Synthesis of Cyclic Hexapeptides via the Hydrazide Method and Evaluation of Their Antibacterial Activities
Abstract
1. Introduction
2. Results
2.1. Synthesis of Cyclic Hexapeptides
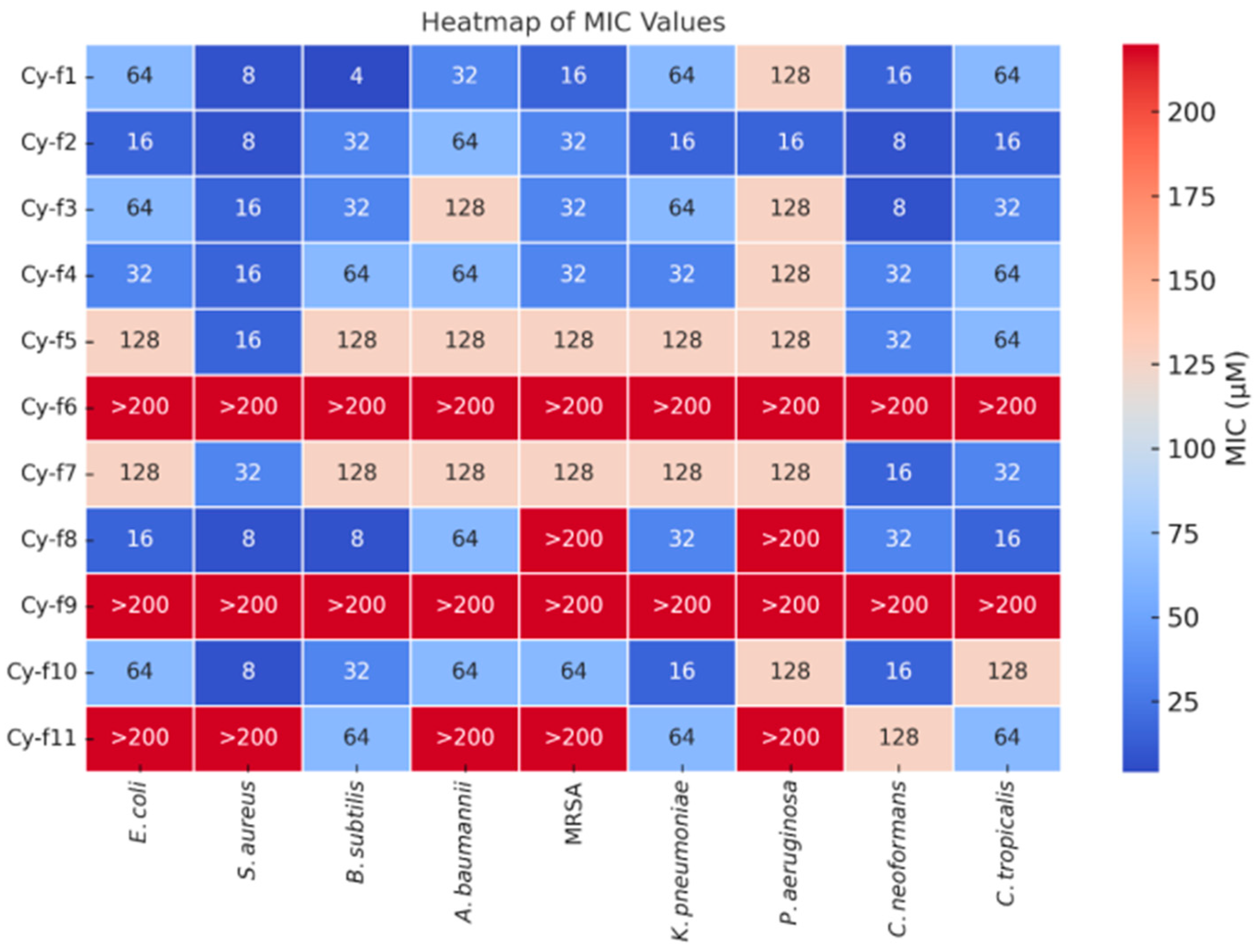
2.2. Antimicrobial Assay of 11 Cyclic Hexapeptides
Comparative Analysis of Antimicrobial Peptide Efficacy
2.3. Hemolytic Assay
2.4. Time-Dependent Growth Kinetics of Cy-f2 Against Klebsiella pneumoniae
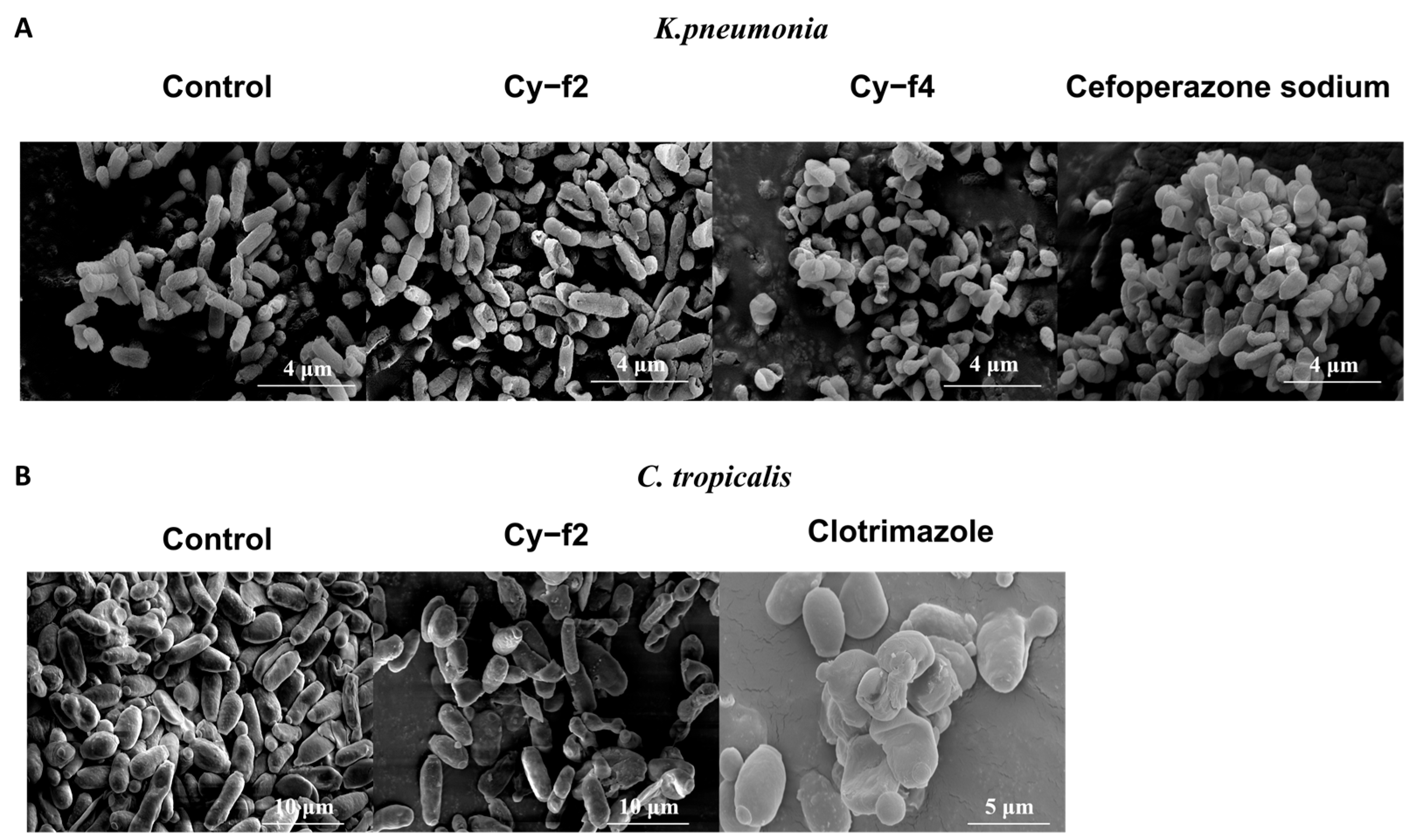
2.5. Scanning Electron Microscopy Analysis of Cy-f2 and Cy-f4 Effects on Klebsiella pneumoniae and Candida tropicalis
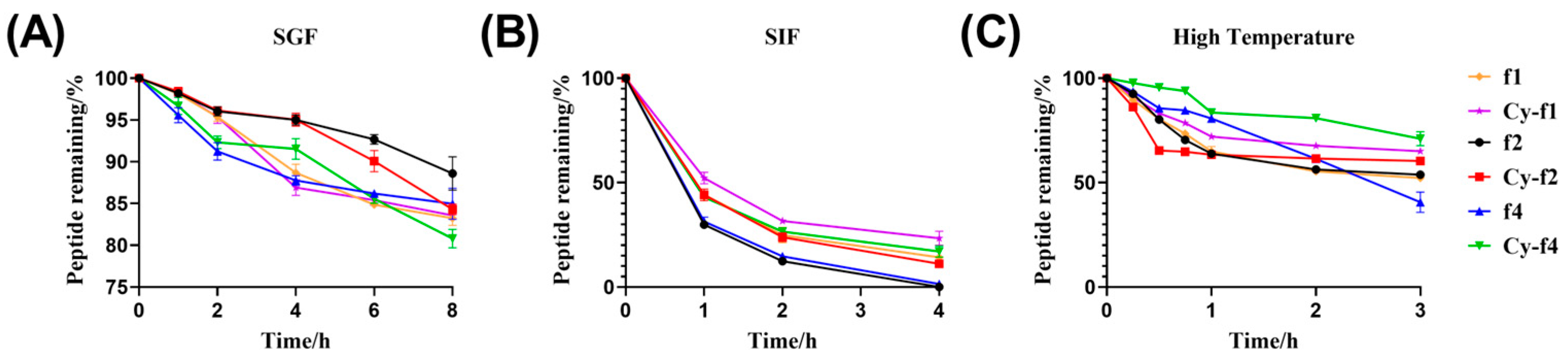
2.6. Stability of Cy-f1, Cy-f2, and Cy-f4
3. Discussion
4. Materials and Methods
4.1. Reagents and Instruments
4.2. Peptides Synthesis and Characterization
4.2.1. Synthesis of Cyclic Hexapeptides
4.2.2. Peptide Molecular Weight Identification
4.3. Biological Studies
4.3.1. Antibacterial Assay
4.3.2. Hemolytic Assay
− O.D of PBS control) × 100%
4.3.3. Time-Dependent Growth Kinetics
4.3.4. Scanning Electron Microscope (SEM)
4.4. Stability
4.4.1. Simulated Gastric Fluid Stability
4.4.2. Simulated Intestinal Fluid Stability
4.4.3. Thermal Stability
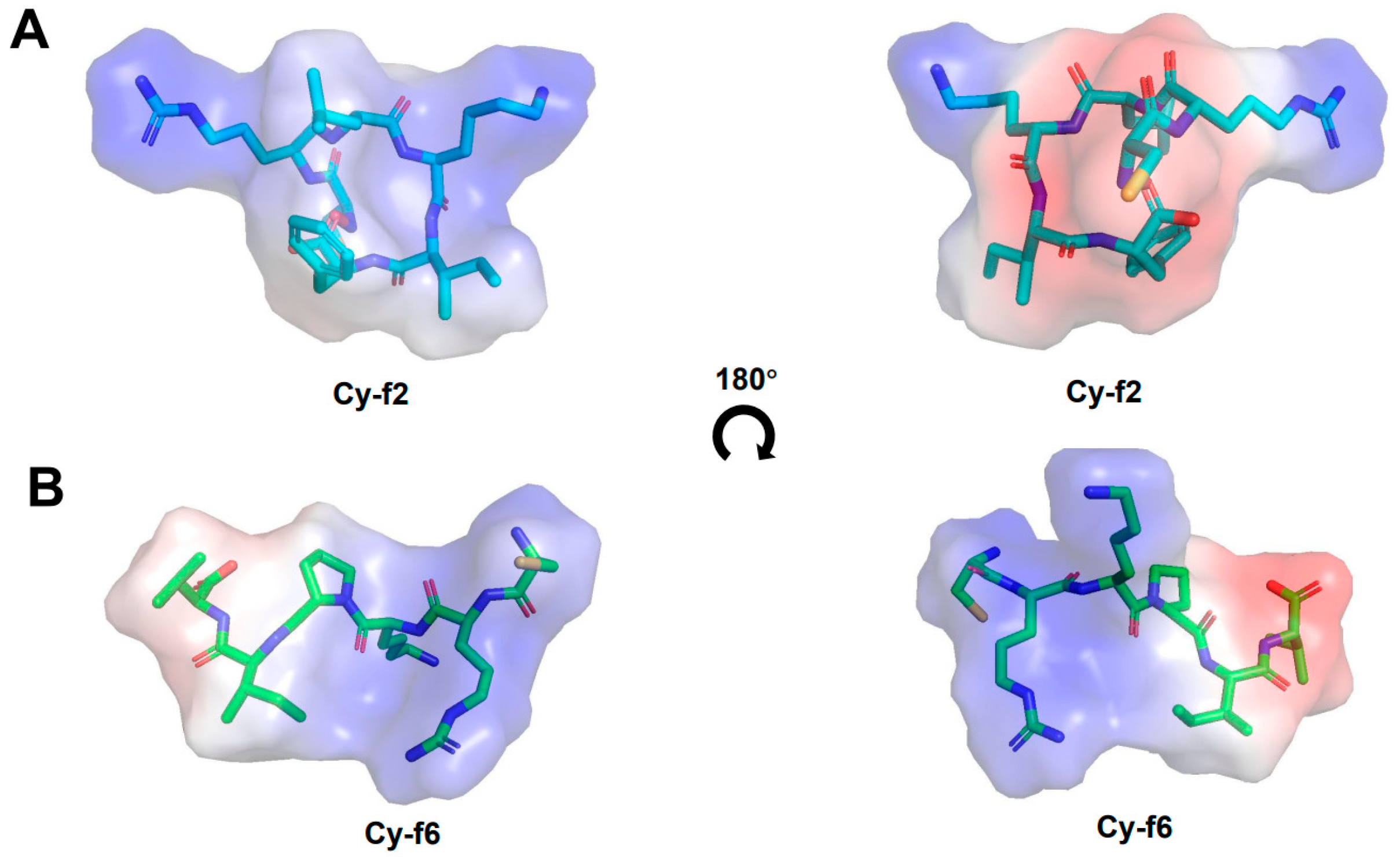
4.5. Structural Prediction by AlphaFold2
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Nang, S.C.; Azad, M.A.K.; Velkov, T.; Zhou, Q.T.; Li, J. Rescuing the Last-Line Polymyxins: Achievements and Challenges. Pharmacol. Rev. 2021, 73, 679–728. [Google Scholar] [CrossRef] [PubMed]
