Unlocking the Potential of Perillaldehyde: A Novel Mechanism for Chronic Myeloid Leukemia by Targeting HSP70
Abstract
1. Introduction
2. Results
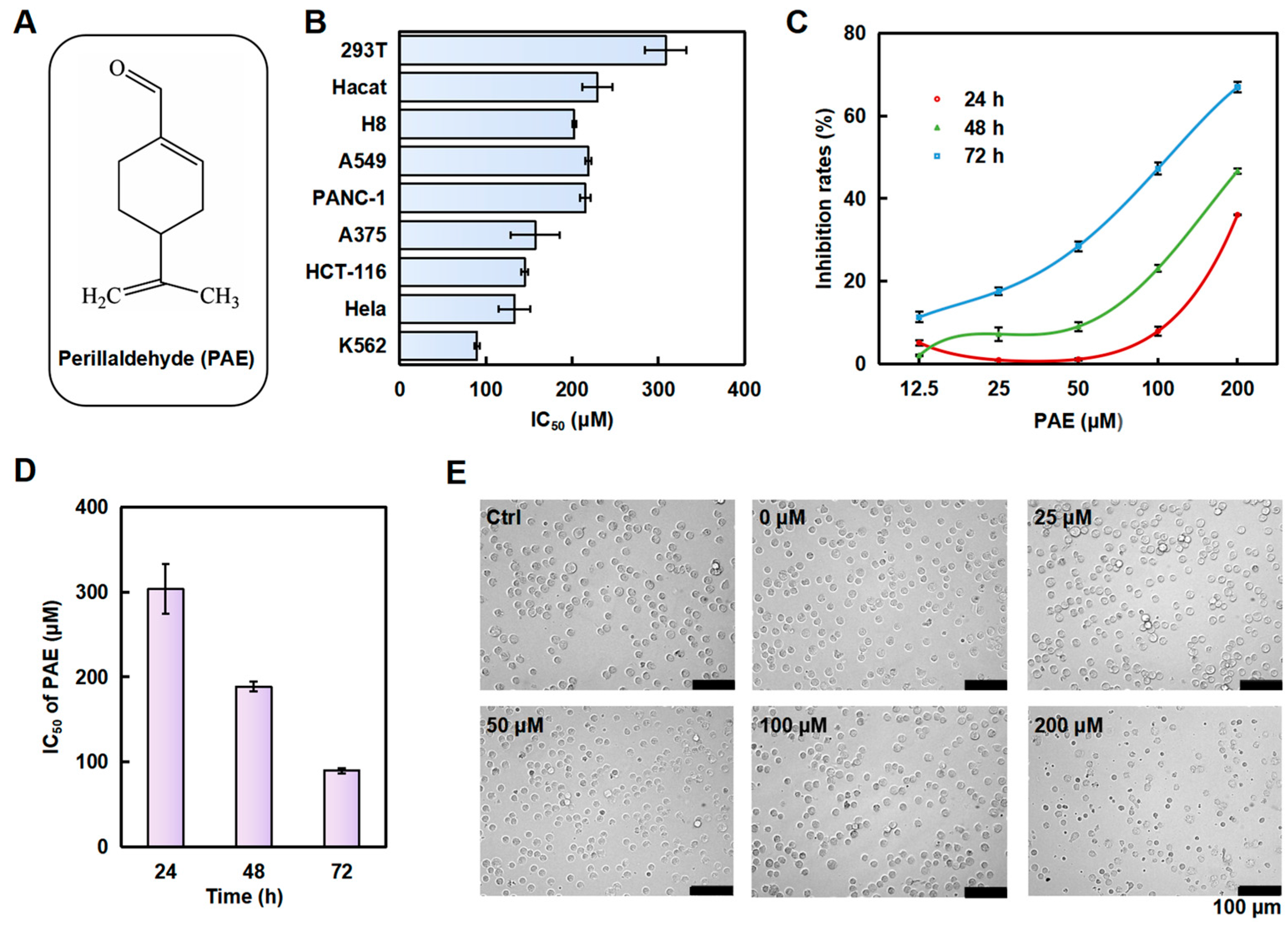
2.1. PAE Inhibited the Growth of K562 Cells In Vitro
2.2. PAE Induced G0/G1 Phase Arrest and Caspase-Dependent Apoptosis in K562 Cells
2.3. PAE Induced Autophagy in K562 Cells
2.4. PAE Disrupted Mitochondrial Function in K562 Cells
2.5. PAE Downregulated the Phosphorylated BCR-ABL Protein and Inhibited Its Downstream Proteins in K562 Cells
2.6. DARTS Assay Reveals That PAE Targets HSP70
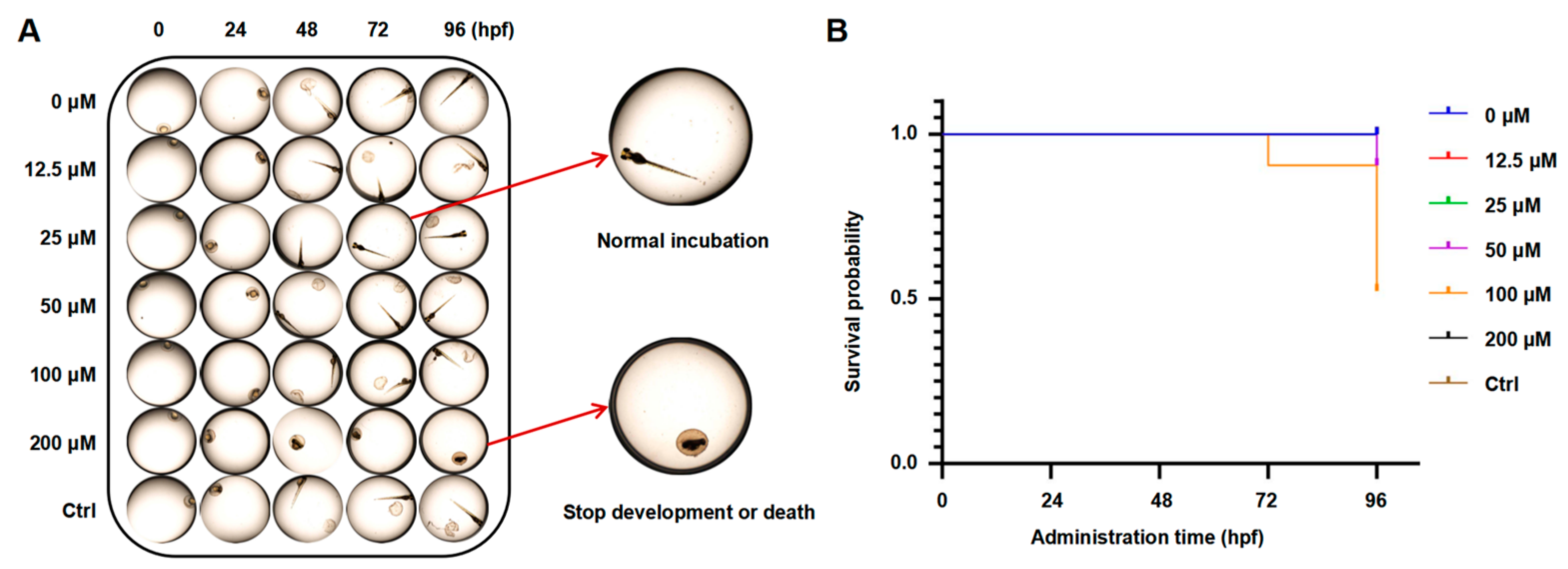
2.7. Effects of PAE on Toxicity in Zebrafish Embryos
3. Discussion
4. Materials and Methods
4.1. Cells and Reagents
4.2. Cell Viability Assessment
4.3. Cell Cycle Analysis
4.4. Monodansylcadaverine (MDC) Staining
4.5. Measurement of the Mitochondrial Membrane Potential (MMP)
4.6. Measurement of Reactive Oxygen Species (ROS) Generation
4.7. Drug Affinity Responsive Target Stability (DARTS)
4.8. Coimmunoprecipitation (Co-IP)
4.9. Molecular Dynamics (MD) Assay and Molecular Docking
4.10. Acute Toxicity Test of Zebrafish Embryos
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2022 update on diagnosis, therapy, and monitoring. Am. J. Hematol. 2022, 97, 1236–1256. [Google Scholar] [CrossRef] [PubMed]
- Al-Rawashde, F.A.; Al-Sanabra, O.M.; Alqaraleh, M.; Jaradat, A.Q.; Al-Wajeeh, A.S.; Johan, M.F.; Wan Taib, W.R.; Ismail, I.; Al-Jamal, H.A.N. Thymoquinone Enhances Apoptosis of K562 Chronic Myeloid Leukemia Cells through Hypomethylation of SHP-1 and Inhibition of JAK/STAT Signaling Pathway. Pharmaceuticals 2023, 16, 884. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2020 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2020, 95, 691–709. [Google Scholar] [CrossRef]
- Szczepanek, E.; Chukwu, O.; Kamińska, M.; Wysogląd, H.; Cenda, A.; Zawada, M.; Jakóbczyk, M.; Wącław, J.; Sacha, T. Long-term outcomes of patients with Chronic Myeloid Leukemia who commenced treatment with imatinib: A 20-year single-centre experience. Leuk. Lymphoma 2022, 63, 2213–2223. [Google Scholar] [CrossRef]
- Alves, R.; Gonçalves, A.C.; Rutella, S.; Almeida, A.M.; De Las Rivas, J.; Trougakos, I.P.; Sarmento Ribeiro, A.B. Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia-From Molecular Mechanisms to Clinical Relevance. Cancers 2021, 13, 4820. [Google Scholar] [CrossRef]
