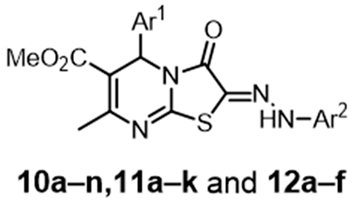
Abstract
Ribonuclease H (RNase H) was identified as an important target for HIV therapy. Currently, no RNase H inhibitors have reached clinical status. Herein, a series of novel thiazolone[3,2-a]pyrimidine-containing RNase H inhibitors were developed, based on the hit compound 10i, identified from screening our in-house compound library. Some of these derivatives exhibited low micromolar inhibitory activity. Among them, compound 12b was identified as the most potent inhibitor of RNase H (IC50 = 2.98 μM). The experiment of magnesium ion coordination was performed to verify that this ligand could coordinate with magnesium ions, indicating its binding ability to the catalytic site of RNase H. Docking studies revealed the main interactions of this ligand with RNase H. A quantitative structure activity relationship (QSAR) was also conducted to disclose several predictive mathematic models. A molecular dynamics simulation was also conducted to determine the stability of the complex. Taken together, thiazolone[3,2-a]pyrimidine can be regarded as a potential scaffold for the further development of RNase H inhibitors.
1. Introduction
Acquired Immunodeficiency Syndrome (AIDS), a severe infectious disease, develops as a consequence of Human Immunodeficiency Virus (HIV) infection [1,2]. The estimated global prevalence of HIV infection in 2022 is 39.0 million, underscoring its enduring and profound impact on public health worldwide [3,4]. The current standard approach for managing HIV infection is antiretroviral therapy (ART) [5,6]. However, the prolonged administration of ART can easily lead to the emergence of drug-resistant viral strains, thereby limiting its long-term efficacy [7,8]. The development of innovative anti-HIV agents is crucial and requires more attention.
In the past, targeting RNase H provided an opportunity for the development of new anti-HIV drugs [9]. The RNase H enzyme is strategically situated within the p66 subunit of reverse transcriptase (RT), enabling the precise hydrolysis of RNA in RNA/DNA heteroduplexes and tRNA precursors, to ensure seamless synthesis during the process of reverse transcription [10,11]. The active site of RNase H contains a conserved DEDD motif, comprising four carboxylate amino acid residues (D443, E478, D498, and D549), which interact with two Mg2+ ions [12]. Currently reported RNase H inhibitors generally function by binding to the active site competitively, through the chelation of trivalent ligands with divalent metals. Such inhibitors include hydroxyisoquinolinedione, β-thujaplicinol, and dihydroxycoumarin [13,14]. The potent and selective RNase H inhibition could be achieved by adopting structurally more elaborate chemotypes, such as diketoacid, pyrimidinolcarboxylic acid, hydroxynaphthyridine, and pyridopyrimidone, which featured a hydrophobic aromatic moiety that was seemingly important for antiviral activity [15]. Most of these small molecule inhibitors chelate with magnesium ions via oxygen atoms, at the active site of the RT RNase H. As an alternative, it is also possible for nitrogen atoms to form a chelation, such as naphthaldehyde hydrazone 1 [16], pyridopyrimidone 2, and pyrrolyl pyrazoles 3, 4 (Figure 1) [17]. Despite extensive efforts to produce RNase H inhibitors, no particular inhibitor has entered clinical trials [18,19,20]. The potential of HIV RT-associated RNase H as a therapeutic target has yet to be fully explored [21,22,23].
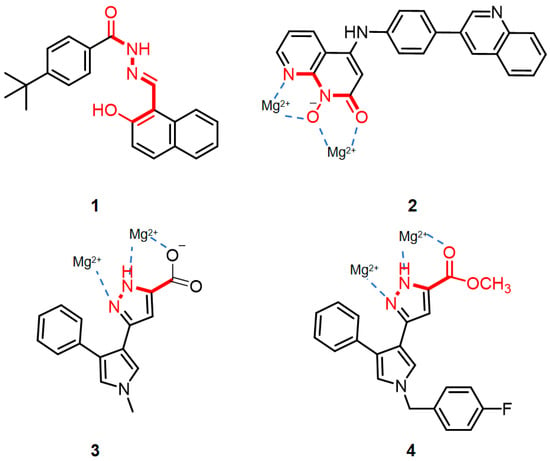
Figure 1.
Previously disclosed representative HIV RNase H inhibitors, chelated with Mg2+. The dotted lines in the diagram showed the interaction of the molecule with the Mg2+.
In this work, a series of novel thiazolone[3,2-a]pyrimidine derivatives were designed and synthesized by using a structure-based design strategy (Figure 2). All of the target compounds were then subjected to in vitro evaluation to assess their ability to inhibit the specific enzymatic activity of recombinant RNase H. By comparing the activity results and analyzing the molecular docking and simulations, we gained some insights into the potential binding models of the compounds.
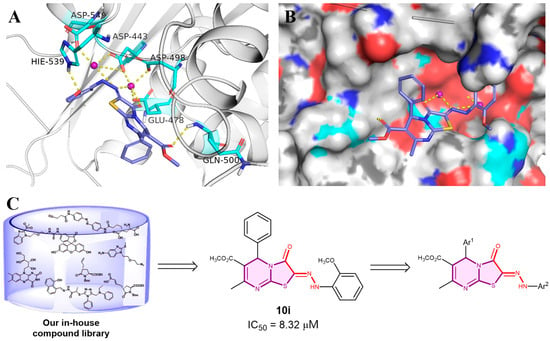
Figure 2.
Docking poses of compounds 10i (A,B) at the binding pocket of the HIV-1 RT (PDB code: 3QIP). (C) The discovery of novel thiazolone[3,2-a]pyrimidine derivatives.
2. Results and Discussion
2.1. Chemistry
The synthetic route of the target compounds 10a–n, 11a–k, and 12a–f is depicted in Scheme 1. Compounds 8a–c were synthesized via the Biginelli reaction, using acetoacetic ethyl ester, thiourea, and different aromatic aldehydes as three components, catalyzed using sulfamic acid in ethanol solvent at 80 °C, yielding 70–85%. Subsequently, compounds 8a–c were reacted with methyl bromoacetate in the presence of pyridine as the base, for 3 h under reflux in ethanol to obtain compounds 9a–c, with yields ranging from 55% to 80%. Compounds 9a–c were reacted with freshly prepared aromatic diazonium salts at 0 °C for 0.5 h in ethanol or dioxane. The reaction was then continued at room temperature for 2 h to obtain the final target compounds 10a–n, 11a–k, and 12a–f, yielding 30–90%.

Scheme 1.
Synthesis of compounds 10a–n, 11a–k, and 12a–f. Reagents and conditions: (a) SO2(NH2)2, EtOH, 80 °C, 2 h, yield: 70–85%; (b) methyl bromoacetate, pyridine, EtOH, reflux, 3 h, yield: 55–80%; (c) 1. ArNH2, NaNO2, HCl, H2O, 0 °C, 30 min; 2. pyridine, EtOH, 0 to 25 °C, 3 h, yield: 30–90%.