- Liscano, Y.; Salamanca, C.H.; Vargas, L.; Cantor, S.; Laverde-Rojas, V.; Oñate-Garzón, J. Increases in Hydrophilicity and Charge on the Polar Face of Alyteserin 1c Helix Change its Selectivity towards Gram-Positive Bacteria. Antibiotics 2019, 8, 238. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef]
- Rozek, A.; Powers, J.P.; Friedrich, C.L.; Hancock, R.E. Structure-based design of an indolicidin peptide analogue with increased protease stability. Biochemistry 2003, 42, 14130–14138. [Google Scholar] [CrossRef]
- Luckett, S.; Garcia, R.S.; Barker, J.J.; Konarev, A.V.; Shewry, P.R.; Clarke, A.R.; Brady, R.L. High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. J. Mol. Biol. 1999, 290, 525–533. [Google Scholar] [CrossRef]
- Tan, P.; Fu, H.; Ma, X. Design, optimization, and nanotechnology of antimicrobial peptides: From exploration to applications. Nano Today 2021, 39, 101229. [Google Scholar] [CrossRef]
- Sakoulas, G.; Bayer, A.S.; Pogliano, J.; Tsuji, B.T.; Yang, S.J.; Mishra, N.N.; Nizet, V.; Yeaman, M.R.; Moise, P.A. Ampicillin enhances daptomycin- and cationic host defense peptide-mediated killing of ampicillin- and vancomycin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 2012, 56, 838–844. [Google Scholar] [CrossRef]
- De León-Borrás, R.; Sánchez-Sergentón, C.; Mayor-Becerra, A.; Laureano-Cuadrado, A.F. Polymyxin B for Gram Negative Multidrug Resistant Bacteria in a Hispanic Population. P. R. Health Sci. J. 2019, 38, 15–21. [Google Scholar]