- Meenakshi Sundaram, D.N.; Jiang, X.; Brandwein, J.M.; Valencia-Serna, J.; Remant, K.C.; Uludağ, H. Current outlook on drug resistance in chronic myeloid leukemia (CML) and potential therapeutic options. Drug Discov. Today 2019, 24, 1355–1369. [Google Scholar] [CrossRef] [PubMed]
- Xiaokaiti, Y.; Li, X. Natural Product Regulates Autophagy in Cancer. Adv. Exp. Med. Biol. 2020, 1207, 709–724. [Google Scholar]
- Masumoto, N.; Nishizaki, Y.; Maruyama, T.; Igarashi, Y.; Nakajima, K.; Yamazaki, T.; Kuroe, M.; Numata, M.; Ihara, T.; Sugimoto, N.; et al. Determination of perillaldehyde in perilla herbs using relative molar sensitivity to single-reference diphenyl sulfone. J. Nat. Med. 2019, 73, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, M.; Pan, S.; Pan, C.; Li, Y.; Tian, J. Perillaldehyde Controls Postharvest Black Rot Caused by Ceratocystis fimbriata in Sweet Potatoes. Front. Microbiol. 2018, 9, 1102. [Google Scholar] [CrossRef]
- Wei, J.; Liu, Z.; Sun, H.; Xu, L. Perillaldehyde ameliorates lipopolysaccharide-induced acute lung injury via suppressing the cGAS/STING signaling pathway. Int. Immunopharmacol. 2024, 130, 111641. [Google Scholar] [CrossRef]
- Lin, Z.; Huang, S.; LingHu, X.; Wang, Y.; Wang, B.; Zhong, S.; Xie, S.; Xu, X.; Yu, A.; Nagai, A.; et al. Perillaldehyde inhibits bone metastasis and receptor activator of nuclear factor-κB ligand (RANKL) signaling-induced osteoclastogenesis in prostate cancer cell lines. Bioengineered 2022, 13, 2710–2719. [Google Scholar] [CrossRef] [PubMed]
- Erhunmwunsee, F.; Pan, C.; Yang, K.; Li, Y.; Liu, M.; Tian, J. Recent development in biological activities and safety concerns of perillaldehyde from perilla plants: A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 6328–6340. [Google Scholar] [CrossRef]
- He, M.; You, J.; Liu, X.; Peng, X.; Li, C.; Yang, S.; Xu, Q.; Lin, J.; Zhao, G. Perillaldehyde Protects Against Aspergillus fumigatus Keratitis by Reducing Fungal Load and Inhibiting Inflammatory Cytokines and LOX-1. Curr. Eye Res. 2022, 47, 1366–1373. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Qu, S.; Yang, K.; Liu, M.; Li, Y.X.; Keller, N.P.; Zeng, X.; Tian, J. Perillaldehyde: A promising antifungal agent to treat oropharyngeal candidiasis. Biochem. Pharmacol. 2020, 180, 114201. [Google Scholar] [CrossRef]
- Wang, C.Y.; Wang, S.Y.; Chen, C. Increasing antioxidant activity and reducing decay of blueberries by essential oils. J. Agric. Food Chem. 2008, 56, 3587–3592. [Google Scholar] [CrossRef]
