 ± 0.03 a | 0.94 ± 0.02 b | 0.71 ± 0.04 a |
| β-xylanase | * 0.43 ± 0.02 a | * 0.77 ± 0.03 b | * 0.57 ± 0.03 c | 0.28 ± 0.02 a | 0.60 ± 0.03 b | 0.28 ± 0.03 a |
| β-glucuronidase | * 0.41 ± 0.02 a | 0.55 ± 0.03 b | * 0.45 ± 0.04 a | 0.31 ± 0.02 a | 0.49 ± 0.03 b | 0.22 ± 0.04 c |
| α-arabinosidase | * 0.28 ± 0.06 a | * 0.61 ± 0.03 b | * 0.38 ± 0.02 c | 0.16 ± 0.06 a | 0.640 ± 0.03 b | 0.24 ± 0.02 a |
| α-mannosidase | 0.18 ± 0.05 a | * 0.45 ± 0.02 b | * 0.31 ± 0.03 c | 0.22 ± 0.05 a | 0.35 ± 0.02 b | 0.23 ± 0.03 a |
| P. eryngii (U/g) | P. ostreatus (U/g) | |||||
|---|---|---|---|---|---|---|
| Substrates | Day 23 | Day 34 | Day 50 | Day 23 | Day 34 | Day 50 |
| C2 | 0.01 ± 0.001 a | * 0.041 ± 0.002 b | * 0.047 ± 0.004 b | 0.008 ± 0.001 a | 0.014 ± 0.002 b | 0.019 ± 0.002 b |
| C4 | 0.020 ± 0.006 | * 0.030 ± 0.001 a | 0.060 ± 0.007 b | 0.016 ± 0.002 a | 0.022 ± 0.004 a | 0.053 ± 0.003 b |
| C10 | * 0.53 ± 0.02 a | 0.58 ± 0.02 b | * 0.62 ± 0.04 b | 0.40 ± 0.02 a | 0.55 ± 0.04 b | 0.50 ± 0.01 b |
| C16 | * 0.29 ± 0.01 a | * 0.41 ± 0.05 b | * 0.44 ± 0.01 b | 0.24 ± 0.01 a | 0.32 ± 0.01 b | 0.30 ± 0.02 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petraglia, T.; Latronico, T.; Liuzzi, G.M.; Fanigliulo, A.; Crescenzi, A.; Rossano, R. Hydrolytic Enzymes in the Secretome of the Mushrooms P. eryngii and P. ostreatus: A Comparison Between the Two Species. Molecules 2025, 30, 2505. https://doi.org/10.3390/molecules30122505
Petraglia T, Latronico T, Liuzzi GM, Fanigliulo A, Crescenzi A, Rossano R. Hydrolytic Enzymes in the Secretome of the Mushrooms P. eryngii and P. ostreatus: A Comparison Between the Two Species. Molecules. 2025; 30(12):2505. https://doi.org/10.3390/molecules30122505
Chicago/Turabian StylePetraglia, Tania, Tiziana Latronico, Grazia Maria Liuzzi, Angela Fanigliulo, Aniello Crescenzi, and Rocco Rossano. 2025. "Hydrolytic Enzymes in the Secretome of the Mushrooms P. eryngii and P. ostreatus: A Comparison Between the Two Species" Molecules 30, no. 12: 2505. https://doi.org/10.3390/molecules30122505
APA StylePetraglia, T., Latronico, T., Liuzzi, G. M., Fanigliulo, A., Crescenzi, A., & Rossano, R. (2025). Hydrolytic Enzymes in the Secretome of the Mushrooms P. eryngii and P. ostreatus: A Comparison Between the Two Species. Molecules, 30(12), 2505. https://doi.org/10.3390/molecules30122505