2.2. Anti-HIV-1 Activity of Compounds 10a–n, 11a–k, and 12a–f
We initiated this project by screening our-in-house compound library and compound 10i was identified as one of the potentially active RNase H inhibitors (IC50 = 8.32 μM). Results from the docking study in Figure 2A,B indicated that 10i was well positioned into the binding pocket of RNase H. The nitrogen atoms of the azo group of the compounds participate in the chelation of magnesium ions with the oxygen atoms of the thiazolone structure. The Ar1 and Ar2 aromatic segments of the molecule also approach the protein surface, through hydrophobic interactions. These findings provide important information for further structural optimization. All these newly designed thiazolone[3,2-a]pyrimidine derivatives were evaluated for their inhibitory activity against HIV RT-associated RNase H, using the diketo acid RDS1643 as positive control. The results are reported in Table 1. Among this series, compounds 12a, 12b, and 12f were found to be the most active derivatives, with IC50 values ranging from 2 to 3 μM. The inhibitory activity of compounds 10d (IC50 = 8.27 μM), 10g (IC50 = 9.01 μM), 12b (IC50 = 2.97 μM), and 12f (IC50 = 3.275 μM) was stronger than others, when the polar substituents were present at the 2-position of the aryl group. The presence of polar substituted groups at the 2-position of the aromatic moiety Ar2 showed a significantly enhanced inhibitory activity, compared to Ar2 with a 3-substituted group or without any substituents on the aromatic ring. The presence of a methyl group at the 2-position of 12d led to a minor decrease in activity (IC50 = 44.86 μM), suggesting that non-polar substituents on Ar2’s aromatic moiety may negatively impact its inhibitory activity. The decrease in activity observed in compounds containing a sulfonic acid group might be attributed to the excessively strong acidity of the sulfonic acid group, which could protonate the diazo group within the molecule. The strong solvent effect of compounds induced by large polar sulfonic acid groups might also contribute to the reduced inhibitory activity of these compounds. When Ar2 was changed from a phenyl group to a naphthyl group, the activity of compound 10f gained a great improvement over that of compound 10k. Generally, by comparing the compounds from series 10, 11, and 12, we found compounds belonging to series 12 exhibited higher inhibitory activity, indicating that the Ar1 fragment with a single methoxy substitution at the 4-position was favorable for increasing their biological activities. And when a negatively charged polar group was present at the 2-position of Ar2, the inhibitory activity is enhanced.

Table 1.
RNase H inhibition of the synthesized compounds 10a–n, 11a–k, and 12a–f.
2.3. QSAR Model
To further design HIV-1 RNase H inhibitors with improved activity, based on the molecular scaffold, we trained a QSAR model for this series of molecules to investigate the structure–activity relationship. We utilized the QSAR module in Maestro. We used pIC50 as the predicted value, with the unit of IC50 activity converted to moles. Among the obtained models, a QSAR model based on the Kernel Partial Least Squares algorithm demonstrated a good fit to both the training and test datasets. The training and prediction outcomes were depicted in Figure 3. For this mathematical model, the training set’s S.D. (standard deviation) = 0.3783, r2 = 0.7242, while the test Set’s rmse = 0.3287 and q2 = 0.6857. These results indicated that the model had a certain degree of reliability, which can guide further structural optimization.
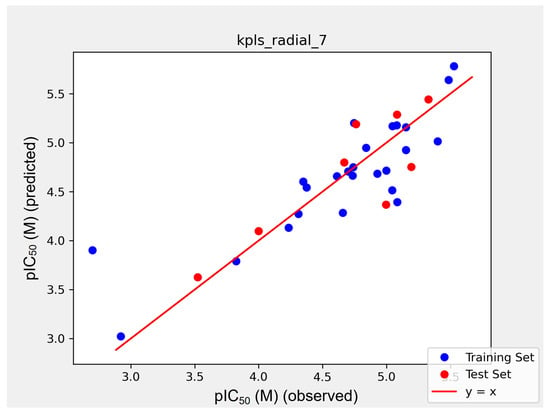
Figure 3.
Scatter plot of the QSAR model. The blue dots in the figure are from the training set and the red dots are from the independent test set. pIC50 (observer) is the experimental value and pIC50 (predicted) is the model predicted value. The IC50 values used for training are from Table 1, converted to moles.
2.4. Prediction of Physicochemical Properties
The values of the physicochemical parameters of compounds with good inhibitory activity were assessed using a free online forecasting tool (www.swissadme.ch/ (accessed on 31 December 2023)); they are summarized in Table 2. The compounds tested did not penetrate the blood–brain barrier, act as P-gp substrates, or inhibit CYP1A2 or CYP2D6. They were not identified as pain-inducing, indicating no interaction with multiple targets and avoiding false positive results. Notably, the representative compound 12b complied with the Lipinski rule. Our findings confirm their safety and favorable physicochemical properties.

Table 2.
Prediction of pharmacokinetics of representative compounds with good inhibitory activity.
2.5. Molecular Docking
We performed a molecular docking study of compounds 10i and 12b with the RNase H active site (PDB code: 3QIP) [25]. The docking results for compounds 10i and 12b are shown in Figure 4. The predicted binding modes of compounds 10i and 12b at the active site of RNase H suggested an interaction between the amide group and diazo group with Mg2+. On one side, Mg2+ ions were connected to the protein through the carboxyl groups of the protein residues Asp443, Asp549, Asp498, and GLu478. On the other side, one Mg2+ ion was involved in a five-membered ring chelation through the oxygen of the amide group and the nitrogen of the diazo group, while another Mg2+ ion interacted with one nitrogen atom in the azo group. The 2-position substituent on the Ar2 fragment could participate in hydrogen bonding interactions with residue HIE539. In addition, the oxygen substituent at the 2-position may interact with the Mg2+ ions. The significant increase in activity may be attributed to the presence of polar substituents at the 2-position of the aromatic moiety Ar2. The ester group on the thiazolopyrimidine core also exhibited a hydrogen bonding interaction with GLN500. The entire diazo group was situated in the hydrophilic region of the protein. The Ar1 and Ar2 fragments of compounds 10i and 12b were located in the hydrophobic region of the protein. The compound 12b coordinated with one magnesium ion through the amide and azo groups, while the nitrogen atom of the azo group interacted with the other magnesium ion. Meanwhile, 5,6-dihydroxy-2-[(2-phenyl-1H-indol-3-yl)methyl]pyrimidine-4-carboxylic acid interacted with one magnesium ion through the carboxyl and hydroxyl groups, and its carbonyl and hydroxyl groups also interacted with the magnesium ion. Both compounds exhibited similar interactions with residue HIE539 via their carbonyl groups.

Figure 4.
(A–C) Molecular docking of compound 12b with RNase H (PDB code: 3QIP); (D) overlay of 12b and 10i within the binding pocket of RNase H; (E) the crystal structure of 5,6-dihydroxy-2-[(2-phenyl-1H-indol-3-yl)methyl]pyrimidine-4-carboxylic acid with RNase H (PDB code: 3QIP); (F) overlay of 12b and 5,6-dihydroxy-2-[(2-phenyl-1H-indol-3-yl)methyl]pyrimidine-4-carboxylic acid within the binding pocket of RNase H; the Mg2+ ions are represented by the purple sphere. The interactions between residues, ligands, and Mg2+ ions are indicated by the yellow lines. The molecular scaffold of 5,6-dihydroxy-2-[(2-phenyl-1H-indol-3-yl)methyl]pyrimidine-4-carboxylic acid is green and the molecular scaffold of compound 12b is orange. Important amino acid residues are represented in cyan.
2.6. Dynamic Simulation
We aimed to observe the dynamic binding of the small molecule 12b to the protein, through molecular dynamics simulations. The RMSF values of Chain were within the range of 0.1 to 0.4 nm (Figure 5A,B). The RMSD fluctuation plot (Figure 5C) indicated that the compound 12b rapidly reached equilibrium during the dynamic binding process. The RMSD fluctuation range was within 0.25 to 0.45 nm, which was within a reasonable range. Comparison plots of three dynamic simulations are shown in the Supplementary Information (SI). Compound 12b did not dissociate from the docking pocket during the simulation that affirmed compound 12b’s reliability as an inhibitor for the RNase H. The hydrogen bonds (Figure 5D) showed that, apart from the coordination interactions with Mg2+, the dynamic changes in hydrogen bond formation between compound 12b and the protein generally ranged from 1 to 2.