- Catanzaro, E.; Turrini, E.; Kerre, T.; Sioen, S.; Baeyens, A.; Guerrini, A.; Bellau, M.L.A.; Sacchetti, G.; Paganetto, G.; Krysko, D.V.; et al. Perillaldehyde is a new ferroptosis inducer with a relevant clinical potential for acute myeloid leukemia therapy. Biomed. Pharmacother. 2022, 154, 113662. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zhou, G.; Liu, Y.; Zhang, J.; Chen, Y.; Liu, L.; Zhang, G. HSP70 Family in Cancer: Signaling Mechanisms and Therapeutic Advances. Biomolecules 2023, 13, 601. [Google Scholar] [CrossRef]
- Werner, C.; Stangl, S.; Salvermoser, L.; Schwab, M.; Shevtsov, M.; Xanthopoulos, A.; Wang, F.; Dezfouli, A.B.; Thölke, D.; Ostheimer, C.; et al. Hsp70 in Liquid Biopsies-A Tumor-Specific Biomarker for Detection and Response Monitoring in Cancer. Cancers 2021, 13, 3706. [Google Scholar] [CrossRef]
- Singh, M.K.; Han, S.; Ju, S.; Ranbhise, J.S.; Ha, J.; Yeo, S.G.; Kim, S.S.; Kang, I. Hsp70: A Multifunctional Chaperone in Maintaining Proteostasis and Its Implications in Human Disease. Cells 2025, 14, 509. [Google Scholar] [CrossRef]
- Zorzi, E.; Bonvini, P. Inducible hsp70 in the regulation of cancer cell survival: Analysis of chaperone induction, expression and activity. Cancers 2011, 3, 3921–3956. [Google Scholar] [CrossRef]
- Song, S.; Lee, J.Y.; Ermolenko, L.; Mazumder, A.; Ji, S.; Ryu, H.; Kim, H.; Kim, D.W.; Lee, J.W.; Dicato, M.; et al. Tetrahydrobenzimidazole TMQ0153 triggers apoptosis, autophagy and necroptosis crosstalk in chronic myeloid leukemia. Cell Death Dis. 2020, 11, 109. [Google Scholar] [CrossRef] [PubMed]
- Abate, M.; Festa, A.; Falco, M.; Lombardi, A.; Luce, A.; Grimaldi, A.; Zappavigna, S.; Sperlongano, P.; Irace, C.; Caraglia, M.; et al. Mitochondria as playmakers of apoptosis, autophagy and senescence. Semin. Cell Dev. Biol. 2020, 98, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Kalpage, H.A.; Bazylianska, V.; Recanati, M.A.; Fite, A.; Liu, J.; Wan, J.; Mantena, N.; Malek, M.H.; Podgorski, I.; Heath, E.I.; et al. Tissue-specific regulation of cytochrome c by post-translational modifications: Respiration, the mitochondrial membrane potential, ROS, and apoptosis. FASEB J. 2019, 33, 1540–1553. [Google Scholar] [CrossRef]
- Song, Y.; Wang, K.; Loor, J.J.; Jiang, Q.; Yang, Y.; Jiang, S.; Liu, S.; He, J.; Feng, X.; Du, X.; et al. β-Hydroxybutyrate inhibits apoptosis in bovine neutrophils through activating ERK1/2 and AKT signaling pathways. J. Dairy. Sci. 2022, 105, 3477–3489. [Google Scholar] [CrossRef]
