Antimicrobial, Antioxidant, and α-Glucosidase-Inhibitory Activities of Prenylated p-Hydroxybenzoic Acid Derivatives from Oberonia ensiformis
Abstract
1. Introduction
2. Results
2.1. Structural Elucidation of the Isolated Compound
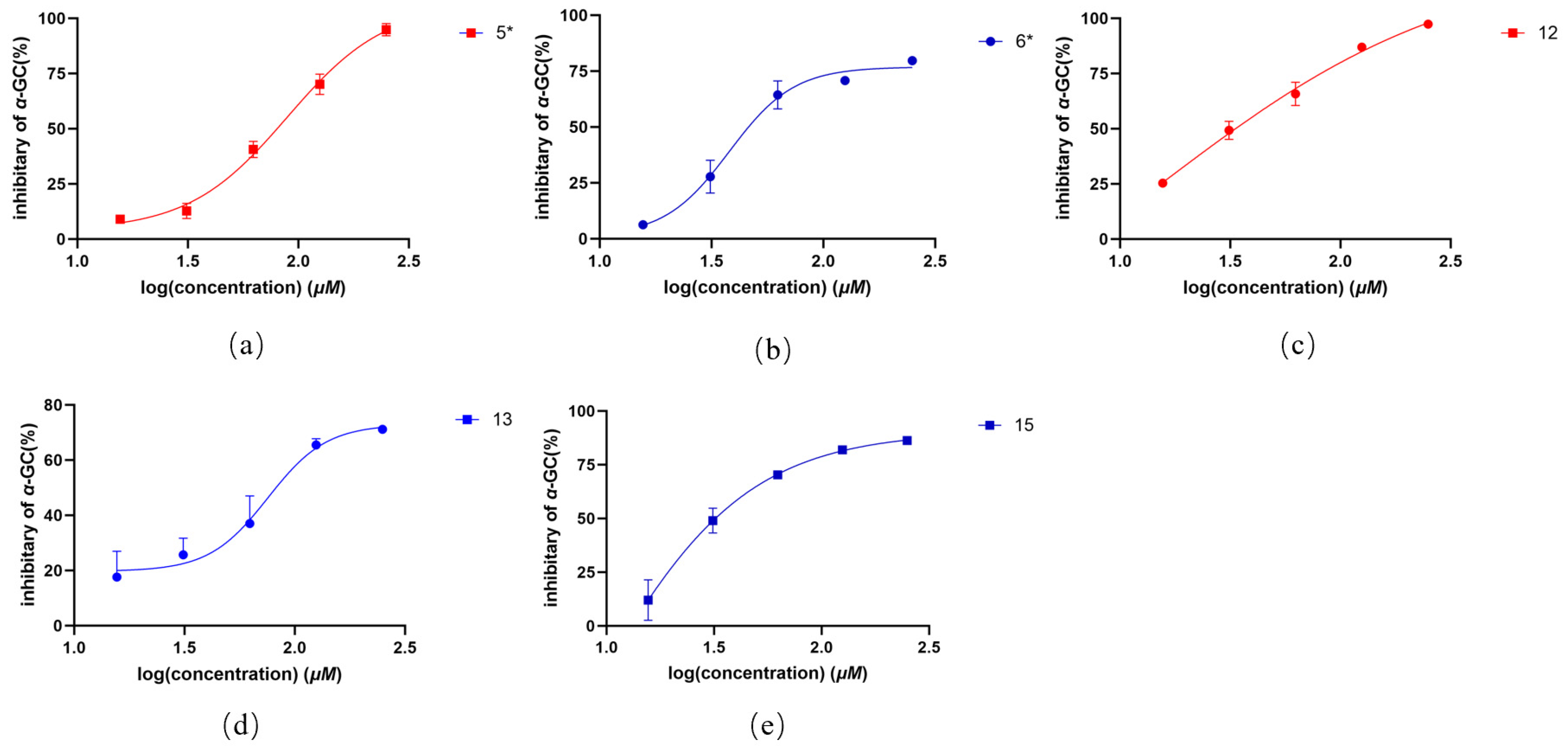
2.2. In Vitro Antioxidant, Antibacterial, and α-Glucosidase Activity of Compounds 1–18
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Plant Material
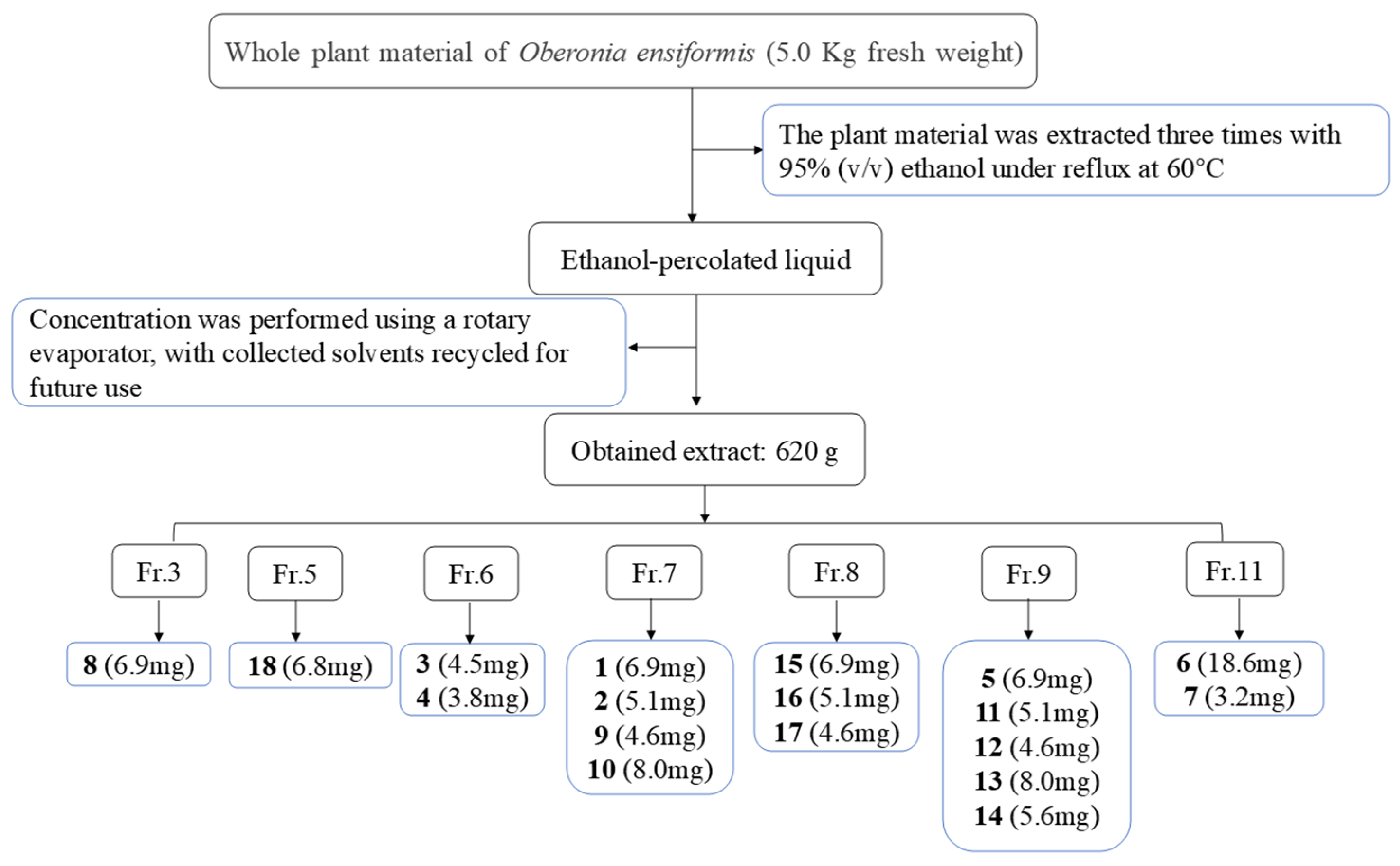
4.3. Extraction and Isolation
4.4. Spectroscopic Data
4.5. Quantum Chemical Calculations
4.6. Antibacterial Assay
4.7. Antioxidant Assay
4.8. α-Glucosidase Inhibition Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chase, M.W.; Cameron, K.M.; Freudenstein, J.V.; Pridgeon, A.M.; Salazar, G.; van den Berg, C.; Schuiteman, A. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef]
- State Administration of Traditional Chinese Medicine. Zhong Hua Ben Cao [Chinese Materia Medica]; Shanghai Science and Technology Press: Shanghai, China, 1999. [Google Scholar]
- National Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China, 2020 ed.; China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- Ren, F.C.; Liu, L.; Lv, Y.F.; Bai, X.; Kang, Q.J.; Hu, X.J.; Zhuang, H.D.; Yang, L.; Hu, J.M.; Zhou, J. Antibacterial Prenylated p-Hydroxybenzoic Acid Derivatives from Oberonia myosurus and Identification of Putative Prenyltransferases. J. Nat. Prod. 2021, 84, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Jong, T.-T.; Chau, S.-W. Antioxidative activities of constituents isolated from Pandanus odoratissimus. Phytochemistry 1998, 49, 2145–2148. [Google Scholar] [CrossRef]
