Recent Advances in the Development of Functional Nucleic Acid Biosensors Based on Aptamer-Rolling Circle Amplification
Abstract
1. Introduction
2. Aptamer
3. The Regulation of RCA Reaction
3.1. The Design of Primers
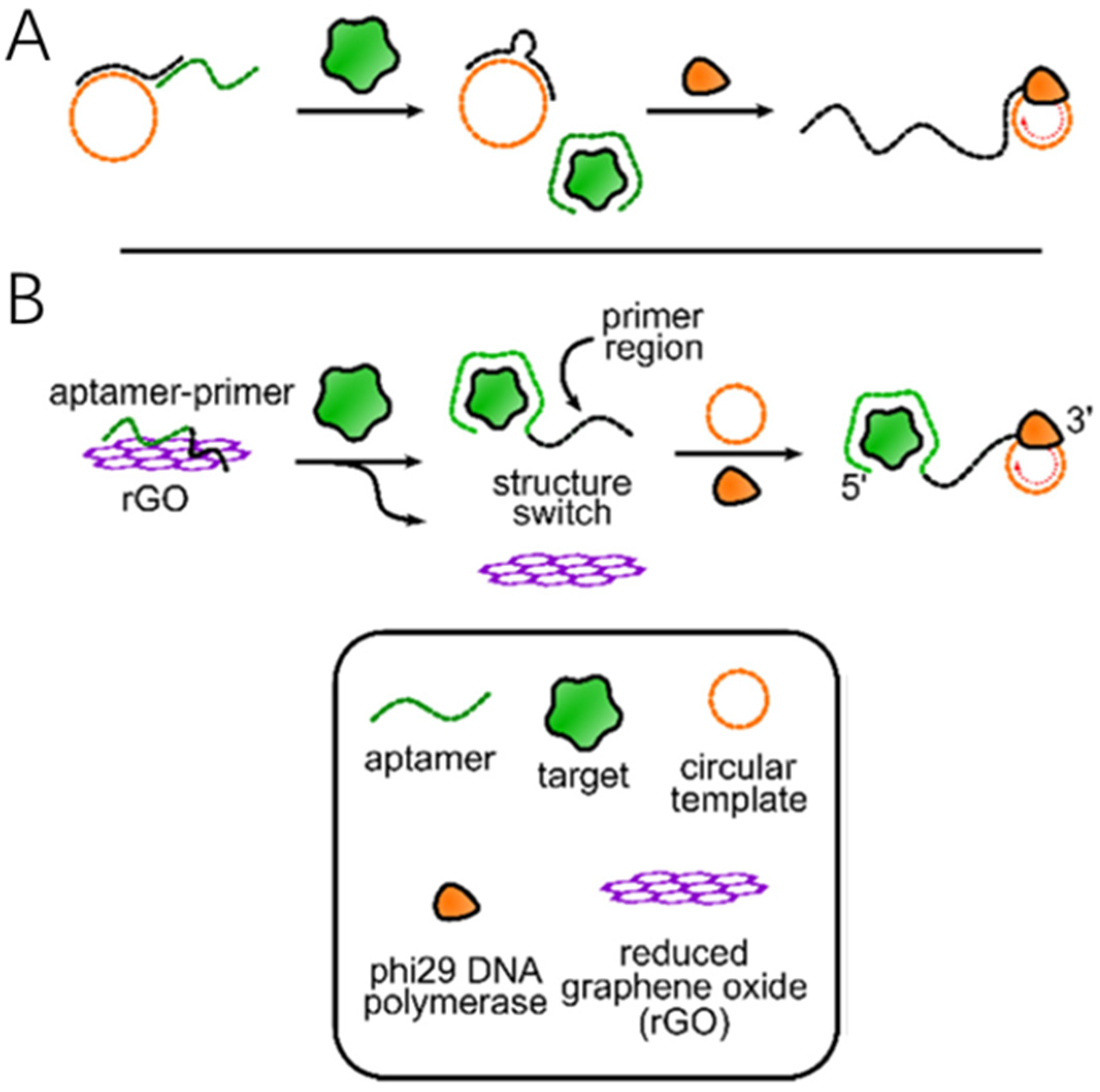
3.1.1. The Primer Is Directly Connected to the Aptamer
3.1.2. RNA-Cleaving DNAzyme (RCD) Mediated Primer Regulation
(1) RCD Cleavage for Primer Release
(2) RCD Regulation of Primer Conformation
(3) RCD-Mediated Primer Generation
(4) RCD Regulation of Primer Competition
(5) RCD Control of Primer Recycling
3.2. The Design of Circular Template
3.2.1. Ligate the Circular Template
3.2.2. Design Circular Aptamers
4. The Amplification Methods of the RCA Reaction
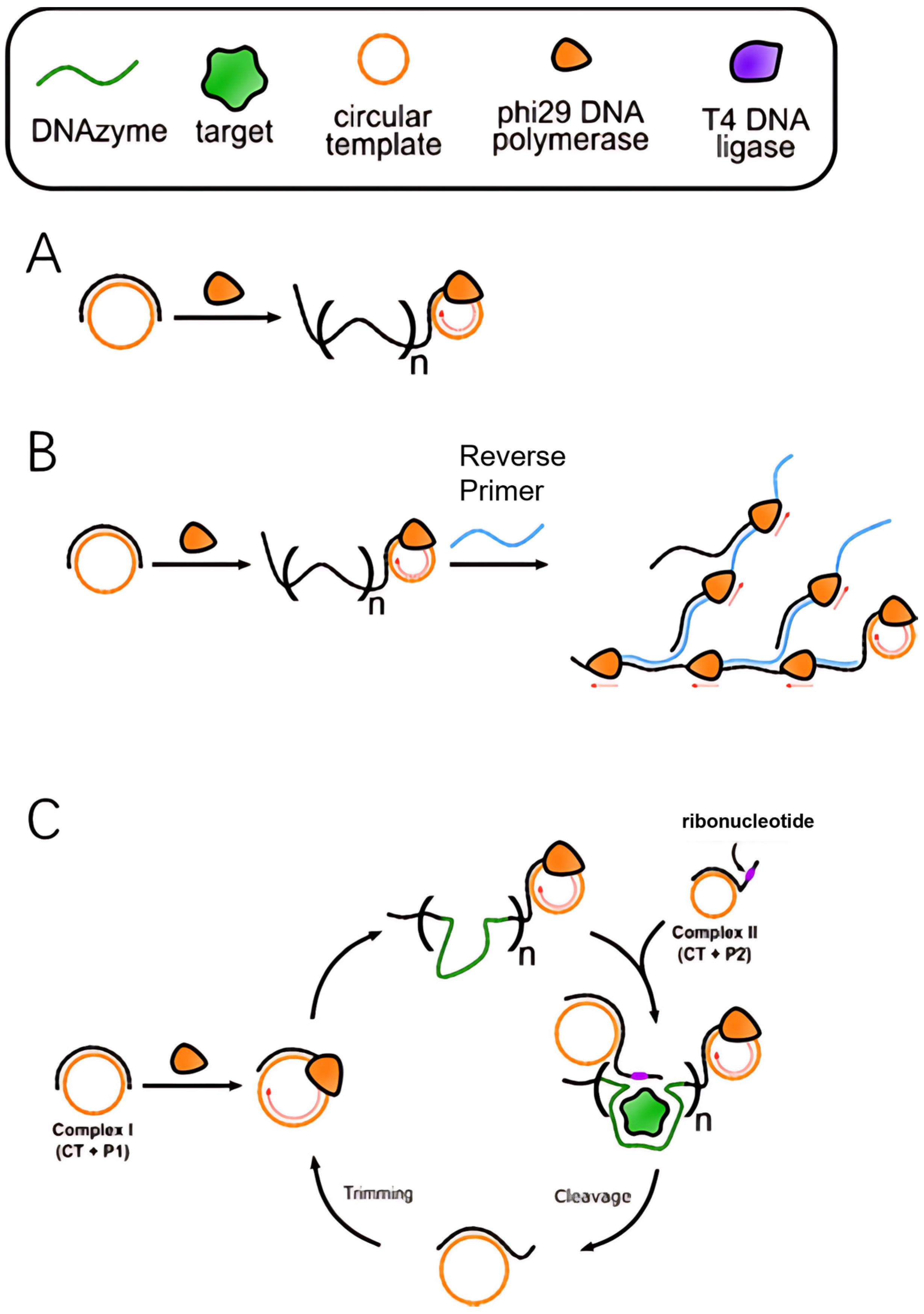
4.1. Linear RCA
4.2. Exponential RCA
- (1)
- Hyperbranched RCA: Reverse primer sites are designed on the initial RCA products, and cascade amplification mediated by multiple primers leads to the formation of a three-dimensional branched structure (Figure 2B). This method can increase the amplification factor to 106–109 times, but precise control of primer concentration is necessary to avoid non-specific amplification [38].
- (2)
- Primer regeneration RCA38: Nucleic acid endonucleases, such as Nickase or RCD, cyclically cleave RCA products, releasing new primers that initiate secondary amplification cycles (Figure 2C). This dynamic regulation approach is particularly useful for real-time quantitative detection, such as the ultrasensitive analysis of viral nucleic acids.
5. Applications of the Combination of Aptamers and RCA
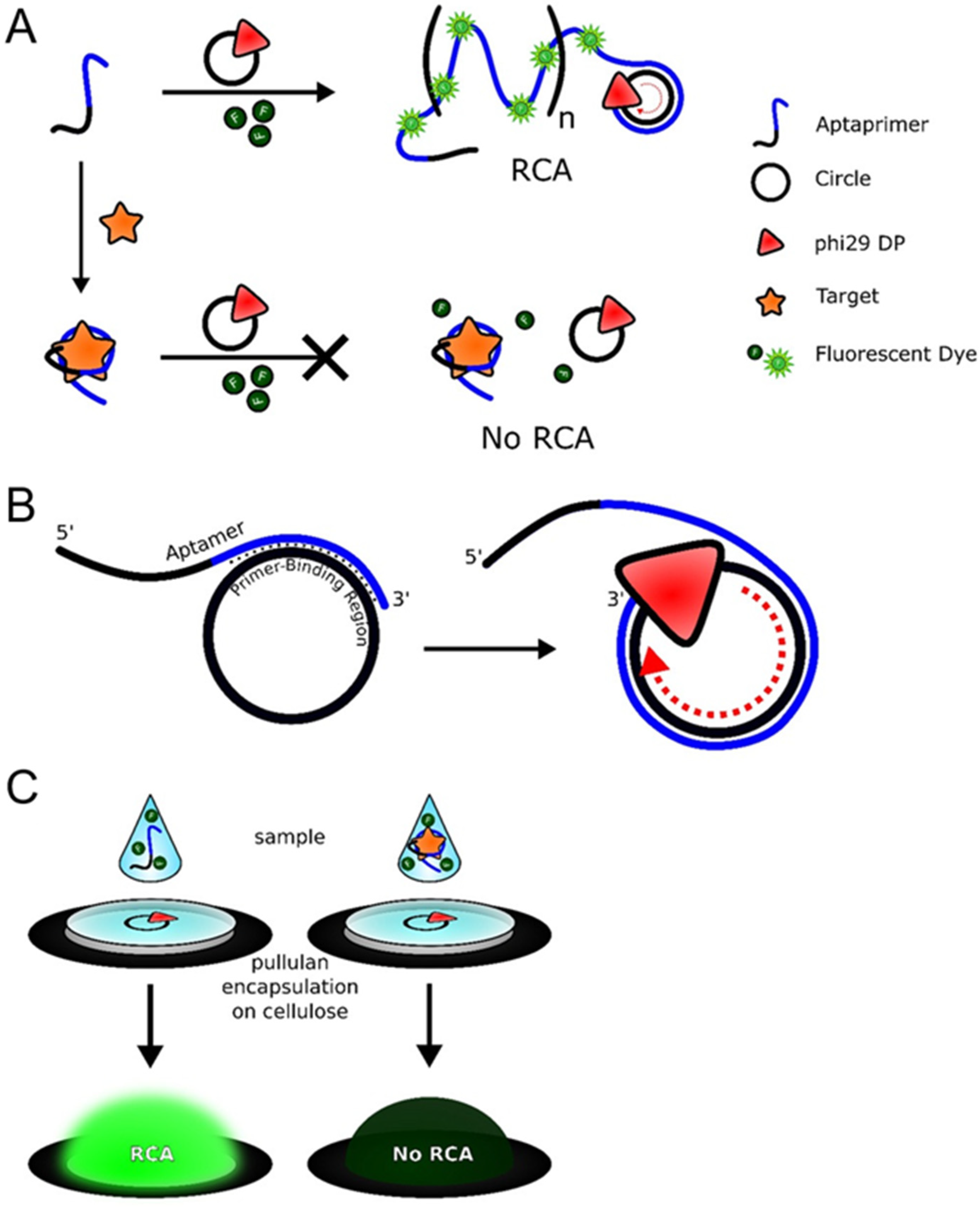
5.1. Fluorescence-Based Detection
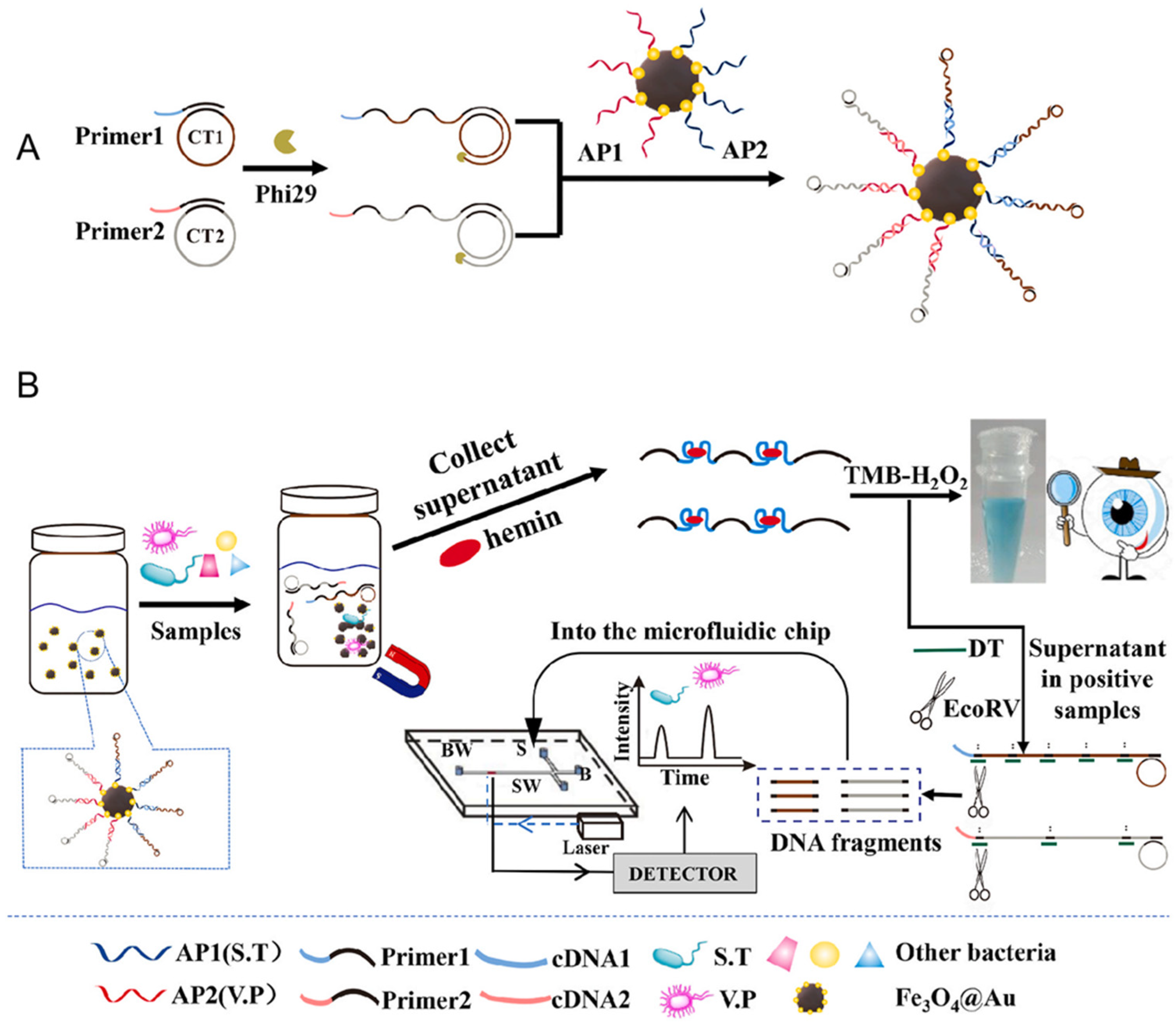
5.2. Microfluidic Systems
5.3. Visual Detection
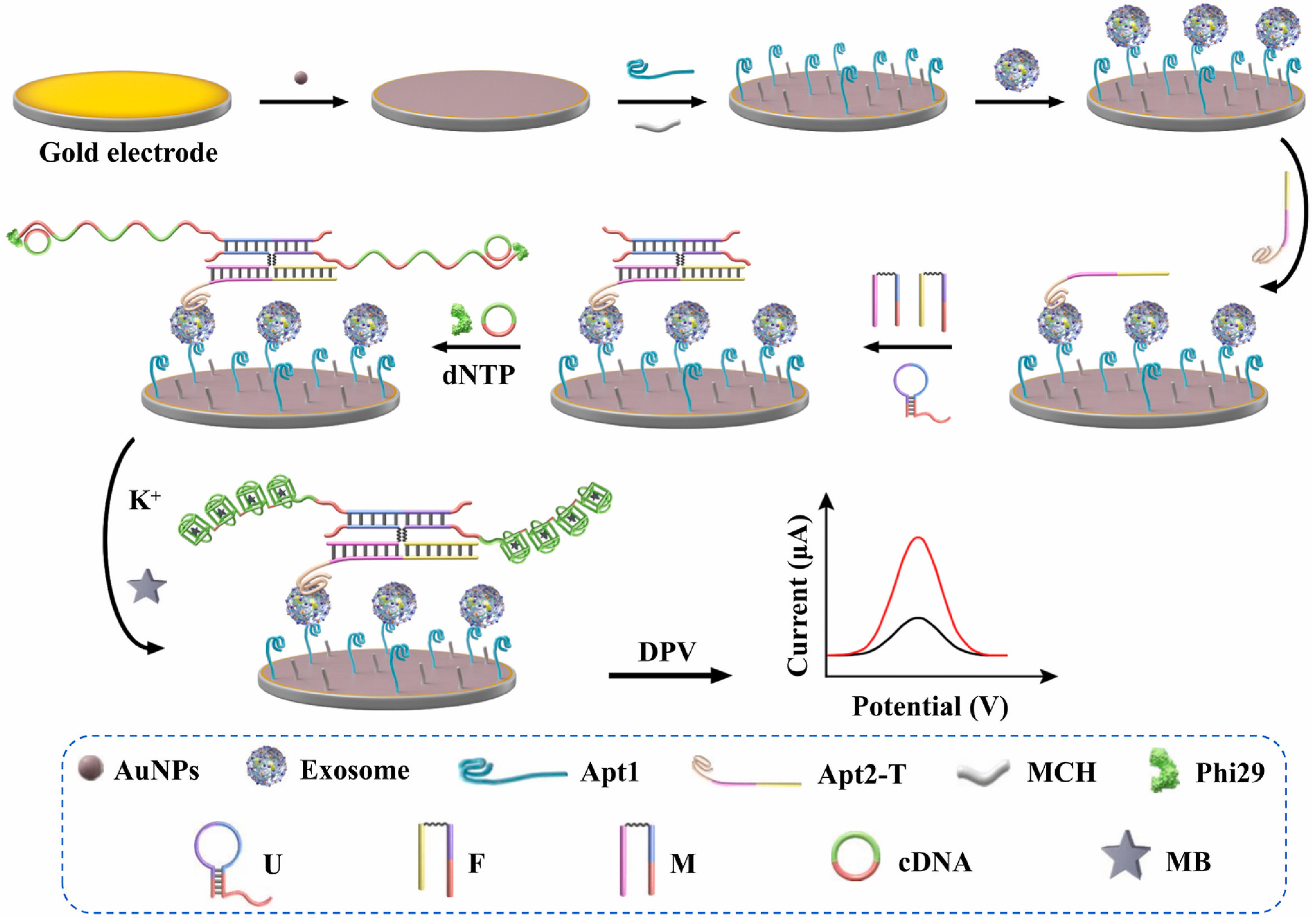
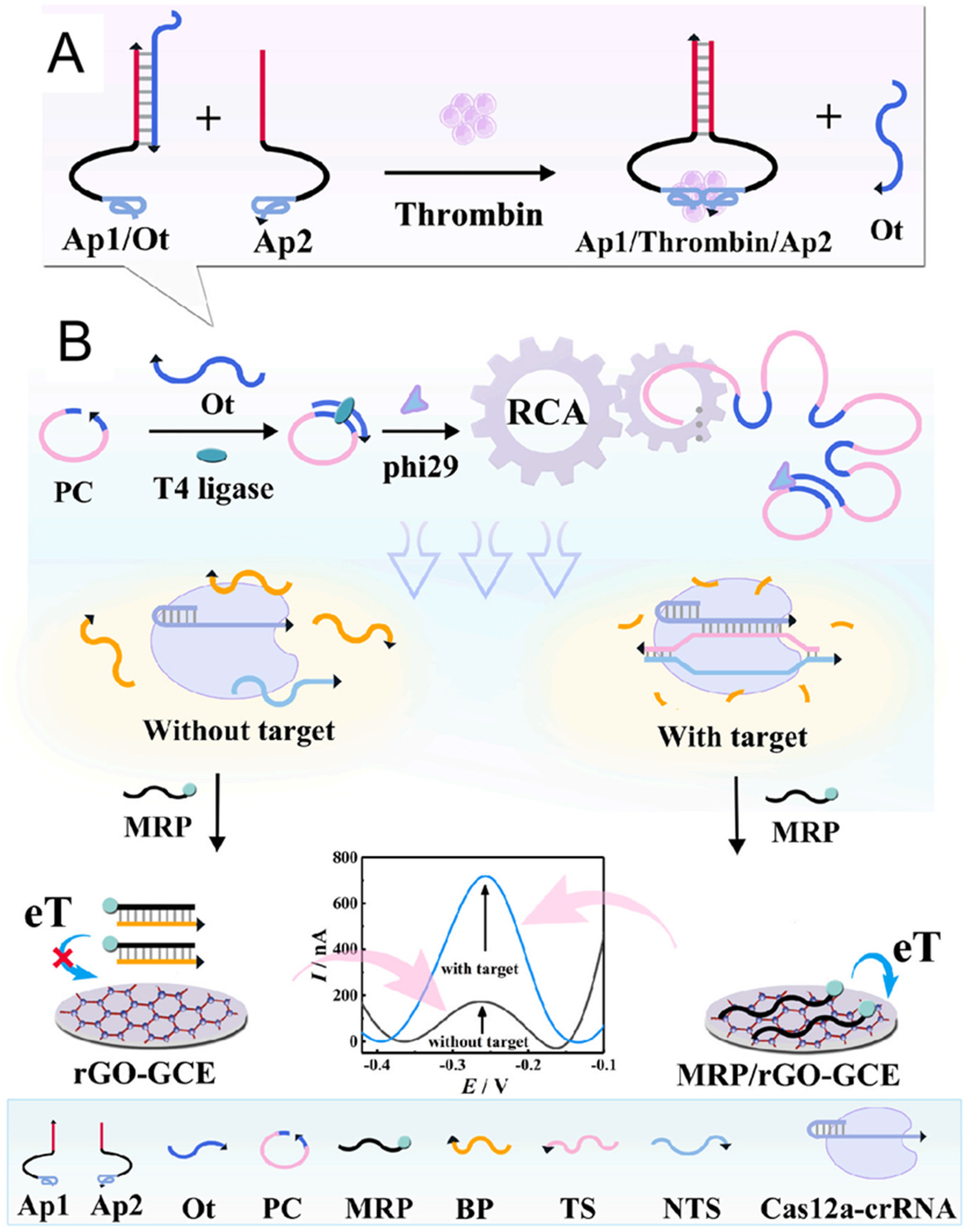
5.4. Electrochemical Sensing Techniques
6. Summary and Outlook
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| functional nucleic acids | FNA |
| systematic evolution of ligands by exponential enrichment | SELEX |
| rolling circle amplification | RCA |
| capillary electrophoresis–SELEX | CE-SELEX |
| protein microarray system–SELEX | PMM-SELEX |
| circular DNA template | CDT |