- Weng, Q.; Zhang, F.; Zheng, Q. Zosurabalpin: A novel tethered macrocyclic peptide antibiotic that kills carbapenem-resistant Acinetobacter baumannii. MedComm 2024, 5, e696. [Google Scholar] [CrossRef] [PubMed]
- Novozhilova, P.O.; Bakina, O.V.; Spirina, L.V.; Tarasenko, N.V. Epigenetic Mechanisms of Antibiotic Resistance. Curr. Mol. Med. 2025, 1875, 5666. [Google Scholar] [CrossRef]
- Liu, S.; Pentelute, B.L.; Kent, S.B. Convergent chemical synthesis of [lysine(24,38,83)] human erythropoietin. Angew. Chem. Int. Ed. Engl. 2012, 51, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.M.; Li, Y.M.; Shen, F.; Huang, Y.C.; Li, J.B.; Lin, Y.; Cui, H.K.; Liu, L. Protein chemical synthesis by ligation of peptide hydrazides. Angew. Chem. Int. Ed. Engl. 2011, 50, 7645–7649. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Liang, L.J.; Si, Y.Y.; Gao, S.; Wang, J.X.; Liang, J.; Mei, Z.; Zheng, J.S.; Liu, L. Practical Chemical Synthesis of Atypical Ubiquitin Chains by Using an Isopeptide-Linked Ub Isomer. Angew. Chem. Int. Ed. Engl. 2017, 56, 13333–13337. [Google Scholar] [CrossRef]
- Lee, C.L.; Liu, H.; Wong, C.T.; Chow, H.Y.; Li, X. Enabling N-to-C Ser/Thr Ligation for Convergent Protein Synthesis via Combining Chemical Ligation Approaches. J. Am. Chem. Soc. 2016, 138, 10477–10484. [Google Scholar] [CrossRef]
- Murar, C.E.; Thuaud, F.; Bode, J.W. KAHA ligations that form aspartyl aldehyde residues as synthetic handles for protein modification and purification. J. Am. Chem. Soc. 2014, 136, 18140–18148. [Google Scholar] [CrossRef]