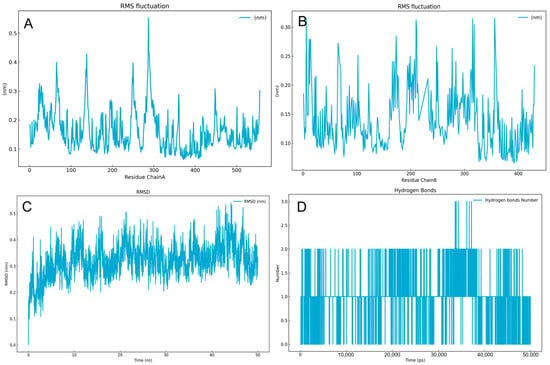
Figure 5.
(A,B): Protein-RMSF during the process of the molecular dynamic simulation, ChainA: p66, ChainB: p51. (C): Ligand-RMSD. (D): The number of hydrogen bonds with a time range from 0 ps to 50,000 ps.
2.7. Magnesium Complexation
To investigate the potential importance of the interaction between the active compounds and Mg2+, spectrophotometric complexation studies were carried out on the good active derivatives, 12a and 12b. In Figure 6, it can be observed that, upon the addition of magnesium ions, the absorbance of the solution increased around the wavelength range of 400–450 nm, indicating a certain interaction between compounds 12a and 12b with magnesium ions.
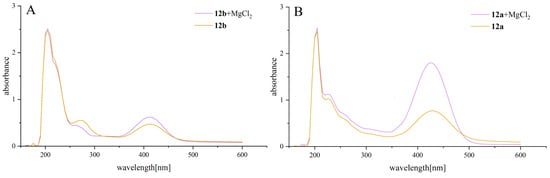
Figure 6.
(A) UV spectra of 12b in EtOH:dioxane:water = 1:1:1 = 5.21 × 10−5 M (orange trace) and 12b = 5.21 × 10−5 M + MgCl2 = 10−3 M (purple trace). (B) UV spectra of 12a in EtOH:dioxane:water = 1:1:1 = 1.24 × 10−4 M (orange trace) and 12a = 1.24 × 10−4 M + MgCl2 = 10−3 M (purple trace).
3. Conclusions
In this study, we have identified a novel class of thiazolone[3,2-a]pyrimidine-containing RNase H inhibitors, derived from the hit compound 10i, by using a structure-based design strategy. Some of these compounds had IC50 values in the low micromolar range. The structure–activity relationship studies suggested that series 12 exhibited the best activity, with compound 12b displaying the highest inhibitory activity against RNase H (IC50 = 2.98 μM). Furthermore, the presence of electron-rich substituents at the 2-position of Ar2 was favorable for increasing the ligand activity. The presence of aromatic fused-ring substituents appears to enhance RNase H inhibition activity, suggesting a direction for further design of inhibitor molecules. The QSAR model was also established to give a predictive guide, which inferred their potential MOA with RNase H. We further validated the reliability of this series of molecules in binding to the RNase H site through molecular docking, dynamic simulations, and Mg2+ coordination experiments. In summary, compound 12b can be used as a lead compound for the further development of potent thiazolone[3,2-a]pyrimidine-based RNase H inhibitors.
4. Experimental
4.1. Chemistry
Commercially available reagents and solvents were procured for use. Thin-layer chromatography (TLC) (Yantai Xinnuo new material Technology Co., Ltd., Yantai, China) analysis was conducted using a 0.25 mm silica gel plate, followed by visualization under UV light with a wavelength of 254 nm. The 1H NMR and 13C NMR spectra were analyzed in DMSO-d6 or CDCl3, using a Bruker AV-400 spectrometer, with TMS serving as the internal standard for characterization. Due to the low solubility of certain compounds, we had to select relatively more favorable deuterated solvents for different compounds. The determination of the melting point was conducted using the Hanon MP-430 digital melting point instrument. The HRMS data were acquired using the Agilent 1290II-6545 spectrometer (Q-TOF). Analysis of sample purity was performed on a Shimadzu SPD-20A/20AV HPLC system using an Inertsil ODS-SP, 5 μm C18 column (150 mm × 4.6 mm).
4.1.1. General Procedure for the Synthesis of Compounds 8a–c
To a mixture of sulfourea (55 mmol, 1.1 eq), methyl acetoacetate (60 mmol, 1.2 eq), and the proper aromatic aldehyde (50 mmol, 1 eq) in EtOH (100 mL), 5 mmol (0.1 eq) sulfonamide was added as catalyst. The mixture was heated to 50 °C for 2 h, which was monitored using TLC. In total, 100 mL of water was added to terminate the reaction and then solid precipitates were observed. The precipitated solid was filtered, recrystallized twice using 30 mL ethanol each time, and dried to obtain the pure product.
4.1.2. General Procedure for the Synthesis of Compounds 9a and 9c
The proper methyl 2-mercapto-4-methyl-6-aryl-1,6-dihydropyrimidine-5-carboxylate (32 mmol, 1 eq) and pyridine (35.2 mmol, 1.1 eq) were added to 100 mL EtOH. Then, 35.2 mmol (1.1 eq) methyl bromoacetate was added to the system, which was stirred under reflux for 3 h. Upon completion of the reaction, monitored using TLC, 100 mL water was added to terminate the reaction, affording solid precipitates, which were then filtrated and purified using gel column chromatography to give the pure products.
- Methyl 7-methyl-3-oxo-5-phenyl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (9a). Yellow solid; 1H NMR (CDCl3, 400 MHz): δ [ppm] 7.23–6.98 (m, 5H), 5.84 (s, 1H), 3.63 (d, J = 17.5 Hz, 1H), 3.51 (d, J = 17.5 Hz, 1H), 3.42 (s, 3H), 2.26 (s, 3H).
- Methyl 5-(4-methoxyphenyl)-7-methyl-3-oxo-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (9c). Yellow solid; 1H NMR (CDCl3, 400 MHz): δ 7.29 (d, J = 8.6 Hz, 2H), 6.83 (d, J = 8.7 Hz, 2H), 6.02 (s, 1H), 3.84 (d, J = 17.4 Hz, 1H), 3.78 (s, 3H), 3.73 (d, J = 17.5 Hz, 1H), 3.64 (s, 3H), 2.48 (s, 3H).
4.1.3. General Procedure for the Synthesis of Compound 9b
Methyl 2-mercapto-4-methyl-6-(2,3,4-trimethoxyphenyl)-1,6-dihydropyrimidine-5-carboxylate (16mmol, 1 eq) and pyridine (17.6 mmol, 1.1 eq) were added to 50 mL dioxane. Then, 17.6 mmol (1.1 eq) methyl bromoacetate was added into the system. The resulting mixtures was stirred under reflux for 3 h and then TLC was used to monitor the reaction. A total of 50 mL water was added to terminate the reaction, then an extraction with ethyl acetate (3 × 50 mL) was performed. The organic layer was dried over anhydrous sodium sulfate and was concentrated under vacuum. The pure product was then obtained through purification using silica gel column chromatography.
- Methyl 7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (9b). Yellow solid; 1H NMR (CDCl3, 400 MHz): δ [ppm] 7.03 (d, J = 8.6 Hz, 1H), 6.58 (d, J = 8.7 Hz, 1H), 6.09 (s, 1H), 3.88 (s, 3H), 3.83 (s, 3H), 3.81 (s, 3H), 3.64 (s, 3H), 2.39 (s, 3H).
4.1.4. General Procedure for the Synthesis of Compounds 10a–n and 12a–f
The proper methyl 7-methyl-3-oxo-5-aryl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (1 mmol, 1 eq) and pyridine (4 mmol, 4 eq) were dissolved in 5 mL EtOH and stirred at 0 °C. The appropriate aromatic amine (1.2 mmol, 1.1 eq) was dissolved in hydrochloric acid (3 mmol, 1 eq) aqueous solution and stirred at 0 °C. Then, a solution of sodium nitrite (1.44 mmol, 1.44 eq) was added to the hydrochloric acid–aromatic amine mixture. The above aqueous solution was stirred for 0.5 h at 0 °C to obtain the aromatic diazonium salt. The resulting aqueous solution was then added dropwise into the ethanol system and stirred for 1 h at 0 °C, followed by 3 h at room temperature, before being quenched with water (10 mL). The crude product obtained from precipitation of a solid was purified using silica gel column chromatography to yield the final product.