| deoxyribonucleoside triphosphates | dNTPs |
| RNA-cleaving DNAzyme | RCD |
| ochratoxin A | OTA |
| G-quadruplex | G4 |
| aptamer probe–hairpin primer probe | APH |
| time-resolved fluorescence nanoparticles | TRFNPs |
| silver nanoclusters | AgNCs |
| Salmonella typhimurium | S.T. |
| Vibrio parahaemolyticus | V.P. |
References
- He, W.; Liu, C.; Han, J.; Wang, S.; Lv, Y.; Li, X.; Guo, X. MNPs-aptazymes-PtNPs molecular motor biosensor mediated by circular cleavage reactions for the ultrasensitive detection of aflatoxin B1 in real samples. Microchem. J. 2024, 207, 112007. [Google Scholar] [CrossRef]
- Curtis, E.A.; Liu, D.R. A naturally occurring, noncanonical GTP aptamer made of simple tandem repeats. RNA Biol. 2014, 11, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.X.; Kwon, Y.J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. [Google Scholar] [CrossRef]
- Singer, B.S.; Shtatland, T.; Brown, D.; Gold, L. Libraries for genomic SELEX. Nucleic Acids Res. 1997, 25, 781–786. [Google Scholar] [CrossRef]
- Hu, Y.-Y.; Yang, G.; Qu, F. Research advances in non-immobilized aptamer screening techniques for small-molecule targets. Chin. J. Chromatogr. 2025, 43, 297–308. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tsuji, S.; Hirabayashi, N.; Kato, S.; Akitomi, J.; Egashira, H.; Tanaka, T.; Waga, I.; Ohtsu, T. Effective isolation of RNA aptamer through suppression of PCR bias. Biochem. Biophys. Res. Commun. 2009, 386, 223–226. [Google Scholar] [CrossRef]
- Yang, X.; Chan, C.H.; Yao, S.; Chu, H.Y.; Lyu, M.; Chen, Z.; Xiao, H.; Ma, Y.; Yu, S.; Li, F.; et al. DeepAptamer: Advancing high-affinity aptamer discovery with a hybrid deep learning model. Mol. Ther. Nucleic Acids 2025, 36, 102436. [Google Scholar] [CrossRef]
- Wang, W.; Jia, L.Y. Progress in aptamer screening methods. Chin. J. Anal. Chem. 2009, 37, 454–460. [Google Scholar] [CrossRef]
- Hatch, A.; Sano, T.; Misasi, J.; Smith, C.L. Rolling circle amplification of DNA immobilized on solid surfaces and its application to multiplex mutation detection. Genet. Anal. Biomol. Eng. 1999, 15, 35–40. [Google Scholar] [CrossRef]
- Boonbanjong, P.; Treerattrakoon, K.; Waiwinya, W.; Pitikultham, P.; Japrung, D. Isothermal amplification technology for disease diagnosis. Biosensors 2022, 12, 677. [Google Scholar] [CrossRef] [PubMed]