- Lan, H.; Wu, K.; Zheng, Y.; Pan, M.; Huang, Y.C.; Gao, S.; Zheng, Q.Y.; Zheng, J.S.; Li, Y.M.; Xiao, B.; et al. Total synthesis of mambalgin-1/2/3 by two-segment hydrazide-based native chemical ligation. J. Pept. Sci. 2016, 22, 320–326. [Google Scholar] [CrossRef]
- Duggan, N.M.; Saez, N.J.; Clayton, D.; Budusan, E.; Watson, E.E.; Tucker, I.J.; Rash, L.D.; King, G.F.; Payne, R.J. Total Synthesis of the Spider-Venom Peptide Hi1a. Org. Lett. 2021, 23, 8375–8379. [Google Scholar] [CrossRef]
- Manzo, G.; Scorciapino, M.A.; Wadhwani, P.; Bürck, J.; Montaldo, N.P.; Pintus, M.; Sanna, R.; Casu, M.; Giuliani, A.; Pirri, G.; et al. Enhanced amphiphilic profile of a short β-stranded peptide improves its antimicrobial activity. PLoS ONE 2015, 10, e0116379. [Google Scholar] [CrossRef]
- Zhu, X.; Dong, N.; Wang, Z.; Ma, Z.; Zhang, L.; Ma, Q.; Shan, A. Design of imperfectly amphipathic α-helical antimicrobial peptides with enhanced cell selectivity. Acta Biomater. 2014, 10, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Vicente, F.E.M.; González-Garcia, M.; Diaz Pico, E.; Moreno-Castillo, E.; Garay, H.E.; Rosi, P.E.; Jimenez, A.M.; Campos-Delgado, J.A.; Rivera, D.G.; Chinea, G.; et al. Design of a Helical-Stabilized, Cyclic, and Nontoxic Analogue of the Peptide Cm-p5 with Improved Antifungal Activity. ACS Omega 2019, 4, 19081–19095. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, J.; Zhan, N.; Sun, T.; Yu, W.; Zhang, L.; Shan, A.; Zhang, A. Therapeutic Potential of Trp-Rich Engineered Amphiphiles by Single Hydrophobic Amino Acid End-Tagging. ACS Appl. Mater. Interfaces 2019, 11, 43820–43834. [Google Scholar] [CrossRef]