- Methyl 2-(2-(2-hydroxyphenyl)hydrazineylidene)-7-methyl-3-oxo-5-phenyl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10a). Yellow solid, yield: 75%, mp: 227–229 °C; 1H NMR (400 MHz, CDCl3) δ 7.43 (d, J = 7.0 Hz, 2H), 7.34 (t, J = 8.1 Hz, 4H), 6.90 (dd, J = 17.3, 7.9 Hz, 3H), 6.25 (s, 1H), 3.69 (s, 3H), 2.56 (s, 3H). 13C NMR (101 MHz, CDCl3 + CD3OD) δ 169.99, 165.12, 158.27, 155.99, 148.44, 143.42, 132.78, 132.67, 131.74, 131.64, 127.66, 124.38, 119.38, 119.03, 117.17, 113.45, 59.07, 55.51, 26.23. HRMS (ESI) m/z: calcd for C21H18N4O4S, [M − H]−: 421.0969; found: 421.0972.
- (2-(6-(Methoxycarbonyl)-7-methyl-3-oxo-5-phenyl-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)-3-methylbenzoic acid (10b). Yellow solid, yield: 88%, mp: 261–263 °C; 1H NMR (400 MHz, DMSO-d6) δ 11.60 (s, 1H), 7.77 (d, J = 6.4 Hz, 1H), 7.45 (d, J = 7.4 Hz, 1H), 7.40–7.27 (m, 5H), 7.10 (t, J = 7.7 Hz, 1H), 6.02 (s, 1H), 3.60 (s, 3H), 2.40 (s, 3H), 2.39 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 170.42, 166.07, 160.50, 155.59, 152.61, 145.89, 141.48, 135.24, 129.77, 129.22, 128.87, 127.63, 126.41, 123.47, 121.49, 118.56, 108.42, 54.54, 51.86, 23.08, 22.26. HRMS (ESI) m/z: calcd for C23H20N4O5S, [M + H]+: 465.1255; found: 465.1262.
- Methyl 7-methyl-3-oxo-5-phenyl-2-(2-(3,4,5-trimethoxyphenyl)hydrazineylidene)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10c). Yellow solid, yield: 85%, mp: 211–213 °C; 1H NMR (400 MHz, DMSO-d6) δ 10.90 (s, 1H), 7.38–7.26 (m, 5H), 6.52 (s, 2H), 6.01 (s, 1H), 3.74 (s, 6H), 3.59 (d, J = 2.7 Hz, 6H), 2.38 (s, 3H). 13C NMR (101 MHz, CDCl3 + CD3OD) δ 169.98, 165.53, 159.24, 157.73, 155.80, 143.54, 143.16, 137.62, 132.77, 132.65, 131.74, 131.68, 123.04, 113.28, 96.06, 64.82, 59.83, 59.05, 55.52, 26.23. HRMS (ESI) m/z: calcd for C24H24N4O6S, [M + H]+: 497.1551; found: 497.1567.
- Methyl 2-((2-(methoxycarbonyl)phenyl)diazenyl)-7-methyl-3-oxo-5-phenyl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10d). Yellow solid, yield: 65%, mp: 184–186 °C; 1H NMR (400 MHz, CDCl3) δ 11.35 (s, 1H), 7.95 (d, J = 6.5 Hz, 1H), 7.76 (d, J = 8.6 Hz, 1H), 7.50 (t, J = 7.1 Hz, 1H), 7.44–7.38 (m, 2H), 7.33–7.27 (m, 3H), 7.00 (t, J = 7.9 Hz, 1H), 6.24 (s, 1H), 3.94 (s, 3H), 3.66 (s, 3H), 2.53 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 168.77, 165.79, 160.71, 152.63, 152.34, 145.05, 139.43, 134.95, 130.80, 128.86, 128.79, 127.99, 125.98, 121.59, 115.00, 112.12, 109.92, 55.27, 52.51, 51.65, 22.75. HRMS (ESI) m/z: calcd for C23H20N4O5S, [M + H]+: 465.1254; found: 465.1274.
- 5-Methoxy-2-(2-(6-(methoxycarbonyl)-7-methyl-3-oxo-5-phenyl-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)benzenesulfonic acid (10e). Yellow solid, yield: 86%, mp: 247–249 °C; 1H NMR (400 MHz, DMSO-d6) δ 10.89 (s, 1H), 7.38–7.25 (m, 6H), 6.91 (d, J = 8.8 Hz, 1H), 6.86 (d, J = 2.5 Hz, 1H), 6.01 (s, 1H), 3.70 (s, 3H), 3.60 (s, 3H), 2.38 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 165.88, 160.45, 154.77, 153.42, 151.61, 140.83, 133.90, 132.26, 129.29, 129.04, 127.65, 122.12, 117.29, 115.59, 112.36, 109.68, 55.90, 55.05, 52.01, 22.87. HRMS (ESI) m/z: calcd for C22H20N4O7S2, [M + H]+: 517.0854; found: 517.0848.
- (6-(Methoxycarbonyl)-7-methyl-3-oxo-5-phenyl-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)naphthalene-1-sulfonic acid (10f). Yellow solid, yield: 80%, mp: 289–293 °C; 1H NMR (400 MHz, DMSO-d6) δ 12.84 (s, 1H), 8.83 (d, J = 9.1 Hz, 1H), 7.91 (d, J = 9.2 Hz, 1H), 7.80 (d, J = 7.6 Hz, 1H), 7.71 (d, J = 9.1 Hz, 1H), 7.51–7.45 (m, 1H), 7.41–7.28 (m, 6H), 6.07 (s, 1H), 3.62 (s, 3H), 2.40 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 165.87, 160.55, 153.48, 151.54, 146.57, 142.85, 140.77, 136.52, 131.87, 130.90, 130.18, 129.30, 129.06, 128.44, 127.71, 127.60, 127.11, 124.75, 124.56, 123.61, 114.57, 109.77, 55.11, 52.03, 22.87. HRMS (ESI) m/z: calcd for C25H20N4O6S2, [M − H]−: 535.0744; found: 535.0749.
- Methyl 2-(2-(2-acetylphenyl)hydrazineylidene)-7-methyl-3-oxo-5-phenyl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10g). Yellow solid, yield: 78%, mp: 199–202 °C; 1H NMR (400 MHz, CDCl3) δ 12.07 (s, 1H), 7.73 (t, J = 7.4 Hz, 2H), 7.43 (t, J = 7.1 Hz, 1H), 7.34 (d, J = 6.5 Hz, 2H), 7.26–7.17 (m, 3H), 6.94 (t, J = 7.6 Hz, 1H), 6.15 (s, 1H), 3.59 (s, 3H), 2.58 (s, 3H), 2.46 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 202.76, 165.80, 160.71, 152.67, 152.32, 145.11, 139.38, 135.46, 131.71, 128.89, 128.81, 128.01, 126.48, 121.37, 118.97, 115.25, 109.95, 55.28, 51.70, 27.99, 22.74. HRMS (ESI) m/z: calcd for C23H20N4O4S, [M − H]−: 447.1125; found: 447.1126.