- Ou, L.; Sun, A.; Liu, K. Rolling circle amplification-based biosensors. Anal. Lett. 2015, 48, 1199–1216. [Google Scholar] [CrossRef]
- Zhan, Y.; Mao, Y.; Sun, P.; Liu, C.; Gou, H.; Qi, H.; Chen, G.; Hu, S.; Tian, B. Tumor-associated antigen-specific cell imaging based on upconversion luminescence and nucleic acid rolling circle amplification. Microchim. Acta 2024, 191, 248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-W.; Qi, G.-X.; Chen, S.; Yu, Y.-L.; Wang, J.-H. Ultrasensitive and wash-free detection of tumor extracellular vesicles by aptamer-proximity-ligation-activated rolling circle amplification coupled to single particle ICP-MS. Anal. Chem. 2024, 96, 10800–10808. [Google Scholar] [CrossRef]
- Tang, J.; Yu, Y.; Shi, H.; He, X.; Lei, Y.; Shangguan, J.; Yang, X.; Qiao, Z.; Wang, K. Polyvalent and thermosensitive DNA nanoensembles for cancer cell detection and manipulation. Anal. Chem. 2017, 89, 6637–6644. [Google Scholar] [CrossRef]
- Zhu, D.; Huang, T.; Zhou, Q.; Yang, Z.; Liu, B.; Li, M.; Li, C.; Chen, J.-X.; Dai, Z.; Chen, J. A label-free fluorescent aptasensor based on a novel exponential rolling circle amplification for highly sensitive ochratoxin A detection. Food Chem. 2023, 410, 135427. [Google Scholar] [CrossRef]
- Xing, Y.; Wang, P.; Zang, Y.; Ge, Y.; Jin, Q.; Zhao, J.; Xu, X.; Zhao, G.; Mao, H. A colorimetric method for H1N1 DNA detection using rolling circle amplification. Analyst 2013, 138, 3457–3462. [Google Scholar] [CrossRef]
- Ding, C.; Liu, H.; Wang, N.; Wang, Z. Cascade signal amplification strategy for the detection of cancer cells by rolling circle amplification and nanoparticles tagging. Chem. Commun. 2012, 48, 5019–5021. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, H.; Wang, L.; Zhu, J.; Jiang, W. Cascade signal amplification based on copper nanoparticle-reported rolling circle amplification for ultrasensitive electrochemical detection of the prostate cancer biomarker. ACS Appl. Mater. Interfaces 2016, 8, 2573–2581. [Google Scholar] [CrossRef]
- Kumar, Y. Isothermal amplification-based methods for assessment of microbiological safety and authenticity of meat and meat products. Food Control 2021, 121, 107679. [Google Scholar] [CrossRef]
