Docking-Based Classification of SGLT2 Inhibitors
Abstract
1. Introduction
2. Results and Discussion
2.1. Suitability Analyses
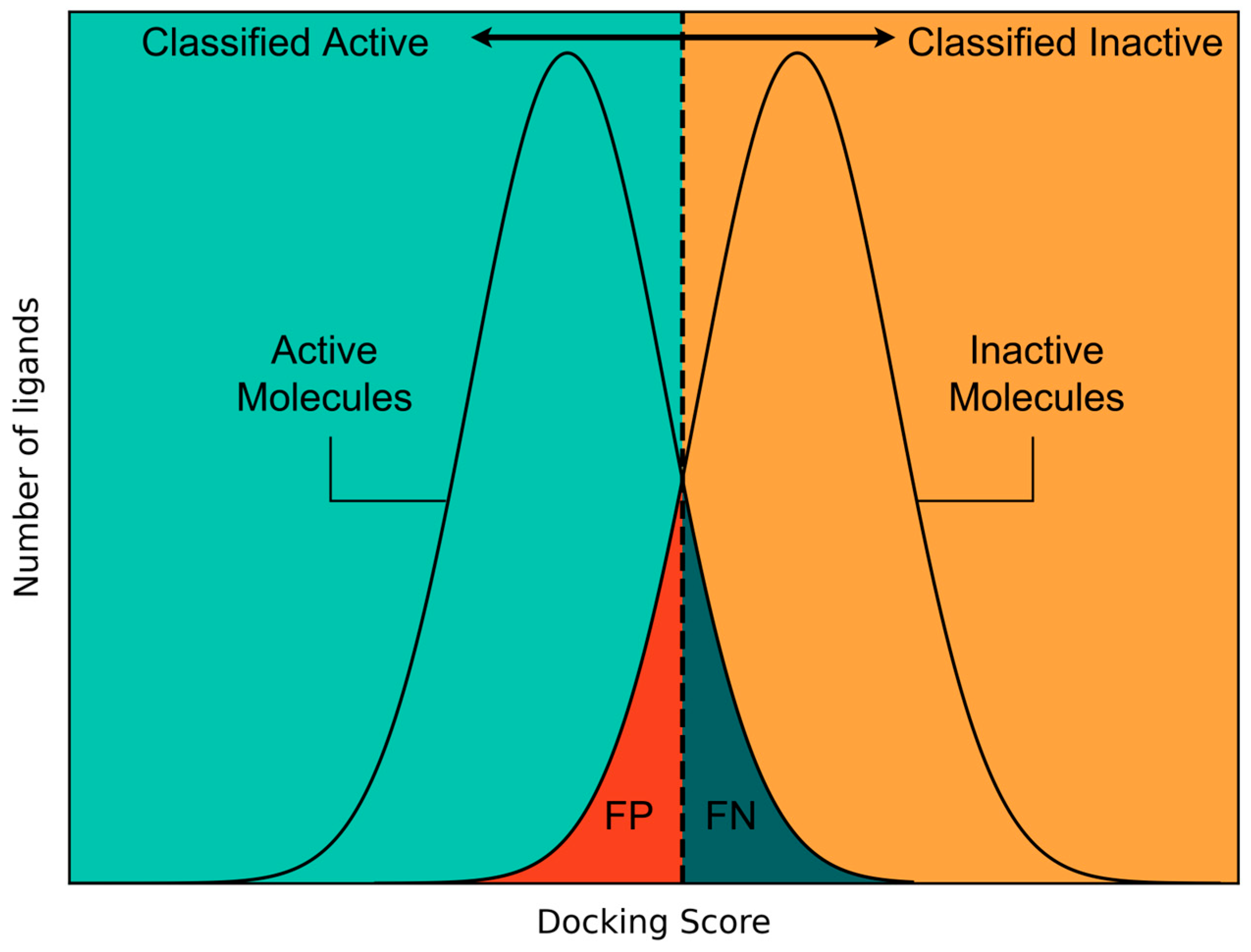
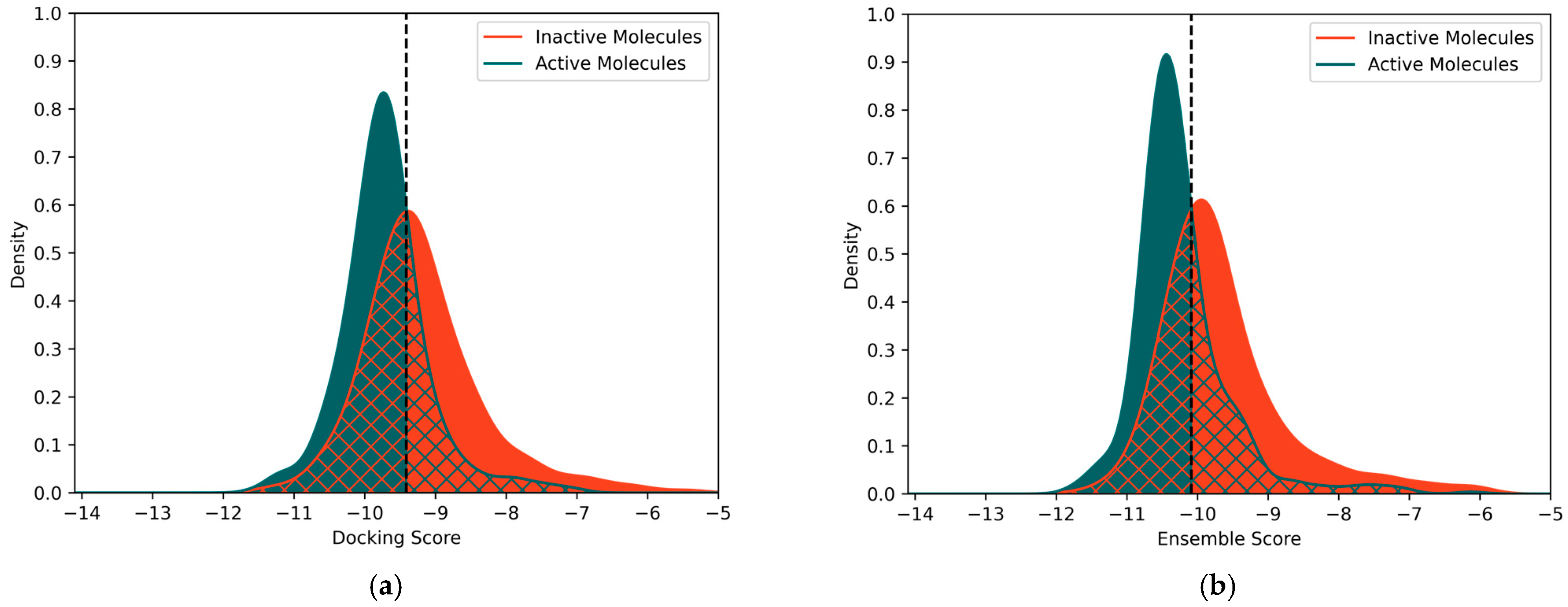
2.2. Docking Score-Based Classification
2.3. Ensemble Docking-Based Classification