- Niu, S.; Liu, Q.; Xia, J.-M.; Xie, C.-L.; Luo, Z.-H.; Shao, Z.; Liu, G.; Yang, X.-W. Polyketides from the deep-sea-derived fungus Graphostroma sp. MCCC 3A00421 showed potent antifood allergic activities. J. Agric. Food Chem. 2018, 66, 1369–1376. [Google Scholar] [CrossRef]
- Hansson, D.; Wubshet, S.; Olson, Å.; Karlsson, M.; Staerk, D.; Broberg, A. Secondary metabolite comparison of the species within the Heterobasidion annosum s.l. complex. Phytochemistry 2014, 108, 243–251. [Google Scholar] [CrossRef]
- Gao, Y.; Yu, A.L.; Li, G.T.; Hai, P.; Li, Y.; Liu, J.K.; Wang, F. Hexacyclic monoterpenoid indole alkaloids from Rauvolfia verticillata. Fitoterapia 2015, 107, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Kutrzeba, L.M.; Ferreira, D.; Zjawiony, J.K. Salvinorins J from Salvia divinorum: Mutarotation in the neoclerodane system. J. Nat. Prod. 2009, 72, 1361–1363. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.A.; Varela, R.M.; Torres, A.; Molinillo, J.M.; Gualtieri, S.C.; Macías, F.A. Phytotoxins from Tithonia diversifolia. J. Nat. Prod. 2015, 78, 1083–1092. [Google Scholar] [CrossRef]
- Flores, N.; Jiménez, I.A.; Giménez, A.; Ruiz, G.; Gutiérrez, D.; Bourdy, G.; Bazzocchi, I.L. Benzoic acid derivatives from Piper species and their antiparasitic activity. J. Nat. Prod. 2008, 71, 1538–1543. [Google Scholar] [CrossRef]