- Chow, H.Y.; Zhang, Y.; Matheson, E.; Li, X. Ligation Technologies for the Synthesis of Cyclic Peptides. Chem. Rev. 2019, 119, 9971–10001. [Google Scholar] [CrossRef]
- Xie, M.; Gao, M.; Yun, Y.; Malmsten, M.; Rotello, V.M.; Zboril, R.; Akhavan, O.; Kraskouski, A.; Amalraj, J.; Cai, X.; et al. Antibacterial Nanomaterials: Mechanisms, Impacts on Antimicrobial Resistance and Design Principles. Angew. Chem. Int. Ed. Engl. 2023, 62, e202217345. [Google Scholar] [CrossRef]
- Li, S.; Wang, Y.; Xue, Z.; Jia, Y.; Li, R.; He, C.; Chen, H. The structure-mechanism relationship and mode of actions of antimicrobial peptides: A review. Trends Food Sci. Technol. 2021, 109, 103–115. [Google Scholar] [CrossRef]
- Oddo, A.; Hansen, P.R. Hemolytic Activity of Antimicrobial Peptides. Methods Mol. Biol. 2017, 1548, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Yuan, X.; Chen, H.; Zhu, Y.; Dong, N.; Shan, A. Strategies employed in the design of antimicrobial peptides with enhanced proteolytic stability. Biotechnol. Adv. 2022, 59, 107962. [Google Scholar] [CrossRef]
- Huang, Y.C.; Guan, C.J.; Tan, X.L.; Chen, C.C.; Guo, Q.X.; Li, Y.M. Accelerated Fmoc solid-phase synthesis of peptides with aggregation-disrupting backbones. Org. Biomol. Chem. 2015, 13, 1500–1506. [Google Scholar] [CrossRef]
- Miao, X.; Zhou, T.; Zhang, J.; Xu, J.; Guo, X.; Hu, H.; Zhang, X.; Hu, M.; Li, J.; Yang, W.; et al. Enhanced cell selectivity of hybrid peptides with potential antimicrobial activity and immunomodulatory effect. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129532. [Google Scholar] [CrossRef]