2.4. Structural Similarity-Based Classification
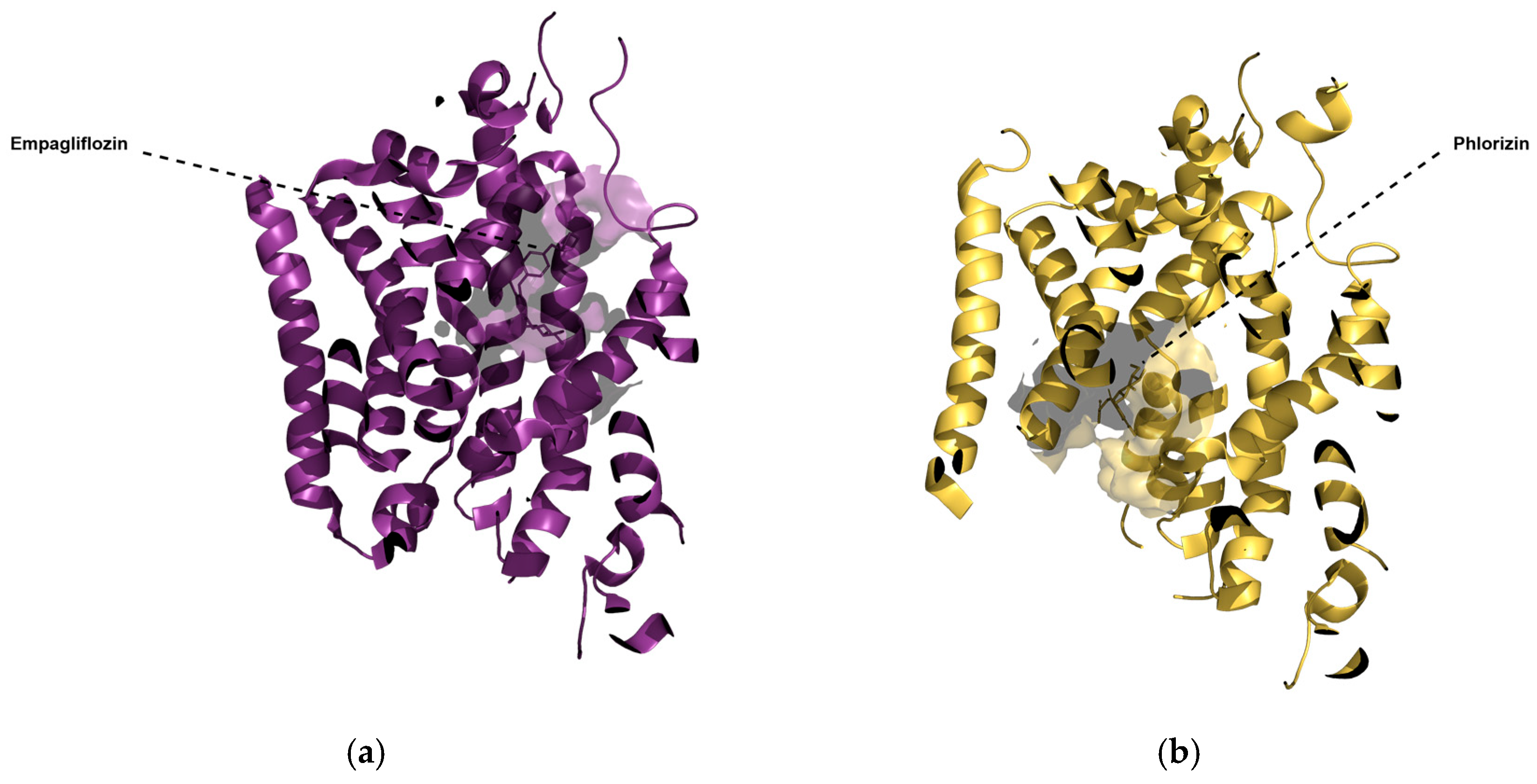
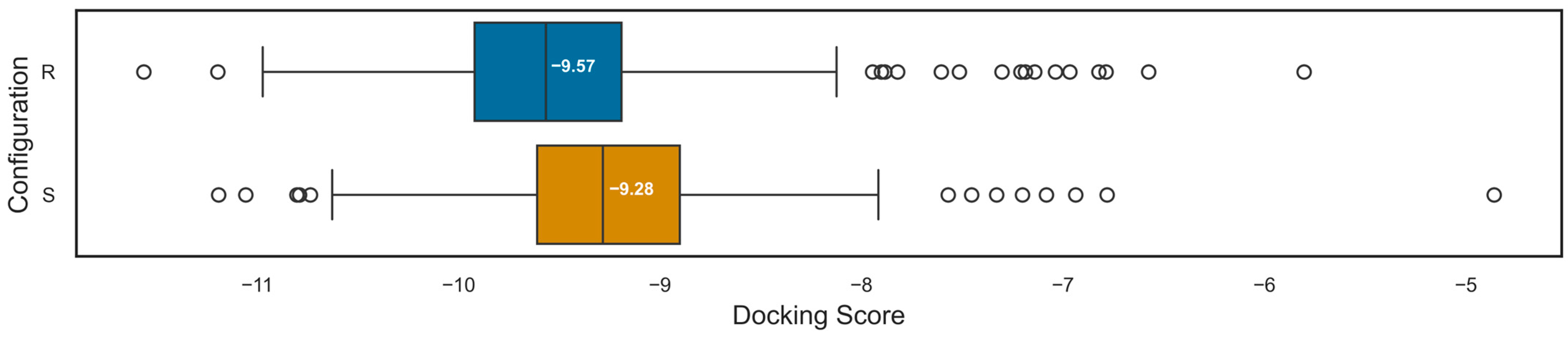
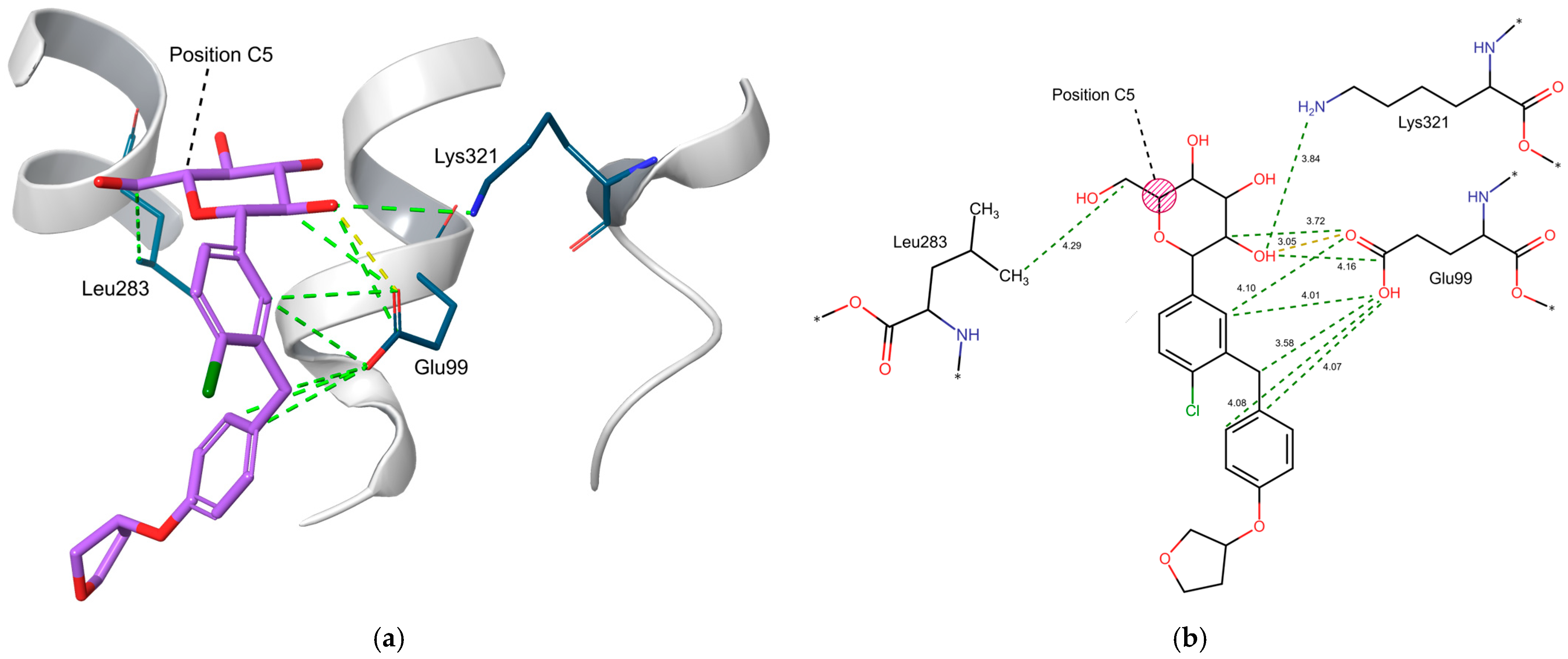
2.5. Three-Dimensional Influence
3. Materials and Methods
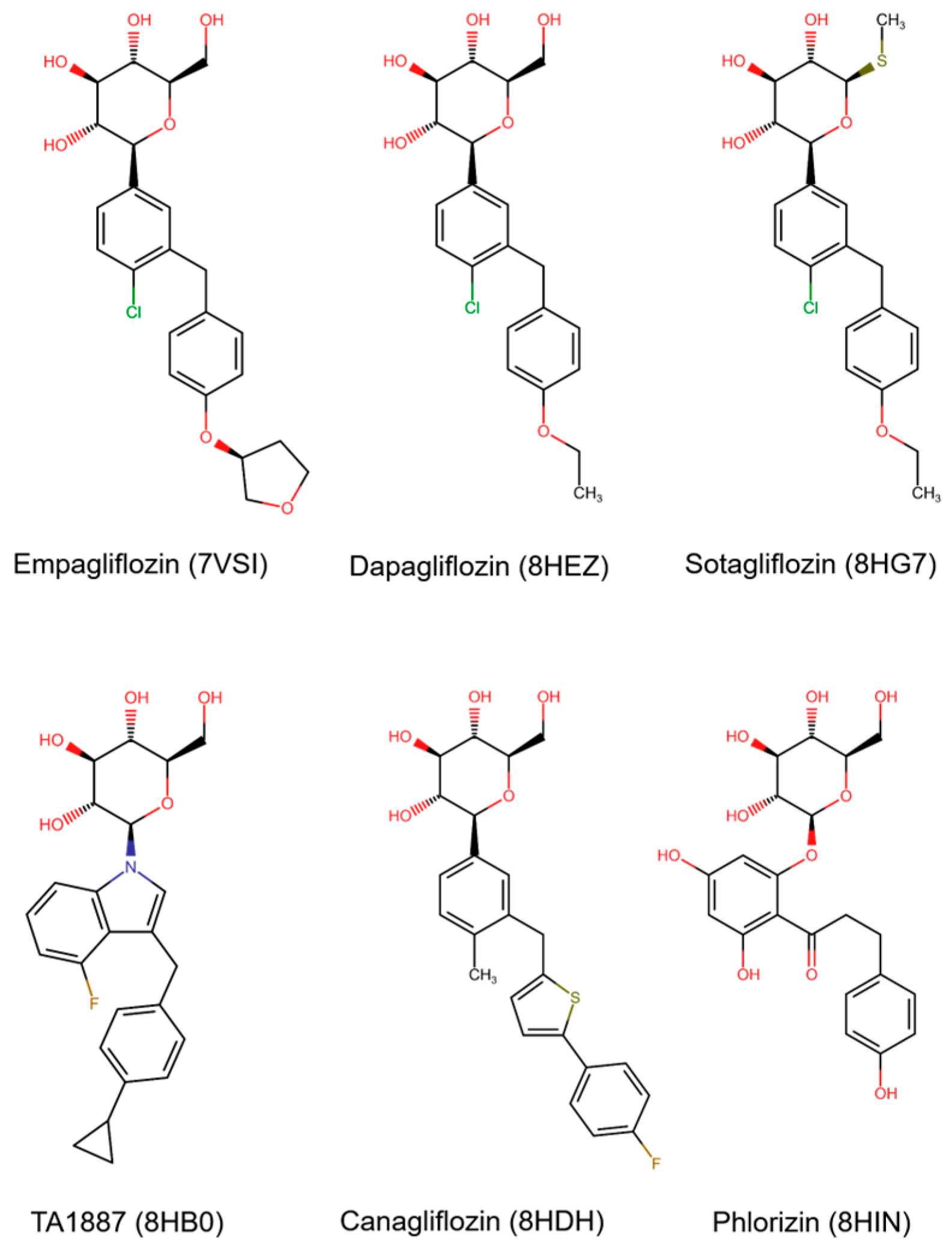
3.1. Data Retrieval
3.2. Molecular Docking