- Fukuoka, M. Chemical and toxicological studies on bracken fern, Pteridium aquilinum var. latiusculum. VI. Isolation of 5-O-caffeoylshikimic acid as an antithiamine factor. Chem. Pharm. Bull. 1982, 30, 3219–3224. [Google Scholar] [CrossRef]
- Brown Sean, P.; Dransfield, P.; Du, X.; Fu, Z.; Houze, J.; Jiao, X.; Lai, S.; Li, A.-R.; Liu, J.; Ma, Z.; et al. Spirocyclic GPR40 Modulators. U.S. Patent US20110190330 A1, 10 June 2014. [Google Scholar]
- Kuo, W.L.; Huang, Y.L.; Shen, C.C.; Shieh, B.J.; Chen, C.C. Prenylated benzoic acids and phenanthrenes from Liparis nakaharai. J. Chin. Chem. Soc. 2007, 54, 1359–1362. [Google Scholar] [CrossRef]
- Orjala, J.; Erdelmeier, C.A.; Wright, A.D.; Rali, T.; Sticher, O. Five new prenylated p-hydroxybenzoic acid derivatives with antimicrobial and molluscicidal activity from Piper aduncum leaves. Planta Med. 1993, 59, 546–551. [Google Scholar] [CrossRef]
- Dlamini, B.S.; Chen, C.R.; Chen, Y.K.; Hsu, J.L.; Shih, W.L.; Chang, C.I. Mechanistic Insights into the Inhibitory Activities of Chemical Constituents from the Fruits of Terminalia boivinii on α-Glucosidase. Chem. Biodivers. 2022, 19, e202200137. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Rodríguez, J.A.; Lévuok-Mena, K.P.; Cardozo-Muñoz, J.C.; Parra-Amin, J.E.; Lopez-Vallejo, F.; Cuca-Suárez, L.E.; Patiño-Ladino, O.J. In Vitro and In Silico Study of the α-Glucosidase and Lipase Inhibitory Activities of Chemical Constituents from Piper cumanense (Piperaceae) and Synthetic Analogs. Plants 2022, 