- Kalia, V.; Miglani, R.; Purnapatre, K.P.; Mathur, T.; Singhal, S.; Khan, S.; Voleti, S.R.; Upadhyay, D.J.; Saini, K.S.; Rattan, A.; et al. Mode of action of Ranbezolid against staphylococci and structural modeling studies of its interaction with ribosomes. Antimicrob. Agents Chemother. 2009, 53, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Lai, Z.; Jian, Q.; Shao, C.; Zhu, Y.; Li, G.; Shan, A. Design of Heptad Repeat Amphiphiles Based on Database Filtering and Structure-Function Relationships to Combat Drug-Resistant Fungi and Biofilms. ACS Appl. Mater. Interfaces 2020, 12, 2129–2144. [Google Scholar] [CrossRef]
- Huang, H.Y.; Wang, Q.; Zhang, C.Y.; Chen, Z.X.; Wang, J.T.; Liao, X.W.; Yu, R.J.; Xiong, Y.S. Synthesis and biological evaluation of ruthenium complexes containing phenylseleny against Gram-positive bacterial infection by damage membrane integrity and avoid drug-resistance. J. Inorg. Biochem. 2023, 242, 112175. [Google Scholar] [CrossRef]
- Sadelaji, S.; Ghaznavi-Rad, E.; Sadoogh Abbasian, S.; Fahimirad, S.; Abtahi, H. Ib-AMP4 antimicrobial peptide as a treatment for skin and systematic infection of methicillin-resistant Staphylococcus aureus (MRSA). Iran. J. Basic. Med. Sci. 2022, 25, 232–238. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Xu, L.; Hu, Y.; Tang, X.; Qu, R.; Zhao, X.; Bai, H.; Li, L.; Chen, W.; Luo, G.; et al. Lysine-Tethered Stable Bicyclic Cationic Antimicrobial Peptide Combats Bacterial Infection in Vivo. J. Med. Chem. 2022, 65, 10523–10533. [Google Scholar] [CrossRef]