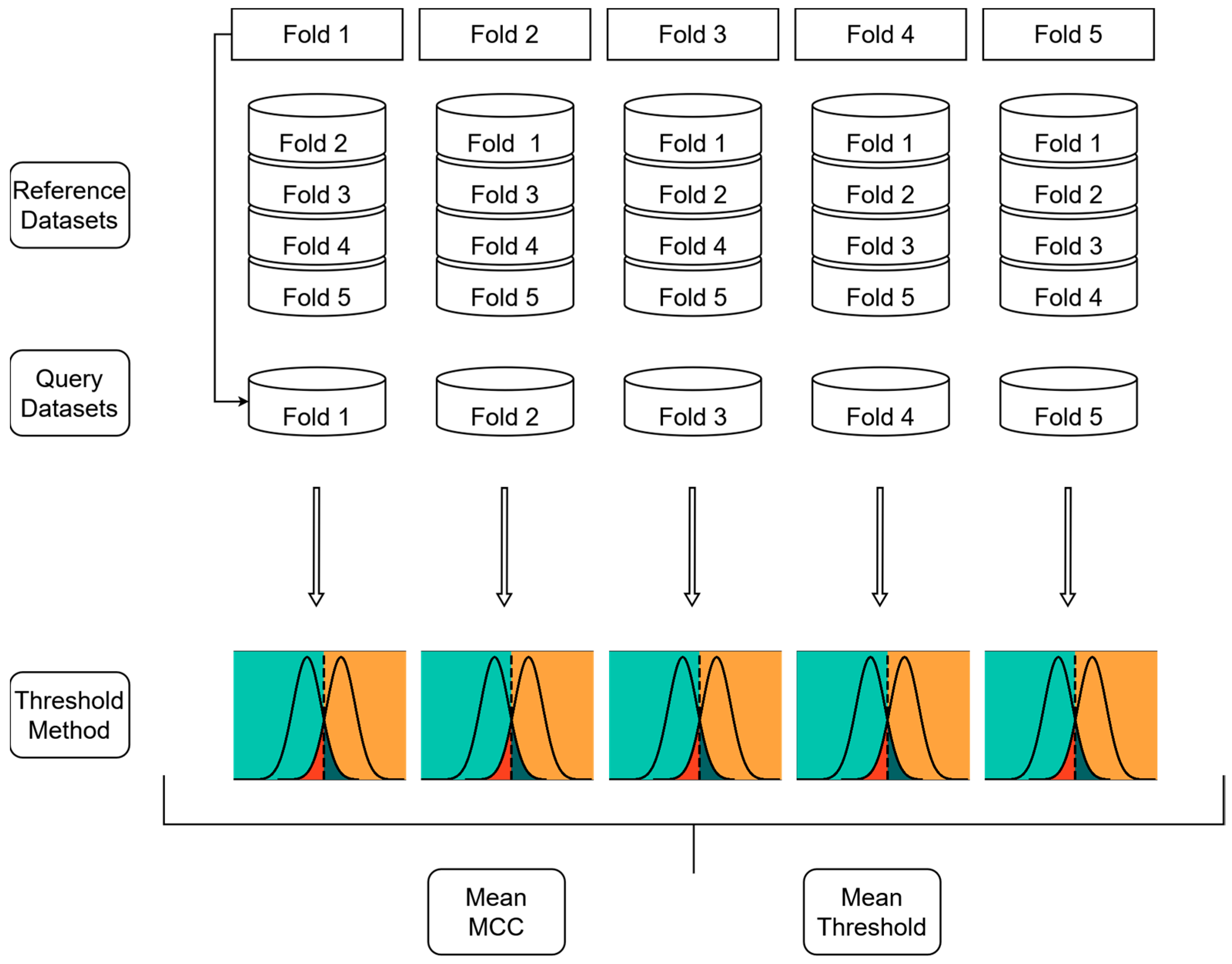
3.3. Classification Models
3.3.1. Docking Score-Based Classification Model
3.3.2. Ensemble Docking-Based Classification Models
3.3.3. Structural Similarity-Based Classification Models
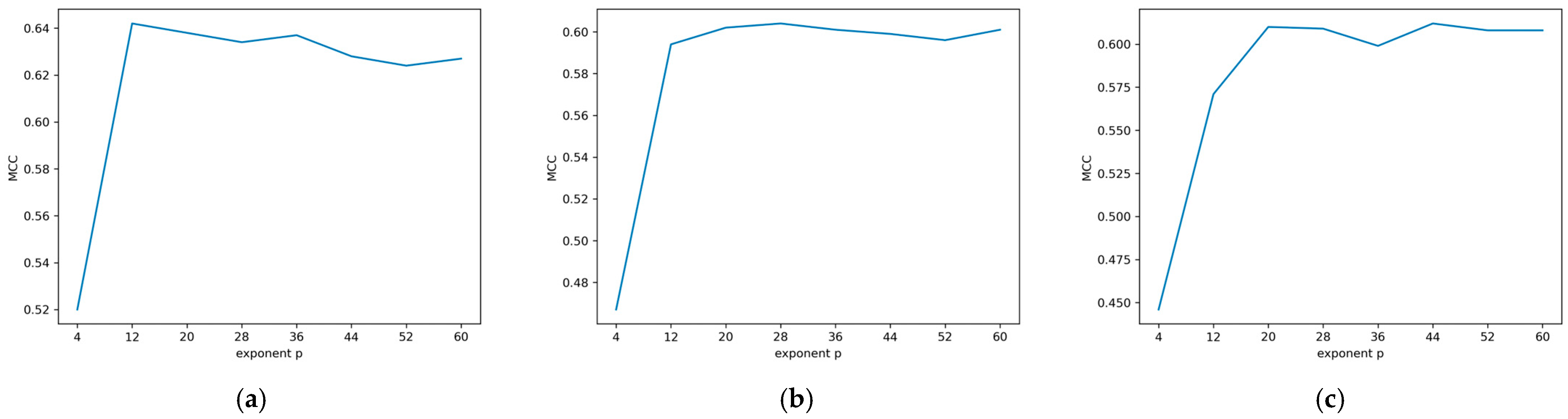
“where and are the docking score of the jth query compound before and after the calibration. is the structural similarity between the jth query compound and the ith reference ligand. The exponent p is treated as an integer constant with its value varying from 1 to 4 in this study, for the exploration of the developed formula. We referred as compound similarity effect (CSE) function for convenience of discussion. n is the total number of reference ligands in the reference dataset. is the docking score of the ith reference ligand. is the experimental binding energy (kcal/mol) of the ith compound in the reference dataset […].”[26].