- Al-Rawashde, F.A.; Wan Taib, W.R.; Ismail, I.; Johan, M.F.; Al-Wajeeh, A.S.; Al-Jamal, H.A.N. Thymoquinone Induces Downregulation of BCR-ABL/JAK/STAT Pathway and Apoptosis in K562 Leukemia Cells. Asian Pac. J. Cancer Prev. 2021, 22, 3959–3965. [Google Scholar] [CrossRef] [PubMed]
- Al-Rawashde, F.A.; Al-Wajeeh, A.S.; Vishkaei, M.N.; Saad, H.K.M.; Johan, M.F.; Taib, W.R.W.; Ismail, I.; Al-Jamal, H.A.N. Thymoquinone Inhibits JAK/STAT and PI3K/Akt/ mTOR Signaling Pathways in MV4-11 and K562 Myeloid Leukemia Cells. Pharmaceuticals 2022, 15, 1123. [Google Scholar] [CrossRef]
- Wang, J.; Liu, B.; Zheng, Q.; Xiao, R.; Chen, J. Newly emerged ROS1 rearrangement in a patient with lung adenocarcinoma following resistance to immune checkpoint inhibitors: A case report. Front. Oncol. 2024, 14, 1507658. [Google Scholar] [CrossRef]
- Blay, J.Y.; von Mehren, M. Nilotinib: A novel, selective tyrosine kinase inhibitor. Semin. Oncol. 2011, 38 (Suppl. 1), S3–S9. [Google Scholar] [CrossRef]
- Shah, N.P.; Tran, C.; Lee, F.Y.; Chen, P.; Norris, D.; Sawyers, C.L. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science 2004, 305, 399–401. [Google Scholar] [CrossRef]
- Ghosh, J.; Kobayashi, M.; Ramdas, B.; Chatterjee, A.; Ma, P.; Mali, R.S.; Carlesso, N.; Liu, Y.; Plas, D.R.; Chan, R.J.; et al. S6K1 regulates hematopoietic stem cell self-renewal and leukemia maintenance. J. Clin. Investig. 2016, 126, 2621–2625. [Google Scholar] [CrossRef]
- Solmaz, S.; Adan Gokbulut, A.; Cincin, B.; Ozdogu, H.; Boga, C.; Cakmakoglu, B.; Kozanoglu, I.; Baran, Y. Therapeutic potential of apigenin, a plant flavonoid, for imatinib-sensitive and resistant chronic myeloid leukemia cells. Nutr. Cancer 2014, 66, 599–612. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feriotto, G.; Tagliati, F.; Giriolo, R.; Casciano, F.; Tabolacci, C.; Beninati, S.; Khan, M.T.H.; Mischiati, C. Caffeic Acid Enhances the Anti-Leukemic Effect of Imatinib on Chronic Myeloid Leukemia Cells and Triggers Apoptosis in Cells Sensitive and Resistant to Imatinib. Int. J. Mol. Sci. 2021, 22, 1644. [Google Scholar] [CrossRef] [PubMed]





| PAE (μM) | Survival Number | Number of Deaths | Number of Deformities | Mortalities | Malformation Rates | |
|---|---|---|---|---|---|---|
| 24 hpf | 0 | 20 | 0 | 0 | 0.00% | 0.00% |
| 12.5 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 25 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 50 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 100 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 200 | 20 | 0 | 0 | 0.00% | 0.00% | |