- Lau, H.Y.; Botella, J.R. Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front. Plant Sci. 2017, 8, 2016. [Google Scholar] [CrossRef] [PubMed]
- Tomas Rozenblum, G.; Gisela Lopez, V.; Daniel Vitullo, A.; Radrizzani, M. Aptamers: Current challenges and future prospects. Expert Opin. Drug Discov. 2016, 11, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on aptamer research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef]
- Du, Y.-L.; Mo, L.-T.; Yi, Y.-S.; Qiu, L.-P.; Tan, W.-H. Aptamers from cell-based selection for bioanalysis and bioimaging. Chin. J. Anal. Chem. 2017, 45, 1757–1764. [Google Scholar] [CrossRef]
- Yang, G.; Han, S.-M.; Zhao, L.-P.; Zhu, C.; Huang, Y.-Y.; Qu, F. Screening aptamer of apo-transferrin via capillary electrophoresis-systematic evolution of ligands by exponential enrichment and environmental factors analysis. Chin. J. Anal. Chem. 2020, 48, 632–641. [Google Scholar] [CrossRef]
- Liu, X.; Li, H.; Jia, W.; Chen, Z.; Xu, D. Selection of aptamers based on a protein microarray integrated with a microfluidic chip. Lab. Chip 2017, 17, 178–185. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, J.; Peng, Y.; Xie, Y.; Xiao, Y. Aptamers: A prospective tool for infectious diseases diagnosis. J. Clin. Lab. Anal. 2022, 36, 24725. [Google Scholar] [CrossRef]
- Rothlisberger, P.; Gasse, C.; Hollenstein, M. Nucleic acid aptamers: Emerging applications in medical imaging, nanotechnology, neurosciences, and drug delivery. Int. J. Mol. Sci. 2017, 18, 2430. [Google Scholar] [CrossRef]
- Catherine, A.T.; Shishido, S.N.; Robbins-Welty, G.A.; Diegelman-Parente, A. Rational design of a structure-switching DNA aptamer for potassium ions. FEBS Open Bio 2014, 4, 788–795. [Google Scholar] [CrossRef]
- Bialy, R.M.; Mainguy, A.; Li, Y.; Brennan, J.D. Functional nucleic acid biosensors utilizing rolling circle amplification. Chem. Soc. Rev. 2022, 51, 9009–9067. [Google Scholar] [CrossRef]