- Methyl 2-(2-(3,4-dimethoxyphenyl)hydrazineylidene)-7-methyl-3-oxo-5-phenyl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10h). Yellow solid, yield: 73%, mp: 204–206 °C; 1H NMR (400 MHz, CDCl3) δ 8.44 (s, 1H), 7.32 (d, J = 6.6 Hz, 2H), 7.26–7.17 (m, 3H), 6.79 (d, J = 2.5 Hz, 1H), 6.65 (d, J = 8.6 Hz, 1H), 6.55 (dd, J = 8.6, 2.5 Hz, 1H), 6.13 (s, 1H), 3.74 (s, 3H), 3.70 (s, 3H), 3.58 (s, 3H), 2.44 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 166.04, 159.82, 150.05, 149.95, 145.88, 145.78, 139.95, 136.04, 128.87, 128.84, 127.99, 127.82, 121.04, 111.74, 111.65, 109.44, 108.11, 106.59, 105.80, 99.27, 98.11, 55.99, 55.37, 54.87, 51.79, 22.91. HRMS (ESI) m/z: calcd for C23H22N4O5S, [M + H]+: 467.1430; found: 467.1438.
- Methyl 2-(2-(2-methoxyphenyl)hydrazineylidene)-7-methyl-3-oxo-5-phenyl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10i). Yellow solid, yield: 55%, mp: 189–192 °C; 1H NMR (400 MHz, DMSO-d6) δ 10.35 (s, 1H), 7.39–7.24 (m, 6H), 7.03 (q, J = 7.9 Hz, 2H), 6.93 (t, J = 6.9 Hz, 1H), 6.02 (s, 1H), 3.86 (s, 3H), 3.60 (s, 3H), 2.38 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 165.73, 160.61, 153.08, 152.02, 146.28, 139.44, 130.96, 128.88, 128.85, 128.79, 127.99, 127.79, 123.35, 121.62, 114.51, 110.40, 109.74, 55.76, 55.23, 51.66, 22.53. HRMS (ESI) m/z: calcd for C22H20N4O4S, [M + H]+: 437.1321; found: 437.1332.
- Methyl 7-methyl-3-oxo-5-phenyl-2-(2-phenylhydrazineylidene)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10j). Yellow solid, yield: 49%, mp: 124–127 °C; 1H NMR (400 MHz, DMSO-d6) δ 10.99 (s, 1H), 7.38–7.29 (m, 7H), 7.23 (d, J = 7.4 Hz, 2H), 6.99 (t, J = 7.3 Hz, 1H), 6.04 (s, 1H), 3.61 (s, 3H), 2.39 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 165.80, 160.84, 152.95, 152.17, 141.94, 139.43, 129.44, 128.89, 128.86, 128.83, 127.97, 123.56, 122.08, 114.62, 109.82, 55.27, 51.70, 22.67. HRMS (ESI) m/z: calcd for C21H18N4O3S, [M − H]−: 405.1020; found: 405.1022.
- (2-(6-(Methoxycarbonyl)-7-methyl-3-oxo-5-phenyl-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)benzenesulfonic acid (10k). Yellow solid, yield: 84%, mp: 262–266 °C; 1H NMR (400 MHz, DMSO-d6) δ 11.24 (s, 1H), 7.99–7.82 (m, 2H), 7.57 (d, J = 7.3 Hz, 1H), 7.37–7.29 (m, 6H), 6.05 (s, 1H), 3.62 (s, 3H), 2.39 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 165.85, 160.46, 153.24, 151.42, 140.71, 138.56, 132.72, 131.07, 129.30, 129.07, 128.00, 127.69, 123.86, 122.28, 113.90, 109.90, 55.14, 52.04, 22.85. HRMS (ESI) m/z: calcd for C21H18N4O6S2, [M − H]−: 485.0588; found: 485.0593.
- Methyl 7-methyl-2-(2-(3-nitrophenyl)hydrazineylidene)-3-oxo-5-phenyl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10l). Yellow solid, yield: 71%, mp: 250–252 °C; 1H NMR (400 MHz, CDCl3) δ 8.00 (s, 1H), 7.88 (d, J = 8.0 Hz, 1H), 7.67 (s, 1H), 7.57 (d, J = 9.5 Hz, 1H), 7.47 (t, J = 8.2 Hz, 1H), 7.41 (d, J = 6.4 Hz, 2H), 7.31 (q, J = 8.5, 7.5 Hz, 3H), 6.24 (s, 1H), 3.67 (s, 3H), 2.54 (s, 3H). 13C NMR (101 MHz, CDCl3 + CD3OD) δ 165.78, 161.05, 154.73, 148.99, 144.07, 139.16, 130.11, 128.94, 128.77, 127.84, 120.16, 117.17, 109.71, 109.31, 55.16, 51.69, 22.22. HRMS (ESI) m/z: calcd for C21H17N5O5S, [M − H]−: 450.0870; found: 450.0879.
- Methyl 2-(2-(3-acetylphenyl)hydrazineylidene)-7-methyl-3-oxo-5-phenyl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10m). Yellow solid, yield: 45%, mp: 213–216 °C; 1H NMR (400 MHz, DMSO-d6) δ 11.12 (s, 1H), 7.76 (s, 1H), 7.59 (d, J = 3.1 Hz, 1H), 7.51–7.41 (m, 2H), 7.40–7.25 (m, 5H), 6.01 (s, 1H), 3.61 (s, 3H), 2.56 (s, 3H), 2.38 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 203.14, 169.95, 165.43, 158.98, 155.78, 147.26, 143.38, 141.80, 133.57, 132.81, 132.77, 132.68, 131.85, 131.77, 126.77, 124.72, 123.21, 118.11, 113.44, 59.05, 55.56, 30.51, 26.25. HRMS (ESI) m/z: calcd for C23H20N4O4S, [M + H]+: 449.1295; found: 449.1286.
- Methyl 2-(2-(3-(methoxycarbonyl)phenyl)hydrazineylidene)-7-methyl-3-oxo-5-phenyl-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (10n). Yellow solid, yield: 42%, mp: 214–216 °C; 1H NMR (400 MHz, DMSO-d6) δ 11.14 (s, 1H), 7.92–7.17 (m, 9H), 6.03 (s, 1H), 3.86 (s, 3H), 3.61 (s, 3H), 2.39 (s, 3H). 13C NMR (101 MHz, CDCl3 + CD3OD) δ 167.07, 165.87, 161.21, 154.60, 151.85, 142.92, 139.45, 130.97, 129.46, 128.86, 128.76, 127.88, 123.97, 121.17, 118.97, 118.92, 115.50, 115.45, 55.11, 52.30, 51.68, 22.45. HRMS (ESI) m/z: calcd for C23H20N4O5S, [M + H]+: 465.1254; found: 465.1252.
- Methyl 2-(2-(2-acetylphenyl)hydrazineylidene)-5-(4-methoxyphenyl)-7-methyl-3-oxo-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (12a). Yellow solid, yield: 64%, mp: 199–201 °C; 1H NMR (400 MHz, CDCl3) δ 12.07 (s, 1H), 7.74 (t, J = 8.5 Hz, 2H), 7.43 (t, J = 8.6 Hz, 1H), 7.26 (d, J = 8.8 Hz, 2H), 7.02–6.89 (m, 1H), 6.75 (d, J = 8.7 Hz, 2H), 6.11 (s, 1H), 3.68 (s, 3H), 3.59 (s, 3H), 2.59 (s, 3H), 2.46 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 202.71, 165.88, 160.81, 159.88, 152.48, 152.03, 145.15, 135.44, 131.70, 131.66, 129.41, 126.67, 121.33, 118.98, 115.28, 114.08, 110.10, 55.27, 54.74, 51.69, 27.98, 22.69. HRMS (ESI) m/z: calcd for C24H22N4O5S, [M − H]−: 477.1231; found: 477.1236.