- Jakobek, L.; Strelec, I.; Kenjerić, D.; Šoher, L.; Tomac, I.; Matić, P. Simulated Gastric and Intestinal Fluid Electrolyte Solutions as an Environment for the Adsorption of Apple Polyphenols onto β-Glucan. Molecules 2022, 27, 6683. [Google Scholar] [CrossRef] [PubMed]
- Braga Emidio, N.; Tran, H.N.T.; Andersson, A.; Dawson, P.E.; Albericio, F.; Vetter, I.; Muttenthaler, M. Improving the Gastrointestinal Stability of Linaclotide. J. Med. Chem. 2021, 64, 8384–8390. [Google Scholar] [CrossRef]
- Ren, J.; Zhu, X.; Xu, P.; Li, R.; Fu, Y.; Dong, S.; Zhangsun, D.; Wu, Y.; Luo, S. d-Amino Acid Substitution of α-Conotoxin RgIA Identifies its Critical Residues and Improves the Enzymatic Stability. Mar. Drugs 2019, 17, 142. [Google Scholar] [CrossRef]
- He, X.H.; You, C.Z.; Jiang, H.L.; Jiang, Y.; Xu, H.E.; Cheng, X. AlphaFold2 versus experimental structures: Evaluation on G protein-coupled receptors. Acta Pharmacol. Sin. 2023, 44, 1–7. [Google Scholar] [CrossRef]
- Yang, Z.; Zeng, X.; Zhao, Y.; Chen, R. AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct. Target. Ther. 2023, 8, 115. [Google Scholar] [CrossRef]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
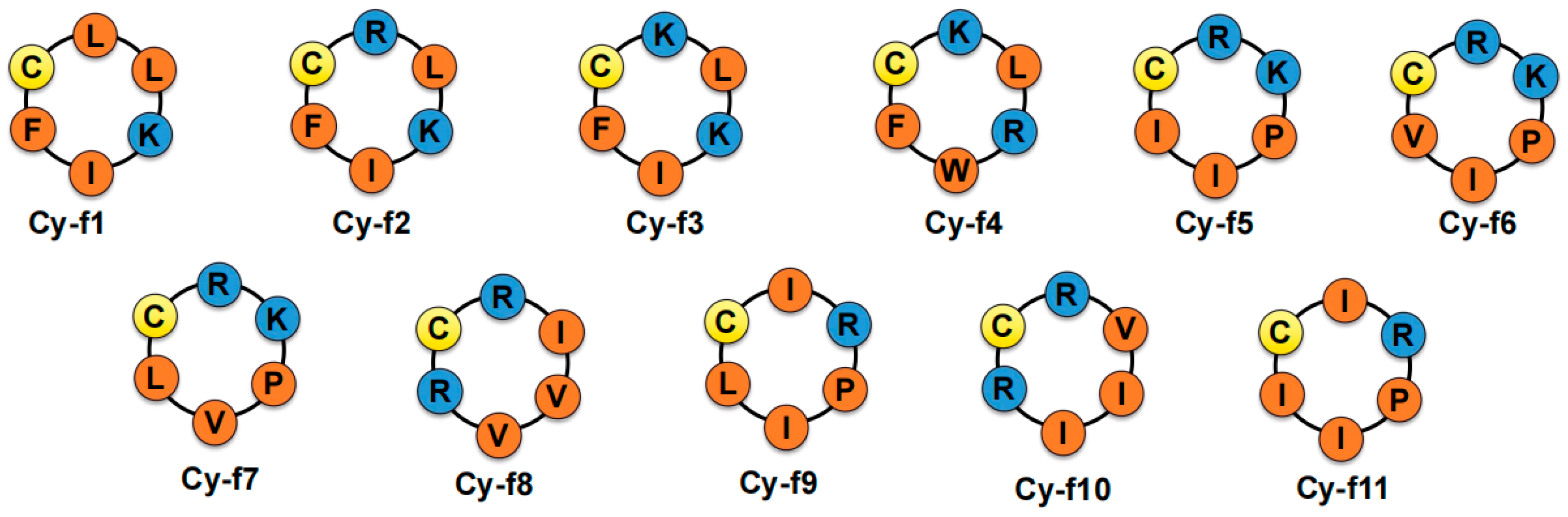

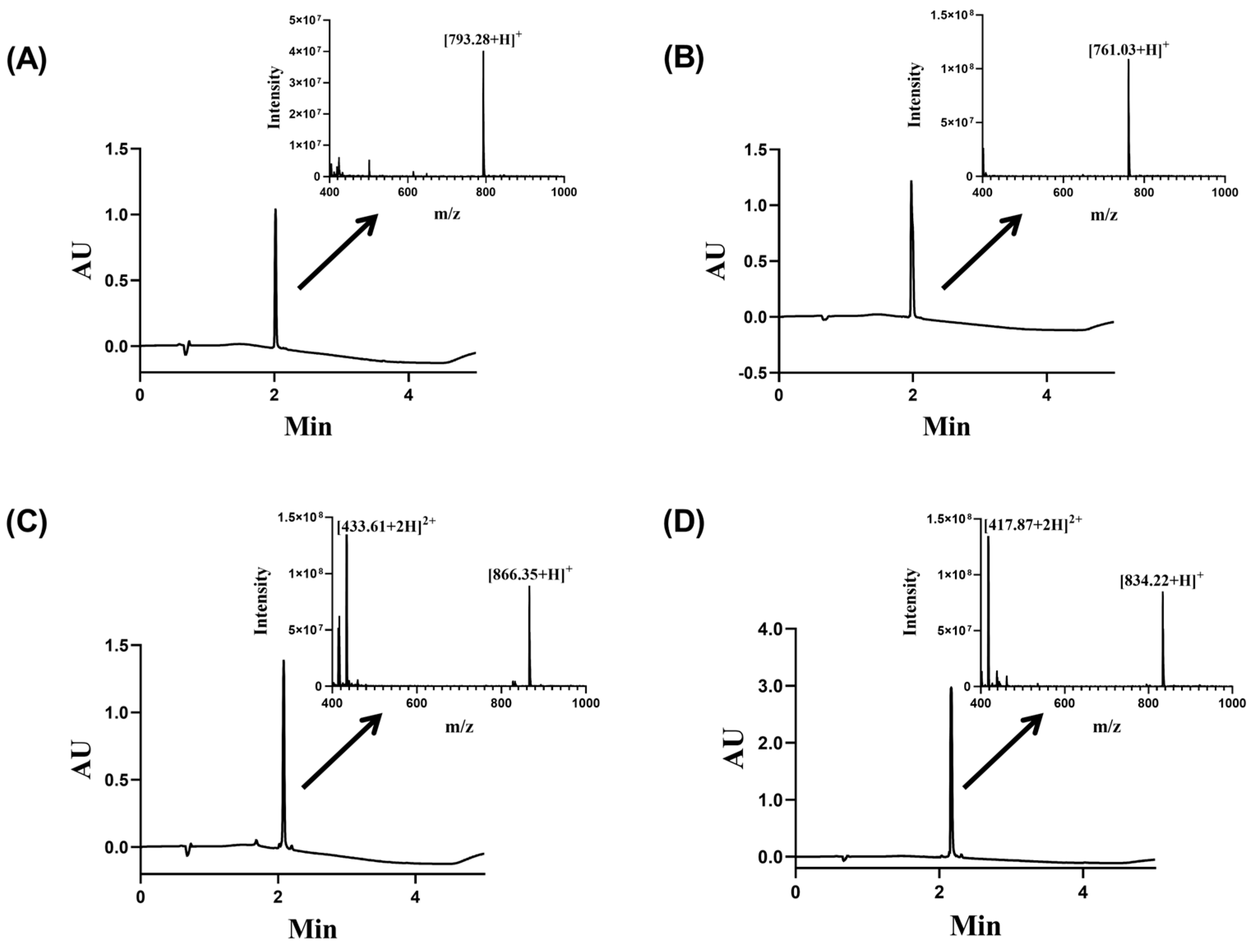


| Peptide Name | Sequence | Theoretical M.W. (g/mol) | Experimental M.W. (g/mol) |
|---|---|---|---|
| Cy-f1 | c-[CLLKIF] | 717.98 | 717.97 |