- Chai, S.; Sun, W.; Hou, X.; Pei, S.; Liu, Y.; Luo, K.; Guan, S.; Lv, W. A primer-regulated rolling circle amplification (RCA) for logic-controlled multiplexed enzyme analysis. Acs Appl. Bio Mater. 2025, 8, 2408–2418. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Song, J.; Shuang, S.; Dong, C.; Brennan, J.D.; Li, Y. A graphene-based biosensing platform based on the release of DNA probes and rolling circle amplification. ACS Nano 2014, 8, 5564–5573. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Q.; Chang, D.; Gu, J.; Brennan, J.D.; Li, Y. A DNAzyme feedback amplification strategy for biosensing. Angew. Chem. Int. Ed. 2017, 56, 6142–6146. [Google Scholar] [CrossRef] [PubMed]
- Sakhabutdinova, A.R.; Maksimova, M.A.; Garafutdinov, R.R. Synthesis of circular DNA templates with T4 RNA ligase for rolling circle amplification. Mol. Biol. 2017, 51, 724–733. [Google Scholar] [CrossRef]
- Di Giusto, D.A.; Wlassoff, W.A.; Gooding, J.J.; Messerle, B.A.; King, G.C. Proximity extension of circular DNA aptamers with real-time protein detection. Nucleic Acids Res. 2005, 33, e64. [Google Scholar] [CrossRef]
- Wang, L.; Tram, K.; Ali, M.M.; Salena, B.J.; Li, J.; Li, Y. Arrest of rolling circle amplification by protein- binding DNA aptamers. Chem. Eur. J. 2014, 20, 2420–2424. [Google Scholar] [CrossRef]
- Bialy, R.M.; Ali, M.M.; Li, Y.; Brennan, J.D. Protein-mediated suppression of rolling circle amplification for biosensing with an aptamer-containing DNA primer. Chem. Eur. J. 2020, 26, 5085–5092. [Google Scholar] [CrossRef]
- Song, J.; Ju, Y.; Kim, S.; Kim, H.; Park, H.G. Palindromic hyperbranched rolling circle amplification enabling ultrasensitive microRNA detection. Chem. Commun. 2022, 58, 6518–6521. [Google Scholar] [CrossRef]
- Lu, P.; Aodeng, G.; Ai, J. Progress in ion detection and clinical diagnosis based on G-quadruplex combined with fluorescence properties. Adv. Sens. Energy Mater. 2024, 3, 100112. [Google Scholar] [CrossRef]
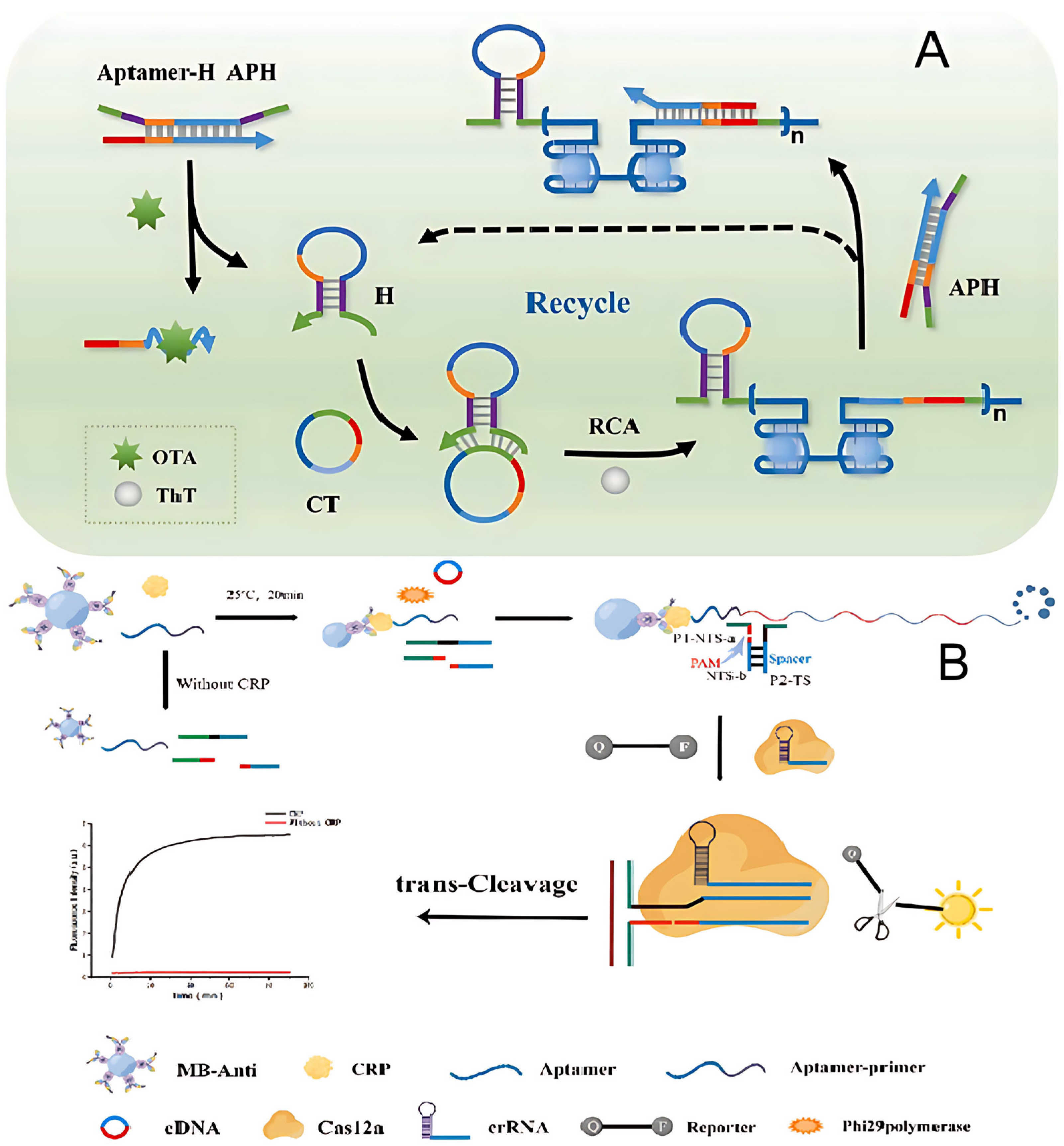
- Wang, W.; Geng, L.; Zhang, Y.; Shen, W.; Bi, M.; Gong, T.; Hu, Z.; Guo, C.; Wang, T.; Sun, T. Development of antibody-aptamer sandwich-like immunosensor based on RCA and Nicked-PAM CRISPR/Cas12a system for the ultra-sensitive detection of a biomarker. Anal. Chim. Acta 2023, 1283, 341849. [Google Scholar] [CrossRef]
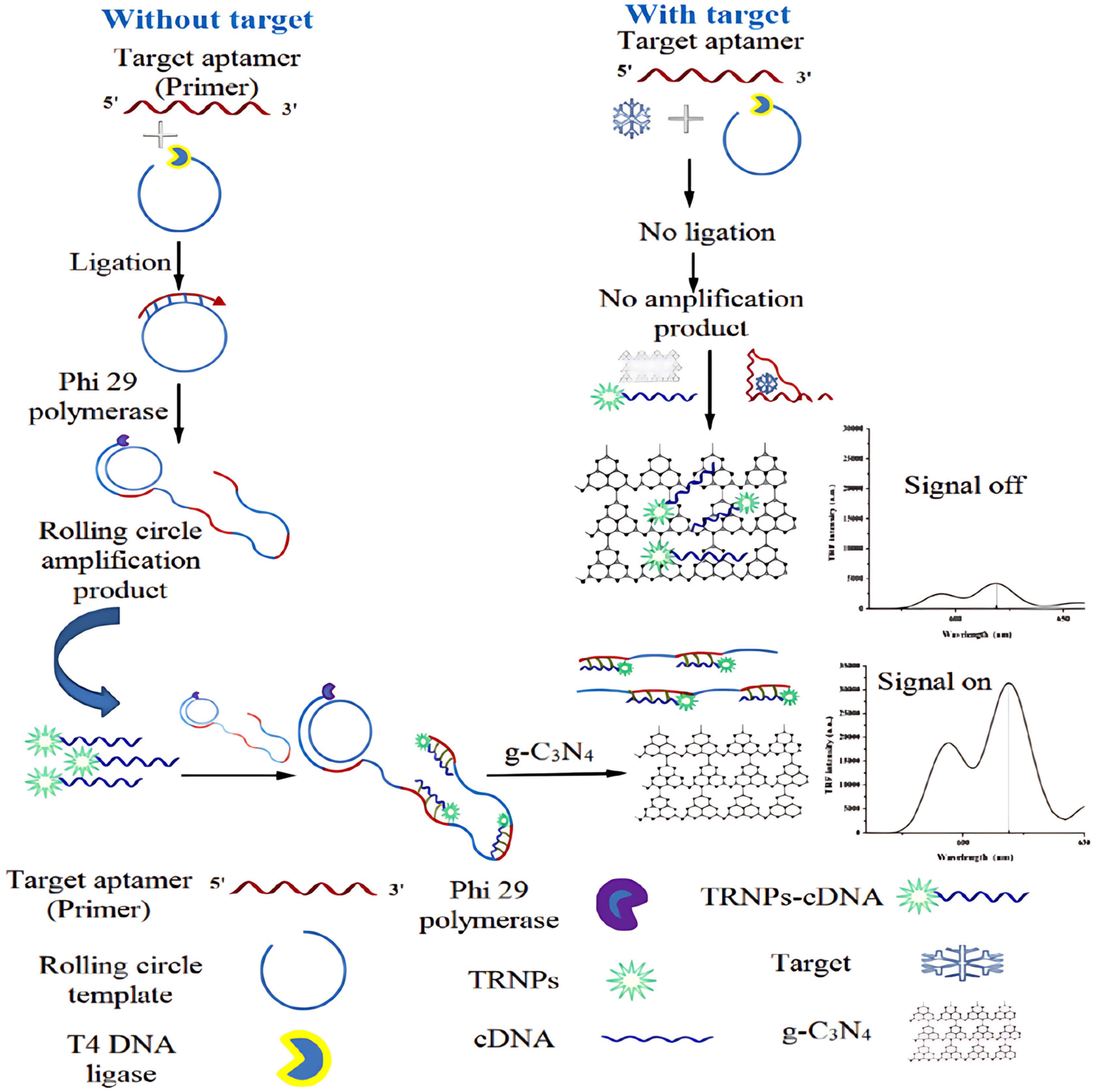