- Methyl 2-(2-(2-(methoxycarbonyl)phenyl)hydrazineylidene)-5-(4-methoxyphenyl)-7-methyl-3-oxo-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (12b). Yellow solid, yield: 75%, mp: 219–221 °C; 1H NMR (400 MHz, CDCl3) δ 11.32 (s, 1H), 7.94 (d, J = 6.4 Hz, 1H), 7.76 (d, J = 7.3 Hz, 1H), 7.49 (t, J = 7.4 Hz, 1H), 7.34 (d, J = 8.8 Hz, 2H), 6.99 (t, J = 8.1 Hz, 1H), 6.82 (d, J = 8.7 Hz, 2H), 6.19 (s, 1H), 3.94 (s, 3H), 3.76 (s, 3H), 3.66 (s, 3H), 2.52 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 168.79, 165.90, 160.87, 159.86, 152.47, 152.10, 145.07, 134.97, 131.68, 130.81, 129.41, 126.12, 121.58, 115.02, 114.08, 112.12, 110.05, 55.28, 54.72, 52.53, 51.69, 22.72. HRMS (ESI) m/z: calcd for C24H22N4O6S, [M + H]+: 495.1348; found: 495.1339.
- Methyl 2-(2-(3-(methoxycarbonyl)phenyl)hydrazineylidene)-5-(4-methoxyphenyl)-7-methyl-3-oxo-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (12c). Yellow solid, yield: 80%, mp: 209–211 °C; 1H NMR (400 MHz, CDCl3) δ 7.78 (s, 1H), 7.59 (d, J = 7.7 Hz, 1H), 7.48 (d, J = 7.7 Hz, 1H), 7.30 (dt, J = 10.7, 3.3 Hz, 2H), 7.24 (s, 1H), 6.77 (d, J = 8.7 Hz, 2H), 6.09 (s, 1H), 3.84 (s, 3H), 3.70 (s, 3H), 3.61 (s, 3H), 2.44 (s, 3H). 13C NMR (101 MHz, CDCl3 + CD3OD) δ 167.17, 165.99, 161.34, 159.78, 154.75, 151.54, 143.05, 131.74, 130.94, 129.42, 129.24, 123.90, 120.95, 118.95, 115.46, 114.00, 109.59, 55.16, 54.50, 52.21, 51.62, 22.29. HRMS (ESI) m/z: calcd for C24H22N4O6S, [M + H]+: 495.1348; found: 495.1341.
- (2-(6-(Methoxycarbonyl)-5-(4-methoxyphenyl)-7-methyl-3-oxo-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)-3-methylbenzoic acid (12d). Yellow solid, yield: 77%, mp: 265–267 °C; 1H NMR (400 MHz, DMSO-d6) δ 7.82 (d, J = 7.7 Hz, 1H), 7.22 (d, J = 8.7 Hz, 2H), 7.17 (d, J = 7.4 Hz, 1H), 6.90 (d, J = 8.8 Hz, 2H), 6.85 (d, J = 7.5 Hz, 1H), 5.98 (s, 1H), 3.72 (s, 3H), 3.60 (s, 3H), 2.43 (s, 3H), 2.38 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 166.07, 160.56, 159.66, 154.97, 152.20, 145.25, 135.40, 133.40, 129.80, 129.10, 126.60, 123.18, 121.74, 119.44, 114.49, 108.80, 55.55, 54.04, 51.90, 23.00, 22.11. HRMS (ESI) m/z: calcd for C24H22N4O6S, [M + H]+: 495.1348; found: 495.1344.
- Methyl 5-(4-methoxyphenyl)-7-methyl-3-oxo-2-(2-(3,4,5-trimethoxyphenyl)hydrazineylidene)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (12e). Yellow solid, yield: 65%, mp: 204–206 °C; 1H NMR (400 MHz, DMSO-d6) δ 10.88 (s, 1H), 7.22 (d, J = 8.7 Hz, 2H), 6.90 (d, J = 8.8 Hz, 2H), 6.54 (s, 2H), 5.97 (s, 1H), 3.76 (s, 6H), 3.72 (s, 3H), 3.61 (s, 3H), 3.60 (s, 3H), 2.39 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 165.93, 160.85, 159.75, 154.21, 154.01, 151.69, 139.69, 133.50, 132.94, 129.21, 120.34, 114.50, 109.39, 92.19, 60.61, 56.13, 55.57, 55.36, 54.36, 51.95, 22.86. HRMS (ESI) m/z: calcd for C25H26N4O7S, [M + H]+: 527.1619; found: 527.1632.
- (6-(Methoxycarbonyl)-5-(4-methoxyphenyl)-7-methyl-3-oxo-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)naphthalene-1-sulfonic acid (12f). Yellow solid, yield: 90%, mp: 271–273 °C; 1H NMR (400 MHz, DMSO-d6) δ 12.81 (s, 1H), 8.84 (d, J = 7.7 Hz, 1H), 7.92 (d, J = 9.1 Hz, 1H), 7.80 (d, J = 6.7 Hz, 1H), 7.73 (d, J = 9.1 Hz, 1H), 7.48 (t, J = 7.0 Hz, 1H), 7.37 (t, J = 6.8 Hz, 1H), 7.24 (d, J = 8.7 Hz, 2H), 6.91 (d, J = 8.8 Hz, 2H), 6.02 (s, 1H), 3.72 (s, 3H), 3.62 (s, 3H), 2.41 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 165.93, 160.60, 159.78, 153.19, 151.35, 136.56, 132.82, 131.90, 130.90, 130.17, 129.16, 128.45, 127.57, 127.13, 124.56, 123.76, 114.58, 109.92, 55.58, 54.53, 52.03, 22.83. HRMS (ESI) m/z: calcd for C26H22N4O7S2, [M + H]+: 567.1016; found: 567.1003.
4.1.5. General Procedure for the Synthesis of Compounds 11a–k
The methyl 7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (1 mmol, 1 eq) and pyridine (4 mmol, 4 eq) were dissolved in 5 mL of dioxane at 0 °C. An appropriate aromatic amine (1.2 mmol, 1.2 eq) was dissolved in an aqueous solution of hydrochloric acid (3 mmol, 3 eq) and stirred at 0 °C. Then, a solution of sodium nitrite (1.44 mmol, 1.44 eq) was added to the hydrochloric acid–aromatic amine mixture to obtain the aromatic diazonium salt by stirring the above aqueous solution for 0.5 h at 0 °C. The resulting solution was added to the dioxane system and stirred at a temperature of -18 °C for 1 h, before raising it to room temperature and stirring for 3 h. The reaction was quenched by adding 10 mL of water, which resulted in precipitation of a solid product that was subsequently purified using silica gel column chromatography.
- Methyl 2-(2-(2-methoxyphenyl)hydrazineylidene)-7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (11a). Yellow solid, yield: 61%, mp: 178–180 °C; 1H NMR (400 MHz, CDCl3) δ 7.89 (s, 1H), 7.49 (dd, J = 7.7, 1.9 Hz, 1H), 7.08 (d, J = 8.7 Hz, 1H), 7.00–6.89 (m, 2H), 6.85 (dd, J = 7.8, 1.6 Hz, 1H), 6.58 (d, J = 8.7 Hz, 1H), 6.22 (s, 1H), 3.90 (s, 6H), 3.81 (s, 3H), 3.78 (s, 3H), 3.67 (s, 3H), 2.43 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 166.24, 160.68, 154.31, 152.64, 146.17, 142.02, 131.14, 125.57, 124.93, 123.04, 121.58, 114.36, 110.32, 109.46, 106.41, 60.68, 60.56, 55.91, 55.75, 53.54, 51.45, 22.79. HRMS (ESI) m/z: calcd for C25H26N4O7S, [M + H]+: 527.1619; found: 527.1634.