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021. [Google Scholar]
- Clar, C.; Gill, J.A.; Court, R.; Waugh, N. Systematic Review of SGLT2 Receptor Inhibitors in Dual or Triple Therapy in Type 2 Diabetes. BMJ Open 2012, 2, e001007. [Google Scholar] [CrossRef]
- Whaley, J.; Tirmenstein, M.; Reilly, T.P.; Poucher, S.M.; Saye, J.; Parikh, S.; List, J.F. Targeting the Kidney and Glucose Excretion with Dapagliflozin: Preclinical and Clinical Evidence for SGLT2 Inhibition as a New Option for Treatment of Type 2 Diabetes Mellitus. Diabetes Metab. Syndr. Obes. Targets Ther. 2012, 5, 135–148. [Google Scholar] [CrossRef]
- Ehrenkranz, J.R.L.; Lewis, N.G.; Ronald Kahn, C.; Roth, J. Phlorizin: A Review. Diabetes Metab. Res. Rev. 2005, 21, 31–38. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Rathore, A.; Parwani, D.; Mallick, C.; Asati, V.; Agarwal, S.; Rajoriya, V.; Das, R.; Kashaw, S.K. An Exhaustive Perspective on Structural Insights of SGLT2 Inhibitors: A Novel Class of Antidiabetic Agent. Eur. J. Med. Chem. 2020, 204, 112523. [Google Scholar] [CrossRef]
- Cai, W.; Jiang, L.; Xie, Y.; Liu, Y.; Liu, W.; Zhao, G. Design of SGLT2 Inhibitors for the Treatment of Type 2 Diabetes: A History Driven by Biology to Chemistry. Med. Chem. 2015, 11, 317–328. [Google Scholar] [CrossRef]
- Niu, Y.; Liu, R.; Guan, C.; Zhang, Y.; Chen, Z.; Hoerer, S.; Nar, H.; Chen, L. Structural Basis of Inhibition of the Human SGLT2–MAP17 Glucose Transporter. Nature 2022, 601, 280–284. [Google Scholar] [CrossRef]
- Hiraizumi, M.; Akashi, T.; Murasaki, K.; Kishida, H.; Kumanomidou, T.; Torimoto, N.; Nureki, O.; Miyaguchi, I. Transport and Inhibition Mechanism of the Human SGLT2–MAP17 Glucose Transporter. Nat. Struct. Mol. Biol. 2024, 31, 159–169. [Google Scholar] [CrossRef]
- Macalino, S.J.Y.; Gosu, V.; Hong, S.; Choi, S. Role of Computer-Aided Drug Design in Modern Drug Discovery. Arch. Pharm. Res. 2015, 38, 1686–1701. [Google Scholar] [CrossRef]
- Paul, S.M.; Mytelka, D.S.; Dunwiddie, C.T.; Persinger, C.C.; Munos, B.H.; Lindborg, S.R.; Schacht, A.L. How to Improve R&D Productivity: The Pharmaceutical Industry’s Grand Challenge. Nat. Rev. Drug Discov. 2010, 9, 203–214. [Google Scholar] [CrossRef]
- Lavecchia, A. Machine-Learning Approaches in Drug Discovery: Methods and Applications. Drug Discov. Today 2015, 20, 318–331. [Google Scholar] [CrossRef]
- Sethi, A.; Joshi, K.; Sasikala, K.; Alvala, M. Molecular Docking in Modern Drug Discovery: Principles and Recent Applications. In Drug Discovery and Development—New Advances; Gaitonde, V., Karmakar, P., Trivedi, A., Eds.; IntechOpen: London, UK, 2020; ISBN 978-1-78923-975-1. [Google Scholar]
- Huang, S.-Y.; Zou, X. Advances and Challenges in Protein-Ligand Docking. Int. J. Mol. Sci. 2010, 11, 3016–3034. [Google Scholar] [CrossRef]
- Keyvanpour, M.R.; Shirzad, M.B. An Analysis of QSAR Research Based on Machine Learning Concepts. Curr. Drug Discov. Technol. 2021, 18, 17–30. [Google Scholar] [CrossRef]
- Shahlaei, M. Descriptor Selection Methods in Quantitative Structure–Activity Relationship Studies: A Review Study. Chem. Rev. 2013, 113, 8093–8103. [Google Scholar] [CrossRef]
- Muratov, E.N.; Bajorath, J.; Sheridan, R.P.; Tetko, I.V.; Filimonov, D.; Poroikov, V.; Oprea, T.I.; Baskin, I.I.; Varnek, A.; Roitberg, A.; et al. QSAR without Borders. Chem. Soc. Rev. 2020, 49, 3525–3564. [Google Scholar] [CrossRef]
- Sharma, M.C.; Sharma, S. Molecular Modeling Studies of Thiophenyl C-Aryl Glucoside SGLT2 Inhibitors as Potential Antidiabetic Agents. Int. J. Med. Chem. 2014, 2014, 739646. [Google Scholar] [CrossRef]
- Gandhi, A.; Masand, V.; Zaki, M.E.A.; Al-Hussain, S.A.; Ghorbal, A.B.; Chapolikar, A. QSAR Analysis of Sodium Glucose Co–Transporter 2 (SGLT2) Inhibitors for Anti-Hyperglycaemic Lead Development. SAR QSAR Environ. Res. 2021, 32, 731–744. [Google Scholar] [CrossRef]
- Dong, L.; Feng, R.; Bi, J.; Shen, S.; Lu, H.; Zhang, J. Insight into the Interaction Mechanism of Human SGLT2 with Its Inhibitors: 3D-QSAR Studies, Homology Modeling, and Molecular Docking and Molecular Dynamics Simulations. J. Mol. Model. 2018, 24, 86. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Asati, V.; Mishra, M.; Das, R.; Kashaw, V.; Kashaw, S.K. Integrated Computational Approach on Sodium-Glucose Co-Transporter 2 (SGLT2) Inhibitors for the Development of Novel Antidiabetic Agents. J. Mol. Struct. 2021, 1227, 129511. [Google Scholar] [CrossRef]