- Niazi, S.; Khan, I.M.; Yu, Y.; Pasha, I.; Lv, Y.; Mohsin, A.; Mushtaq, B.S.; Wang, Z. A novel fluorescent aptasensor for aflatoxin M1 detection using rolling circle amplification and g-C3N4 as fluorescence quencher. Sens. Actuators B Chem. 2020, 315, 128049. [Google Scholar] [CrossRef]
- Zhang, M.; Ye, L. Detection of SARS-CoV-2 receptor binding domain using fluorescence probe and DNA flowers enabled by rolling circle amplification. Microchim. Acta 2023, 190, 163. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Qi, P.; Zhou, Y.; Wang, Y.; Xin, Y.; Sun, Y.; Zhang, D. Multi pathogenic microorganisms determination using DNA composites-encapsulated DNA silver nanocluster/graphene oxide-based system through rolling cycle amplification. Microchim. Acta 2022, 189, 403. [Google Scholar] [CrossRef]
- Jiang, Y.; Qiu, Z.; Le, T.; Zou, S.; Cao, X. Developing a dual-RCA microfluidic platform for sensitive E. coli O 157:H7 whole-cell detections. Anal. Chim. Acta 2020, 1127, 79–88. [Google Scholar] [CrossRef]
- Li, S.; Jiang, Y.; Yang, X.; Lin, M.; Dan, H.; Zou, S.; Cao, X. In situ rolling circle amplification surface modifications to improve E. coli O157:H7 capturing performances for rapid and sensitive microfluidic detection applications. Anal. Chim. Acta 2021, 1150, 338229. [Google Scholar] [CrossRef]
- Yu, J.; Wu, H.; He, L.; Tan, L.; Jia, Z.; Gan, N. The universal dual-mode aptasensor for simultaneous determination of different bacteria based on naked eyes and microfluidic-chip together with magnetic DNA encoded probes. Talanta 2021, 225, 122062. [Google Scholar] [CrossRef]
- Gao, X.; Sun, Z.; Wang, X.; Zhang, W.; Xu, D.; Sun, X.; Guo, Y.; Xu, S.; Li, F. Construction of a dual-model aptasensor based on G-quadruplexes generated via rolling circle amplification for visual/sensitive detection of kanamycin. Sci. Total Environ. 2022, 839, 156276. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Yan, Y.; Zhi, S.; Bi, S. Dumbbell DNA-mediated rolling circle amplification for visual biosensing of intracellular glutathione. Sens. Actuators B Chem. 2022, 373, 132745. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, W.; Hu, Y.; Wei, X.; Huang, L.; Fan, S.; Huang, M.; Song, Y.; Yu, Y.; Fu, F. Visual detection of aflatoxin B1 based on specific aptamer recognition combining with triple amplification strategy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 271, 120862. [Google Scholar] [CrossRef]