- Methyl 7-methyl-3-oxo-2-(2-phenylhydrazineylidene)-5-(2,3,4-trimethoxyphenyl)-2,3-dihydro-5H-thiazolo [3,2-a]pyrimidine-6-carboxylate (11b). Yellow solid, yield: 46%, mp: 189–192 °C; 1H NMR (400 MHz, CDCl3) δ 7.49 (s, 1H), 7.33 (t, J = 7.8 Hz, 2H), 7.22 (d, J = 7.6 Hz, 2H), 7.11 (d, J = 8.6 Hz, 1H), 7.05 (t, J = 7.3 Hz, 1H), 6.61 (d, J = 8.7 Hz, 1H), 6.25 (s, 1H), 3.92 (s, 3H), 3.85 (s, 3H), 3.82 (s, 3H), 3.70 (s, 3H), 2.46 (s, 3H). 13C NMR (101 MHz, CDCl3 + CD3OD) δ 166.53, 161.55, 155.53, 154.35, 152.60, 150.25, 142.88, 141.98, 129.27, 125.73, 125.00, 123.04, 119.80, 114.63, 109.05, 106.56, 60.61, 60.55, 55.90, 53.47, 51.48, 22.45. HRMS (ESI) m/z: calcd for C24H24N4O6S, [M + H]+: 497.1509; found: 497.1519.
- (2-(6-(Methoxycarbonyl)-7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)-3-methylbenzoic acid (11c). Yellow solid, yield: 30%, mp: 209–213 °C; 1H NMR (400 MHz, DMSO-d6) δ 7.81 (s, 1H), 7.15 (d, J = 7.3 Hz, 1H), 6.96 (d, J = 8.6 Hz, 1H), 6.84 (t, J = 7.4 Hz, 1H), 6.75 (d, J = 8.7 Hz, 1H), 6.06 (s, 1H), 3.78 (s, 6H), 3.71 (s, 3H), 3.58 (s, 3H), 2.41 (s, 3H), 2.30 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 166.36, 160.48, 154.96, 154.12, 152.26, 150.82, 145.44, 141.92, 135.32, 129.72, 126.58, 126.26, 125.23, 123.13, 121.53, 119.69, 108.38, 107.56, 61.00, 60.62, 56.24, 52.70, 51.62, 23.04, 22.06. HRMS (ESI) m/z: calcd for C26H26N4O8S, [M + H]+: 554.1570; found: 555.1573.
- Methyl 7-methyl-2-(2-(3-nitrophenyl)hydrazineylidene)-3-oxo-5-(2,3,4-trimethoxyphenyl)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (11d). Yellow solid, yield: 48%, mp: 236–238 °C; 1H NMR (400 MHz, CDCl3) δ 8.20 (s, 1H), 7.97 (t, J = 2.2 Hz, 1H), 7.82 (d, J = 6.8 Hz, 1H), 7.50 (d, J = 9.4 Hz, 1H), 7.41 (t, J = 8.1 Hz, 1H), 7.08 (d, J = 8.6 Hz, 1H), 6.58 (d, J = 8.7 Hz, 1H), 6.21 (s, 1H), 3.88 (s, 3H), 3.82 (s, 3H), 3.79 (s, 3H), 3.68 (s, 3H), 2.43 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 166.05, 160.48, 154.50, 152.60, 149.70, 149.01, 143.49, 142.01, 130.29, 125.72, 124.27, 120.09, 117.56, 109.84, 109.35, 106.46, 60.72, 60.60, 55.91, 54.00, 51.62, 22.59. HRMS (ESI) m/z: calcd for C24H23N5O8S, [M + H]+: 541.1357; found: 542.1361.
- 2-(2-(6-(Methoxycarbonyl)-7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)benzenesulfonic acid (11e). Yellow solid, yield: 74%, mp: 238–240 °C; 1H NMR (400 MHz, DMSO-d6) δ 11.15 (s, 1H), 7.59 (d, J = 7.7 Hz, 1H), 7.35 (d, J = 7.7 Hz, 2H), 6.99 (d, J = 8.6 Hz, 2H), 6.76 (d, J = 8.7 Hz, 1H), 6.07 (s, 1H), 3.78 (s, 6H), 3.70 (s, 3H), 3.60 (s, 3H), 2.31 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 166.17, 160.32, 154.31, 152.88, 152.37, 149.94, 141.89, 138.67, 132.55, 131.04, 127.98, 125.50, 125.35, 124.20, 122.06, 113.76, 109.42, 107.44, 61.02, 60.64, 56.24, 53.38, 51.82, 22.86. HRMS (ESI) m/z: calcd for C24H24N4O9S2, [M + H]+: 577.1067; found: 577.1062.
- 5-Methoxy-2-(2-(6-(methoxycarbonyl)-7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)benzenesulfonic acid (11f). Yellow solid, yield: 76%, mp: 244–246 °C; 1H NMR (400 MHz, DMSO-d6) δ 10.93 (s, 1H), 7.31 (d, J = 8.9 Hz, 1H), 7.13 (s, 1H), 6.98 (d, J = 8.6 Hz, 2H), 6.75 (d, J = 8.6 Hz, 1H), 6.06 (s, 1H), 3.77 (s, 6H), 3.73 (s, 3H), 3.70 (s, 3H), 3.59 (s, 3H), 2.31 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 166.19, 160.34, 154.58, 154.28, 153.06, 152.36, 150.10, 141.87, 133.57, 132.38, 125.47, 122.52, 117.31, 115.45, 112.32, 109.23, 107.43, 61.02, 60.63, 56.22, 55.88, 53.30, 51.79, 22.87. HRMS (ESI) m/z: calcd for C25H26N4O10S2, [M − H]−: 605.1010; found: 605.1006.
- (2-(6-(Methoxycarbonyl)-7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-5H-thiazolo[3,2-a]pyrimidin-2(3H)-ylidene)hydrazineyl)naphthalene-1-sulfonic acid (11g). Yellow solid, yield: 76%, mp: 241–244 °C; 1H NMR (400 MHz, DMSO-d6) δ 12.74 (s, 1H), 8.85 (d, J = 8.9 Hz, 1H), 7.90 (d, J = 9.0 Hz, 1H), 7.79 (d, J = 8.1 Hz, 1H), 7.71 (d, J = 9.0 Hz, 1H), 7.48 (t, J = 7.8 Hz, 1H), 7.36 (t, J = 7.5 Hz, 1H), 7.00 (d, J = 8.7 Hz, 1H), 6.77 (d, J = 8.6 Hz, 1H), 6.09 (s, 1H), 3.78 (d, J = 4.4 Hz, 6H), 3.70 (s, 3H), 3.60 (s, 3H), 2.32 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 166.20, 160.41, 154.32, 153.09, 152.36, 150.03, 141.95, 136.69, 131.82, 130.95, 130.10, 128.42, 127.57, 127.08, 125.49, 125.43, 124.55, 124.46, 123.96, 114.59, 109.41, 107.54, 61.04, 60.64, 56.28, 53.29, 51.77, 22.86. HRMS (ESI) m/z: calcd for C28H26N4O9S2, [M − H]−: 625.1061; found: 625.1056.
- Methyl 2-(2-(2-(methoxycarbonyl)phenyl)hydrazineylidene)-7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (11h). Yellow solid, yield: 55%, mp: 200–202 °C; 1H NMR (400 MHz, CDCl3) δ 11.29 (s, 1H), 7.95 (d, J = 6.4 Hz, 1H), 7.74 (d, J = 8.4 Hz, 1H), 7.48 (t, J = 7.1 Hz, 1H), 7.08 (d, J = 8.6 Hz, 1H), 7.01–6.94 (m, 1H), 6.58 (d, J = 8.6 Hz, 1H), 6.23 (s, 1H), 3.95 (s, 3H), 3.90 (s, 3H), 3.82 (s, 3H), 3.79 (s, 3H), 3.67 (s, 3H), 2.43 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 168.42, 165.89, 160.36, 153.97, 152.29, 144.83, 141.67, 134.57, 130.44, 126.06, 125.20, 124.48, 121.03, 114.60, 111.66, 109.27, 106.03, 60.31, 60.21, 55.56, 53.22, 52.14, 51.10, 22.49. HRMS (ESI) m/z: calcd for C26H26N4O8S, [M + H]+: 554.1570; found: 555.1574.