- Trisciuzzi, D.; Alberga, D.; Mansouri, K.; Judson, R.; Cellamare, S.; Catto, M.; Carotti, A.; Benfenati, E.; Novellino, E.; Mangiatordi, G.F.; et al. Docking-Based Classification Models for Exploratory Toxicology Studies on High-Quality Estrogenic Experimental Data. Future Med. Chem. 2015, 7, 1921–1936. [Google Scholar] [CrossRef]
- Kolšek, K.; Mavri, J.; Sollner Dolenc, M.; Gobec, S.; Turk, S. Endocrine Disruptome—An Open Source Prediction Tool for Assessing Endocrine Disruption Potential through Nuclear Receptor Binding. J. Chem. Inf. Model. 2014, 54, 1254–1267. [Google Scholar] [CrossRef]
- Klepsch, F.; Vasanthanathan, P.; Ecker, G.F. Ligand and Structure-Based Classification Models for Prediction of P-Glycoprotein Inhibitors. J. Chem. Inf. Model. 2014, 54, 218–229. [Google Scholar] [CrossRef]
- Amaro, R.E.; Baudry, J.; Chodera, J.; Demir, Ö.; McCammon, J.A.; Miao, Y.; Smith, J.C. Ensemble Docking in Drug Discovery. Biophys. J. 2018, 114, 2271–2278. [Google Scholar] [CrossRef]
- Carlson, H.A.; Masukawa, K.M.; McCammon, J.A. Method for Including the Dynamic Fluctuations of a Protein in Computer-Aided Drug Design. J. Phys. Chem. A 1999, 103, 10213–10219. [Google Scholar] [CrossRef]
- Ji, B.; He, X.; Zhang, Y.; Zhai, J.; Man, V.H.; Liu, S.; Wang, J. Incorporating Structural Similarity into a Scoring Function to Enhance the Prediction of Binding Affinities. J. Cheminform. 2021, 13, 11. [Google Scholar] [CrossRef]
- Goodwin, N.C.; Mabon, R.; Harrison, B.A.; Shadoan, M.K.; Almstead, Z.Y.; Xie, Y.; Healy, J.; Buhring, L.M.; DaCosta, C.M.; Bardenhagen, J.; et al. Novel L-Xylose Derivatives as Selective Sodium-Dependent Glucose Cotransporter 2 (SGLT2) Inhibitors for the Treatment of Type 2 Diabetes. J. Med. Chem. 2009, 52, 6201–6204. [Google Scholar] [CrossRef]
- Deng, Z.; Chuaqui, C.; Singh, J. Structural Interaction Fingerprint (SIFt): A Novel Method for Analyzing Three-Dimensional Protein−Ligand Binding Interactions. J. Med. Chem. 2004, 47, 337–344. [Google Scholar] [CrossRef]
- Pang, M.-H.; Liu, X.-F.; Tan, X.-G.; Wang, Y.-Q. A Molecular Dynamics Investigation of Drug Dissociation from SGLT and Its Implication in Antidiabetic Medication Development. New J. Chem. 2023, 47, 19933–19942. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Ghezzi, C.; Hirayama, B.A.; Gorraitz, E.; Loo, D.D.F.; Liang, Y.; Wright, E.M. SGLT2 Inhibitors Act from the Extracellular Surface of the Cell Membrane. Physiol. Rep. 2014, 2, e12058. [Google Scholar] [CrossRef]
- Pharmacoinformatics Research Group. PharminfoVienna Sandbox 2021. Available online: https://github.com/PharminfoVienna/sandbox (accessed on 13 May 2025).
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL Database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef]
- Grempler, R.; Thomas, L.; Eckhardt, M.; Himmelsbach, F.; Sauer, A.; Sharp, D.E.; Bakker, R.A.; Mark, M.; Klein, T.; Eickelmann, P. Empagliflozin, a Novel Selective Sodium Glucose Cotransporter-2 (SGLT-2) Inhibitor: Characterisation and Comparison with Other SGLT-2 Inhibitors. Diabetes Obes. Metab. 2012, 14, 83–90. [Google Scholar] [CrossRef]
- Braem, A.; Deshpande, P.P.; Ellsworth, B.A.; Washburn, W.N. Discovery and Development of Selective Renal Sodium-Dependent Glucose Cotransporter 2 (SGLT2) Dapagliflozin for the Treatment of Type 2 Diabetes. In Carbohydrates as Drugs; Seeberger, P.H., Rademacher, C., Eds.; Topics in Medicinal Chemistry; Springer International Publishing: Cham, Switzerland, 2014; Volume 12, pp. 73–94. ISBN 978-3-319-08674-3. [Google Scholar]
- Lapuerta, P.; Zambrowicz, B.; Strumph, P.; Sands, A. Development of Sotagliflozin, a Dual Sodium-Dependent Glucose Transporter 1/2 Inhibitor. Diab. Vasc. Dis. Res. 2015, 12, 101–110. [Google Scholar] [CrossRef]
- Kirchmair, J.; Markt, P.; Distinto, S.; Wolber, G.; Langer, T. Evaluation of the Performance of 3D Virtual Screening Protocols: RMSD Comparisons, Enrichment Assessments, and Decoy Selection—What Can We Learn from Earlier Mistakes? J. Comput. Aided Mol. Des. 2008, 22, 213–228. [Google Scholar] [CrossRef]