11, 2188. [Google Scholar] [CrossRef]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef] [PubMed]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Sensitivity analysis of DP4+ with the probability distribution terms: Development of a universal and customizable method. J. Org. Chem. 2021, 86, 8544–8548. [Google Scholar] [CrossRef]
- Zhao, W.; Xu, L.-L.; Zhang, X.; Gong, X.-W.; Zhu, D.-L.; Xu, X.-H.; Wang, F.; Yang, X.-L. Three new phenanthrenes with antimicrobial activities from the aerial parts of Juncus effusus. Fitoterapia 2018, 130, 247–250. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, H.L.; Hong, L.; Xu, L.L.; Gong, X.W.; Zhu, D.L.; Xu, X.H.; Zhao, W.; Wang, F.; Yang, X.L. Three new hopane-type triterpenoids from the aerial part of Adiantum capillus-veneris and their antimicrobial activities. Fitoterapia 2019, 133, 146–149. [Google Scholar] [CrossRef]
- Lorenzo, M.E.; Casero, C.N.; Gómez, P.E.; Segovia, A.F.; Figueroa, L.C.; Quiroga, A.; Werning, M.L.; Wunderlin, D.A.; Baroni, M.V. Antioxidant characteristics and antibacterial activity of native woody species from Catamarca, Argentina. Nat. Prod. Res. 2022, 36, 885–890. [Google Scholar] [CrossRef]
- Chen, F.; Huang, G.; Yang, Z.; Hou, Y. Antioxidant activity of Momordica charantia polysaccharide and its derivatives. Int. J. Biol. Macromol. 2019, 138, 673–680. [Google Scholar] [CrossRef]