- Yurdusev, E.; Trahan, P.-L.; Perreault, J. Adaptation of a model spike aptamer for isothermal amplification-based sensing. Sensors 2024, 24, 6875. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, S.; Tang, X.; Feng, L.; Ding, Z.; Chen, Z.; Luo, X.; Deng, R.; Sheng, J.; Xie, S.; et al. DNA four-way junction-driven dual-rolling circle amplification sandwich-type aptasensor for ultra-sensitive and specific detection of tumor-derived exosomes. Biosens. Bioelectron. 2024, 246, 115841. [Google Scholar] [CrossRef] [PubMed]
- Qing, M.; Sun, Z.; Wang, L.; Du, S.Z.; Zhou, J.; Tang, Q.; Luo, H.Q.; Li, N.B. CRISPR/Cas12a-regulated homogeneous electrochemical aptasensor for amplified detection of protein. Sens. Actuators B Chem. 2021, 348, 130713. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Alibolandi, M.; Nameghi, M.A.; Gerayelou, G.; Abnous, K. A novel electrochemical aptasensor for ochratoxin a sensing in spiked food using strand-displacement polymerase reaction. Talanta 2021, 223, 121705. [Google Scholar] [CrossRef] [PubMed]

| Detection Method | Sensitivity | Complexity | Instrument Requirement | Key Advantages | Limitations |
|---|---|---|---|---|---|
| Fluorescent | High (fM–pM) | Medium | Fluorescence reader | High sensitivity; visual readout; multiplexing | Prone to background noise; high equipment cost |
| Electrochemical | Very high (aM–fM) | High | Potentiostat | Portable; excellent sensitivity and specificity | Surface modification complexity; reproducibility issues |
| Visual | Moderate (nM–pM) | Low | None/Basic | Simple; naked-eye detection | Lower sensitivity; often qualitative/semi-quantitative |
| Microfluidic | High (pM–fM) | High | Microfluidic chip system | Integrated, fast, miniaturized | High fabrication cost; needs standardization |
| Aspect | Description | Examples/Notes |
|---|---|---|
| Principle | Combination of aptamer-based target recognition with RCA for signal enhancement | RCA generates long DNA products to amplify detection signals upon target recognition by aptamers |