- Gimeno, A.; Ojeda-Montes, M.J.; Tomás-Hernández, S.; Cereto-Massagué, A.; Beltrán-Debón, R.; Mulero, M.; Pujadas, G.; Garcia-Vallvé, S. The Light and Dark Sides of Virtual Screening: What Is There to Know? Int. J. Mol. Sci. 2019, 20, 1375. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Dhanjal, J.K.; Kumar, V.; Garg, S.; Subramani, C.; Agarwal, S.; Wang, J.; Zhang, H.; Kaul, A.; Kalra, R.S.; Kaul, S.C.; et al. Molecular Mechanism of Anti-SARS-CoV2 Activity of Ashwagandha-Derived Withanolides. Int. J. Biol. Macromol. 2021, 184, 297–312. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Cole, J.C.; Murray, C.W.; Nissink, J.W.M.; Taylor, R.D.; Taylor, R. Comparing Protein–Ligand Docking Programs Is Difficult. Proteins Struct. Funct. Bioinforma. 2005, 60, 325–332. [Google Scholar] [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Triballeau, N.; Acher, F.; Brabet, I.; Pin, J.-P.; Bertrand, H.-O. Virtual Screening Workflow Development Guided by the “Receiver Operating Characteristic” Curve Approach. Application to High-Throughput Docking on Metabotropic Glutamate Receptor Subtype 4. J. Med. Chem. 2005, 48, 2534–2547. [Google Scholar] [CrossRef]
- Hubbard, R.; Bayarri, M.J. Confusion Over Measures of Evidence (p’s) Versus Errors (α’s) in Classical Statistical Testing. Am. Stat. 2003, 57, 171–178. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The Advantages of the Matthews Correlation Coefficient (MCC) over F1 Score and Accuracy in Binary Classification Evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Brodersen, K.H.; Ong, C.S.; Stephan, K.E.; Buhmann, J.M. The Balanced Accuracy and Its Posterior Distribution. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 3121–3124. [Google Scholar]
- Wang, R.; Wang, S. How Does Consensus Scoring Work for Virtual Library Screening? An Idealized Computer Experiment. J. Chem. Inf. Comput. Sci. 2001, 41, 1422–1426. [Google Scholar] [CrossRef]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of MDL Keys for Use in Drug Discovery. J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Daylight Chemical Information Systems, Inc. Daylight Theory Manual; Daylight Chemical Information Systems, Inc.: Laguna Niguel, CA, USA, 2011. [Google Scholar]

| Protein (PDB ID) | ROC AUC | 10% EF | Number of Docked Ligands | Number and Percentage of Ligands Not Docked |
|---|---|---|---|---|
| 7VSI | 0.72 | 1.5 | 1147 | 50 (4.18%) |
| 8HEZ | 0.66 | 1.5 | 1120 | 77 (6.43%) |
| 8HG7 | 0.67 | 1.4 | 1158 | 39 (3.26%) |
| 8HB0 | 0.59 | 1.2 | 1112 | 85 (7.10%) |
| 8HDH | 0.60 | 1.1 | 1157 | 39 (3.34%) |
| Metrics | 7VSI, 8HG7 | 7VSI, 8HEZ, 8HG7 | 7VSI, 8HEZ, 8HG7, 8HB0 | 7VSI, 8HEZ, 8HG7, 8HB0, 8HDH |
|---|---|---|---|---|
| Balanced Accuracy | 0.70 | 0.71 | 0.69 | 0.68 |
| MCC | 0.39 | 0.41 | 0.38 | 0.36 |
| Morgan Fingerprint | RDKit Fingerprint | MACCS Keys | |
|---|---|---|---|
| MCC—cross-validation | 0.64 | 0.60 | 0.61 |
| MCC—external dataset | 0.63 | 0.58 | 0.67 |
| Exponent p | 12 | 28 | 44 |
| Average intersection point | 7.82 | 7.82 | 8.04 |
| Structural Similarity Morgan Fingerprint | Ensemble Docking 7VSI, 8HEZ, 8HG7 | Docking Score 7VSI | |
|---|---|---|---|
| MCC | 0.64 | 0.41 | 0.36 |
| Protein (PDB ID) | Resolution (Å) | Ligand |
|---|---|---|
| 7VSI | 2.95 | Empagliflozin |
| 8HEZ | 2.80 | Dapagliflozin |
| 8HG7 | 3.10 | Sotagliflozin |
| 8HB0 | 2.90 | TA1887 |
| 8HDH | 3.10 | Canagliflozin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazoudji, A.; Ecker, G.F. Docking-Based Classification of SGLT2 Inhibitors. Molecules 2025, 30, 2179. https://doi.org/10.3390/molecules30102179
Mazoudji A, Ecker GF. Docking-Based Classification of SGLT2 Inhibitors. Molecules. 2025; 30(10):2179. https://doi.org/10.3390/molecules30102179
Chicago/Turabian StyleMazoudji, Ajouan, and Gerhard F. Ecker. 2025. "Docking-Based Classification of SGLT2 Inhibitors" Molecules 30, no. 10: 2179. https://doi.org/10.3390/molecules30102179
APA StyleMazoudji, A., & Ecker, G. F. (2025). Docking-Based Classification of SGLT2 Inhibitors. Molecules, 30(10), 2179. https://doi.org/10.3390/molecules30102179