- Zhu, J.; Xu, Z.; Liu, X. Chemical composition, antioxidant activities, and enzyme inhibitory effects of Lespedeza bicolour Turcz. essential oil. J. Enzym. Inhib. Med. Chem. 2025, 40, 2460053. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.Z.; Zhu, Y.L.; Yuan, R.W.; Deng, L.; Hou, C.; Li, W.; Liu, T.; Kong, X.Q.; Zhang, L.J.; Liao, H.B. Terpenoids with α-glucosidase inhibitory activity from Rhododendron minutiflorum Hu. Phytochemistry 2022, 196, 113083. [Google Scholar] [CrossRef] [PubMed]
- Daou, M.; Elnaker, N.A.; Ochsenkühn, M.A.; Amin, S.A.; Yousef, A.F.; Yousef, L.F. In vitro α-glucosidase inhibitory activity of Tamarix nilotica shoot extracts and fractions. PLoS ONE 2022, 17, e0264969. [Google Scholar] [CrossRef] [PubMed]
- Benramdane, H.; Benariba, N.; Silva, C.F.M.; Catarino, M.D.; Bartolomeu, M.; Fekhikher, Z.; Pinto, D. Phytochemical Profile, Antioxidant, Anti-Alzheimer, And α-Glucosidase Inhibitory Effect Of Algerian Peganum harmala Seeds Extract. Chem. Biodivers. 2024, 21, e202401308. [Google Scholar] [CrossRef]
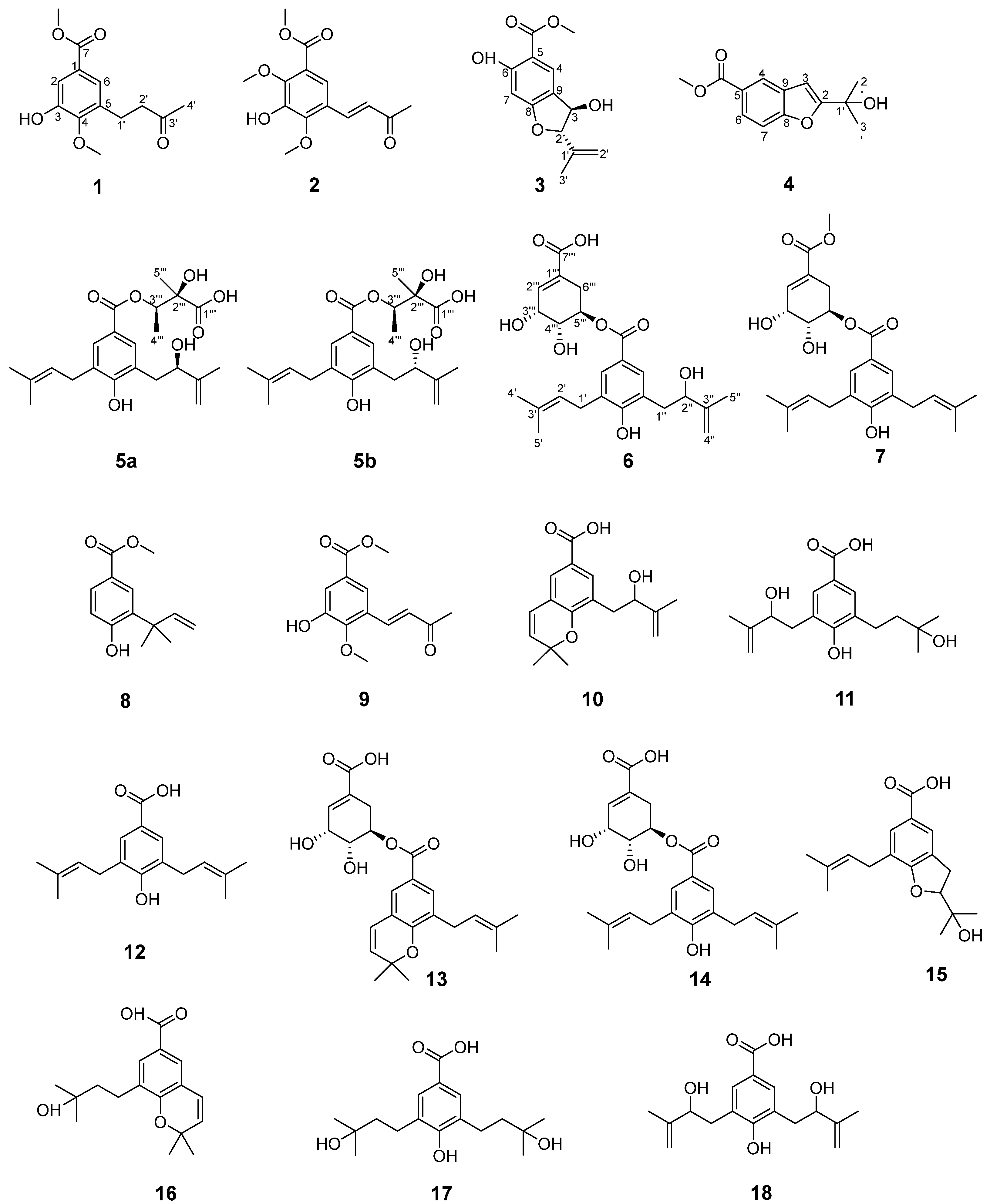
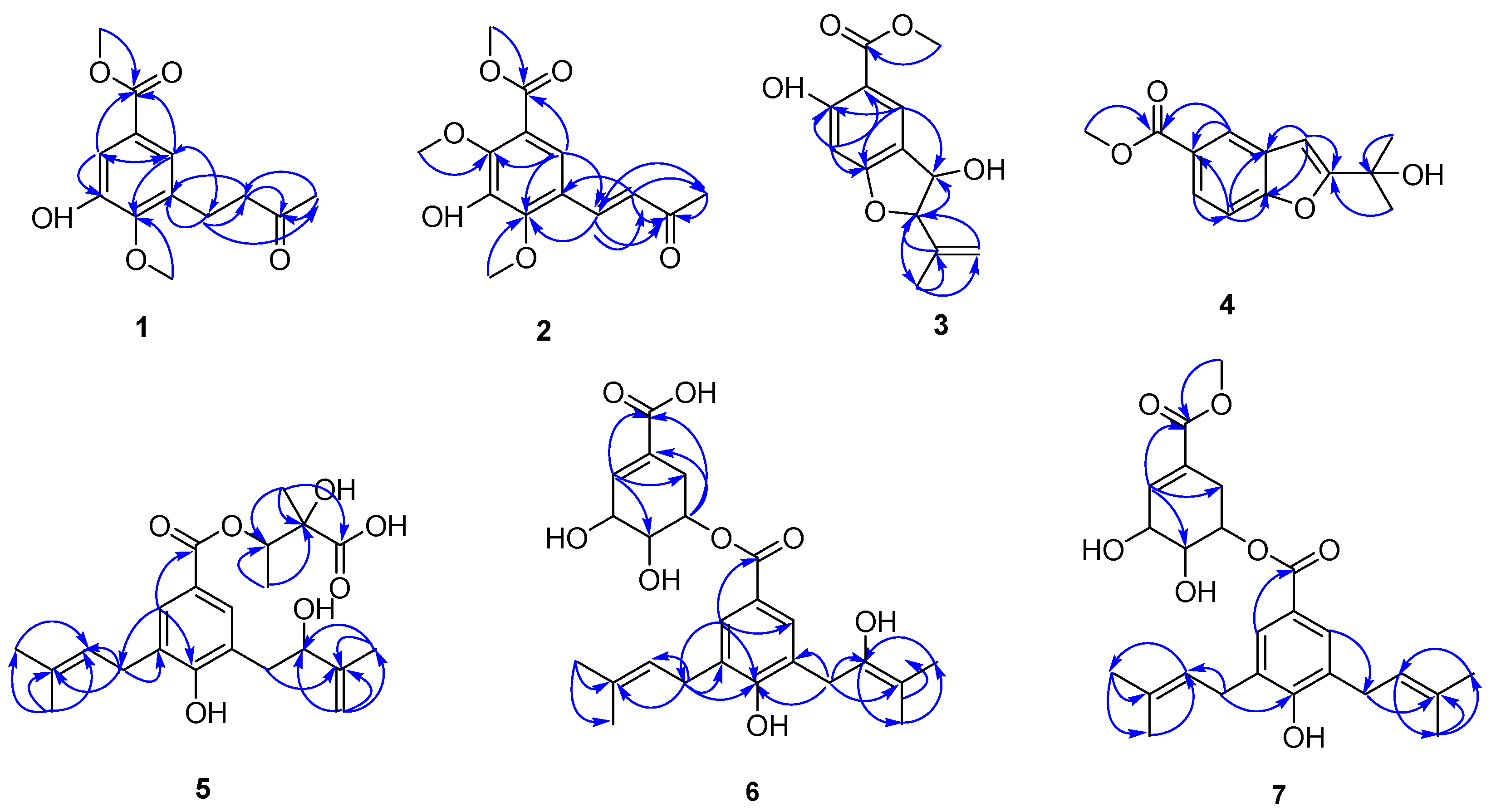
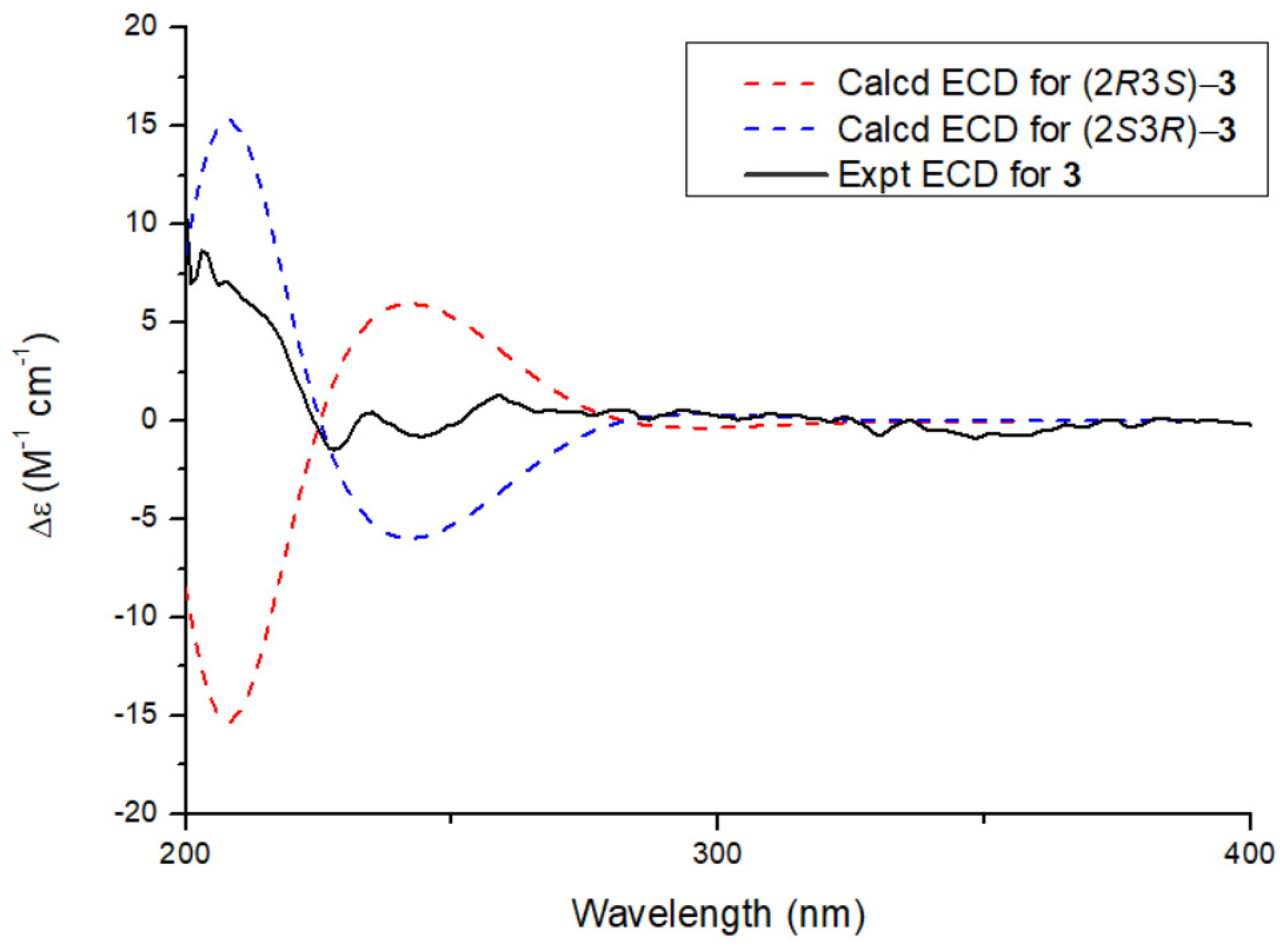

| Position | 1 | 2 | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 126.7 | 119.1 | ||
| 2 | 115.2 | 7.49, d (2.1) | 150.0 | |
| 3 | 148.9 | 7.43, d (2.1) | 142.8 | |
| 4 | 149.2 | 150.0 | ||
| 5 | 134.1 | 123.7 | ||
| 6 | 122.9 | 121.5 | 7.69, s | |
| 7 | 166.6 | 165.3 | ||
| 8 | ||||
| 9 | ||||
| 1′ | 23.7 | 2.93, t (7.8) | 137.1 | 7.73, d (16.4) |
| 2′ | 47.6 | 2.79, t (7.8) | 128.5 | 6.77, d (16.4) |
| 3′ | 207.5 | 198.6 | ||