| Cy-f2 | c-[CRLKIF] | 761.01 | 761.03 |
| Cy-f3 | c-[CKLKIF] | 733.00 | 733.22 |
| Cy-f4 | c-[CKLRWF] | 834.06 | 834.22 |
| Cy-f5 | c-[CRKPII] | 710.95 | 711.35 |
| Cy-f6 | c-[CRKPIV] | 696.92 | 697.25 |
| Cy-f7 | c-[CRKPVL] | 696.92 | 697.53 |
| Cy-f8 | c-[CRIVVR] | 726.95 | 727.34 |
| Cy-f9 | c-[CIRPIL] | 695.94 | 696.75 |
| Cy-f10 | c-[CRVIIR] | 740.98 | 741.27 |
| Cy-f11 | c-[CIRPII] | 695.94 | 696.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Liu, M.; Ruan, B.; Liao, Z.; Tang, X.; Zhangsun, D.; Wu, Y.; Luo, S. Synthesis of Cyclic Hexapeptides via the Hydrazide Method and Evaluation of Their Antibacterial Activities. Molecules 2025, 30, 2444. https://doi.org/10.3390/molecules30112444
Cui Y, Liu M, Ruan B, Liao Z, Tang X, Zhangsun D, Wu Y, Luo S. Synthesis of Cyclic Hexapeptides via the Hydrazide Method and Evaluation of Their Antibacterial Activities. Molecules. 2025; 30(11):2444. https://doi.org/10.3390/molecules30112444
Chicago/Turabian StyleCui, Yunfei, Meng Liu, Binghui Ruan, Zhouyuji Liao, Xue Tang, Dongting Zhangsun, Yong Wu, and Sulan Luo. 2025. "Synthesis of Cyclic Hexapeptides via the Hydrazide Method and Evaluation of Their Antibacterial Activities" Molecules 30, no. 11: 2444. https://doi.org/10.3390/molecules30112444
APA StyleCui, Y., Liu, M., Ruan, B., Liao, Z., Tang, X., Zhangsun, D., Wu, Y., & Luo, S. (2025). Synthesis of Cyclic Hexapeptides via the Hydrazide Method and Evaluation of Their Antibacterial Activities. Molecules, 30(11), 2444. https://doi.org/10.3390/molecules30112444