| Ctrl | 20 | 0 | 0 | 0.00% | 0.00% | |
| 48 hpf | 0 | 20 | 0 | 0 | 0.00% | 0.00% |
| 12.5 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 25 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 50 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 100 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 200 | 20 | 0 | 0 | 0.00% | 0.00% | |
| Ctrl | 20 | 0 | 0 | 0.00% | 0.00% | |
| 72 hpf | 0 | 20 | 0 | 0 | 0.00% | 0.00% |
| 12.5 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 25 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 50 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 100 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 200 | 20 | 0 | 3 | 0.00% | 15.00% | |
| Ctrl | 20 | 0 | 0 | 0.00% | 0.00% | |
| 96 hpf | 0 | 20 | 0 | 0 | 0.00% | 0.00% |
| 12.5 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 25 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 50 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 100 | 20 | 0 | 2 | 0.00% | 10.00% | |
| 200 | 18 | 2 | 7 | 10.00% | 35.00% | |
| Ctrl | 20 | 0 | 0 | 0.00% | 0.00% | |
| 120 hpf | 0 | 20 | 0 | 0 | 0.00% | 0.00% |
| 12.5 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 25 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 50 | 20 | 0 | 0 | 0.00% | 0.00% | |
| 100 | 18 | 2 | 2 | 10.00% | 10.00% | |
| 200 | 10 | 10 | 7 | 50.00% | 35.00% | |
| Ctrl | 20 | 0 | 0 | 0.00% | 0.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Wang, J.; Jiang, R.; Liu, M.; Zhang, W. Unlocking the Potential of Perillaldehyde: A Novel Mechanism for Chronic Myeloid Leukemia by Targeting HSP70. Molecules 2025, 30, 2294. https://doi.org/10.3390/molecules30112294
Zhang M, Wang J, Jiang R, Liu M, Zhang W. Unlocking the Potential of Perillaldehyde: A Novel Mechanism for Chronic Myeloid Leukemia by Targeting HSP70. Molecules. 2025; 30(11):2294. https://doi.org/10.3390/molecules30112294
Chicago/Turabian StyleZhang, Miaomiao, Jinfeng Wang, Rongsong Jiang, Ming Liu, and Weiyi Zhang. 2025. "Unlocking the Potential of Perillaldehyde: A Novel Mechanism for Chronic Myeloid Leukemia by Targeting HSP70" Molecules 30, no. 11: 2294. https://doi.org/10.3390/molecules30112294
APA StyleZhang, M., Wang, J., Jiang, R., Liu, M., & Zhang, W. (2025). Unlocking the Potential of Perillaldehyde: A Novel Mechanism for Chronic Myeloid Leukemia by Targeting HSP70. Molecules, 30(11), 2294. https://doi.org/10.3390/molecules30112294