| 4′ | 30.0 | 2.17, s | 27.7 | 2.39, s |
| COOCH3 | 52.1 | 3.88, s | 52.5 | 3.93, s |
| OCH3-2 | 62.7 | 3.95, s | ||
| OCH3-4 | 61.3 | 3.84, s | 61.5 | 4.00, s |
| Position | 3 a | 4 b | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 2 | 92.7 | 4.85, d (1.0) | 167.0 | |
| 3 | 76.7 | 5.12, br s | 101.8 | 6.77, d (1.0) |
| 4 | 114.3 | 7.01, s | 124.4 | 8.28, d (1.8) |
| 5 | 136.6 | 126.2 | ||
| 6 | 157.0 | 126.7 | 7.98, dd (8.6, 1.8) | |
| 7 | 109.5 | 7.30, s | 111.9 | 7.54, d (8.6) |
| 8 | 152.3 | 158.8 | ||
| 9 | 113.3 | 130.1 | ||
| 1′ | 141.4 | 69.7 | ||
| 2′ | 113.0 | 4.94, br s | 28.9 | 1.65, s |
| 5.10, br s | ||||
| 3′ | 17.6 | 1.75, s | 28.9 | 1.65, s |
| COOCH3 | 52.6 | 3.95, s | 52.6 | 3.94, s |
| -C=O | 170.3 | 168.8 | ||
| Position | 5a | 5b | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 120.6 | 120.6 | ||
| 2 | 129.4 | 7.58, s | 129.4 | 7.58, s |
| 3 | 128.4 | 128.3 | ||
| 4 | 158.0 | 158.1 | ||
| 5 | 125.9 | 126.0 | ||
| 6 | 130.9 | 7.60, s | 131.0 | 7.59, s |
| 7 | 165.1 | 165.1 | ||
| 1′ | 28.2 | 3.26, d (6.9) | 28.2 | 3.26, d (6.9) |
| 2′ | 122.4 | 5.24, t (7.4) | 122.4 | 5.24, t (7.4) |
| 3′ | 131.9 | 131.9 | ||
| 4′ | 25.5 | 1.70, s | 25.5 | 1.70, s |
| 5′ | 17.7 | 1.69, s | 17.7 | 1.68, s |
| 1″ | 37.7 | 2.80, d (6.6) | 37.7 | 2.80, d (6.6) |
| 2″ | 74.9 | 4.23, d (18.8) | 74.7 | 4.23, d (18.8) |
| 3″ | 147.1 | 147.2 | ||
| 4″ | 110.4 | 4.75, d (3.7) | 110.3 | 4.75, d (3.7) |
| 4.90, d (6.2) | 4.90, d (6.2) | |||
| 5″ | 18.0 | 1.73, s | 18.1 | 1.73, s |