| Key targets detected | Viral RNAs Small-molecule toxins Cell surface markers Proteins and metabolites Whole cells or pathogens | SARS-CoV-2 RNA OTA, Aflatoxin B1 CD63 on exosomes Thrombin, Mucin-1, PDGF, ATP, Glutathione E. coli, Salmonella typhimurium |
| Application areas | Disease diagnostics Food safety Environmental monitoring | Effective even in complex biological and environmental samples |
| Advantages | High sensitivity High specificity Signal amplification without thermal cycling Potential for miniaturization and portability | Aptamers provide target recognition; RCA ensures signal amplification |
| Challenges | Reduced aptamer selectivity in complex samples Inefficient RCA in suboptimal reaction conditions Interference from non-target biomolecules | Serum proteins or nucleases may bind non-specifically Low Mg2+ or inhibitors reduce RCA activity DNA-binding proteins or albumin disrupt performance |
| Strategies to Overcome Challenges | Chemically modify aptamers for better stability and binding Engineer polymerases for robust RCA in biological fluids Integrate with complementary technologies | CRISPR/Cas detection Nanomaterials (e.g., gold NPs, graphene oxide) Microfluidic or portable systems |
| Future Directions | Real-time biomarker monitoring Single-cell analysis On-site toxin detection using portable sensors | Emphasis on simplicity, standardization, cost-effectiveness, and clinical applicability |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; He, W. Recent Advances in the Development of Functional Nucleic Acid Biosensors Based on Aptamer-Rolling Circle Amplification. Molecules 2025, 30, 2375. https://doi.org/10.3390/molecules30112375
Liu C, He W. Recent Advances in the Development of Functional Nucleic Acid Biosensors Based on Aptamer-Rolling Circle Amplification. Molecules. 2025; 30(11):2375. https://doi.org/10.3390/molecules30112375
Chicago/Turabian StyleLiu, Ce, and Wanchong He. 2025. "Recent Advances in the Development of Functional Nucleic Acid Biosensors Based on Aptamer-Rolling Circle Amplification" Molecules 30, no. 11: 2375. https://doi.org/10.3390/molecules30112375
APA StyleLiu, C., & He, W. (2025). Recent Advances in the Development of Functional Nucleic Acid Biosensors Based on Aptamer-Rolling Circle Amplification. Molecules, 30(11), 2375. https://doi.org/10.3390/molecules30112375