| 1‴ | 175.8 | 175.8 | ||
| 2‴ | 75.3 | 75.3 | ||
| 3‴ | 74.2 | 5.09, d (6.4) | 74.2 | 5.09, d (6.4) |
| 4‴ | 13.6 | 1.22, d (6.3) | 13.6 | 1.22, d (6.3) |
| 5‴ | 22.3 | 1.27, s | 22.3 | 1.27, s |
| Position | 6 a | 7 b | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 120.3 | 122.1 | ||
| 2 | 129.1 | 7.49, s | 129.9 | 7.57, s |
| 3 | 128.2 | 129.5 | ||
| 4 | 158.1 | 158.7 | ||
| 5 | 126.1 | 129.5 | ||
| 6 | 130.7 | 7.54, s | 129.9 | 7.57, s |
| 7 | 165.3 | 167.7 | ||
| 1′ | 28.0 | 3.25, d (7.6) | 29.2 | 3.30, d (1.6) |
| 2′ | 122.1 | 5.23, t (7.6) | 122.8 | 5.30, m |
| 3′ | 132.2 | 134.3 | ||
| 4′ | 25.4 | 1.68, s | 25.9 | 1.76, s |
| 5′ | 15.6 | 1.64, s | 17.8 | 1.70, s |
| 1″ | 37.4 | 2.75, d (4.3) 2.80, d (8.3) | 29.2 | 3.30, d (1.6) |
| 2″ | 74.6 | 4.19, dd (8.3) | 122.8 | 5.30, m |
| 3″ | 147.1 | 134.3 | ||
| 4″ | 18.1 | 4.88, s | 25.9 | 1.77, s |
| 4.73, s | ||||
| 5″ | 110.2 | 1.71, s | 17.8 | 1.70, s |
| 1‴ | 128.0 | 129.3 | ||
| 2‴ | 138.8 | 6.68, br s | 139.5 | 6.86, br s |
| 3‴ | 65.5 | 4.27, br s | 67.4 | 4.41, br s |
| 4‴ | 67.5 | 3.79, s | 69.2 | 3.98, dd (4.4) |
| 5‴ | 70.2 | 5.15, dt (5.9) | 71.5 | 5.34, dt (4.3) |
| 6‴ | 27.4 | 2.21, br dd (18.3) | 28.3 | 2.41, dd (19.5) |
| 2.62, dd (18.3) | 2.85, dd (18.5, 2.4) | |||
| 7‴ | 167.7 | 168.3 | ||
| COOCH3 | 52.4 | 3.76, s | ||
| No. | Compound | IC50 (μg/mL) * |
|---|---|---|
| positive control | L-ascorbate | 2.37 ± 0.11 |
| 6 | oberoniaensiformisin F | 173.76 ± 23.92 |
| 12 | nervogenic acid | 185.36 ± 1.96 |
| No. | Compound | IC50 (μg/mL) * |
|---|---|---|
| positive control | acarbose | 0.66 ± 0.36 |
| 5 | oberoniaensiformisin F | 106.10 ± 2.12 |
| 6 | oberoniaensiformisin F | 58.07 ± 6.22 |
| 12 | nervogenic acid | 34.03 ± 0.16 |
| 13 | oberoniamyosurusin L | 86.53 ± 1.08 |
| 15 | oberoniamyosurusin I | 38.80 ± 4.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.-L.; Tang, W.; Wang, Z.; Wang, Y.-X.; Li, N.; Ren, F.-C. Antimicrobial, Antioxidant, and α-Glucosidase-Inhibitory Activities of Prenylated p-Hydroxybenzoic Acid Derivatives from Oberonia ensiformis. Molecules 2025, 30, 2132. https://doi.org/10.3390/molecules30102132
Wang L-L, Tang W, Wang Z, Wang Y-X, Li N, Ren F-C. Antimicrobial, Antioxidant, and α-Glucosidase-Inhibitory Activities of Prenylated p-Hydroxybenzoic Acid Derivatives from Oberonia ensiformis. Molecules. 2025; 30(10):2132. https://doi.org/10.3390/molecules30102132
Chicago/Turabian StyleWang, Lu-Lu, Wei Tang, Zhuo Wang, Yi-Xiang Wang, Ning Li, and Fu-Cai Ren. 2025. "Antimicrobial, Antioxidant, and α-Glucosidase-Inhibitory Activities of Prenylated p-Hydroxybenzoic Acid Derivatives from Oberonia ensiformis" Molecules 30, no. 10: 2132. https://doi.org/10.3390/molecules30102132
APA StyleWang, L.-L., Tang, W., Wang, Z., Wang, Y.-X., Li, N., & Ren, F.-C. (2025). Antimicrobial, Antioxidant, and α-Glucosidase-Inhibitory Activities of Prenylated p-Hydroxybenzoic Acid Derivatives from Oberonia ensiformis. Molecules, 30(10), 2132. https://doi.org/10.3390/molecules30102132