- Methyl 2-(2-(2-acetylphenyl)hydrazineylidene)-7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (11i). Yellow solid, yield: 52%, mp: 205–208 °C; 1H NMR (400 MHz, CDCl3) δ 12.12 (s, 1H), 7.81 (dd, J = 15.9, 8.3 Hz, 2H), 7.49 (t, J = 7.2 Hz, 1H), 7.08 (d, J = 8.6 Hz, 1H), 7.01 (t, J = 7.0 Hz, 1H), 6.58 (d, J = 8.7 Hz, 1H), 6.23 (s, 1H), 3.90 (s, 3H), 3.82 (s, 3H), 3.78 (s, 3H), 3.67 (s, 3H), 2.67 (s, 3H), 2.44 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 202.65, 166.25, 160.69, 154.31, 152.63, 152.54, 150.55, 145.27, 142.02, 135.40, 131.69, 127.02, 125.55, 124.83, 121.12, 118.88, 115.20, 109.68, 106.39, 60.68, 60.56, 55.91, 53.55, 51.46, 27.98, 22.88. HRMS (ESI) m/z: calcd for C26H26N4O7S, [M + H]+: 539.1609; found: 539.1605.
- Methyl 2-(2-(3-(methoxycarbonyl)phenyl)hydrazineylidene)-7-methyl-3-oxo-5-(2, 3, 4-trimethoxyphenyl)-2, 3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (11j). Yellow solid, yield: 67%, mp: 213–215 °C; 1H NMR (400 MHz, CDCl3) δ 7.94 (s, 1H), 7.72 (d, J = 7.6 Hz, 1H), 7.60 (d, J = 8.3 Hz, 1H), 7.44 (d, J = 8.4 Hz, 1H), 7.16 (d, J = 8.7 Hz, 1H), 6.69 (d, J = 8.7 Hz, 1H), 6.27 (s, 1H), 3.97 (d, J = 5.4 Hz, 6H), 3.91 (s, 6H), 3.76 (s, 3H), 2.50 (s, 3H). 13C NMR (101 MHz, CDCl3 + CD3OD) δ 167.36, 166.49, 161.39, 155.14, 154.43, 152.66, 150.18, 143.33, 142.05, 131.10, 129.54, 125.80, 124.89, 123.91, 121.30, 119.05, 115.54, 109.27, 60.67, 60.62, 55.97, 53.65, 52.34, 51.56, 22.55. HRMS (ESI) m/z: calcd for C26H26N4O8S, [M + H]+: 661.2733; found: 555.1577.
- Methyl 2-(2-(3-acetylphenyl)hydrazineylidene)-7-methyl-3-oxo-5-(2,3,4-trimethoxyphenyl)-2,3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (11k). Yellow solid, yield: 40%, mp: 216–219 °C; 1H NMR (400 MHz, DMSO-d6) δ 11.00 (s, 1H), 7.75 (s, 1H), 7.59 (dt, J = 6.4, 2.1 Hz, 1H), 7.51–7.40 (m, 2H), 6.97 (d, J = 8.7 Hz, 1H), 6.75 (d, J = 8.8 Hz, 1H), 6.04 (s, 1H), 3.78 (s, 6H), 3.70 (s, 3H), 3.59 (s, 3H), 2.56 (s, 3H), 2.31 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 198.09, 166.19, 160.67, 154.25, 152.31, 141.86, 138.23, 130.31, 125.45, 122.89, 119.04, 109.04, 107.46, 61.05, 60.62, 56.24, 53.08, 51.78, 27.25, 22.90. HRMS (ESI) m/z: calcd for C26H26N4O7S, [M + H]+: 539.1609; found: 539.1604.
4.2. Other Protocols
Additional experimental methods are provided in the Supporting Information.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29092120/s1, Figure S1: Ligand-RMSD comparison of three-time molecular dynamics simulation; Figure S2: Inhibitory profile of compounds on HIV-1 RNase H activity; References citation [26,27,28].
Author Contributions
Conceptualization, G.M. and F.-E.C.; methodology, G.M. and F.-E.C.; investigation, X.-D.Z.; software, X.-D.Z.; Chemical synthesis, X.-D.Z.; Enzyme activity studies, A.C., S.M.; resources, F.-E.C. and E.T.; writing—original draft preparation, X.-D.Z.; writing—review and editing, S.W.; supervision, F.-E.C., S.W., C.P. and E.D.C.; project administration, F.-E.C.; funding acquisition, G.M. and E.T. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to appreciate their gratification for the National Natural Science Foundation of China (No. 21871055) for the financial support. This research was supported by EU funding within the MUR PNRR Extended, Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT, Spoke 5) and RAS LR 7/07 project antivirale-unica F73C22001570002.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, F.-E.C., upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cohen, M.S.; Chen, Y.Q.; McCauley, M.; Gamble, T.; Hosseinipour, M.C.; Kumarasamy, N.; Hakim, J.G.; Kumwenda, J.; Grinsztejn, B.; Pilotto, J.H.S.; et al. Antiretroviral Therapy for the Prevention of HIV-1 Transmission. N. Engl. J. Med. 2016, 375, 830–839. [Google Scholar] [CrossRef]
- De Clercq, E. Antiviral drugs in current clinical use. J. Clin. Virol. 2004, 30, 115–133. [Google Scholar] [CrossRef]
- Singh, A.K.; Das, K. Insights into HIV-1 Reverse Transcriptase (RT) Inhibition and Drug Resistance from Thirty Years of Structural Studies. Viruses 2022, 14, 1027. [Google Scholar] [CrossRef]
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2012, 2, a007161. [Google Scholar] [CrossRef]
- Chun, T.W.; Stuyver, L.; Mizell, S.B.; Ehler, L.A.; Mican, J.A.; Baseler, M.; Lloyd, A.L.; Nowak, M.A.; Fauci, A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 1997, 94, 13193–13197. [Google Scholar] [CrossRef]
- Bagasra, O. A unified concept of HIV latency. Expert Opin. Biol. Ther. 2006, 6, 1135–1149. [Google Scholar] [CrossRef]
- Menéndez-Arias, L. Molecular basis of human immunodeficiency virus type 1 drug resistance: Overview and recent developments. Antivir. Res. 2013, 98, 93–120. [Google Scholar] [CrossRef]
- Zhan, P.; Pannecouque, C.; De Clercq, E.; Liu, X. Anti-HIV Drug Discovery and Development: Current Innovations and Future Trends. J. Med. Chem. 2016, 59, 2849–2878. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, F.; Zhang, J.; Liu, C.; López-Carrobles, N.; Liu, X.; Menéndez-Arias, L.; Zhan, P. Current medicinal chemistry strategies in the discovery of novel HIV-1 ribonuclease H inhibitors. Eur. J. Med. Chem. 2022, 243, 114760. [Google Scholar] [CrossRef]
- Kang, J.X.; Zhao, G.K.; Yang, X.M.; Huang, M.X.; Hui, W.Q.; Zeng, R.; Ouyang, Q. Recent advances on dual inhibitors targeting HIV reverse transcriptase associated polymerase and ribonuclease H. Eur. J. Med. Chem. 2023, 250, 115196. [Google Scholar] [CrossRef]
- Lapkouski, M.; Tian, L.; Miller, J.T.; Le Grice, S.F.J.; Yang, W. Complexes of HIV-1 RT, NNRTI and RNA/DNA hybrid reveal a structure compatible with RNA degradation. Nat. Struct. Mol. Biol. 2013, 20, 230–236. [Google Scholar] [CrossRef]
- Di Santo, R. Inhibiting the HIV Integration Process: Past, Present, and the Future. J. Med. Chem. 2014, 57, 539–566. [Google Scholar] [CrossRef]
- Moianos, D.; Prifti, G.-M.; Makri, M.; Zoidis, G. Targeting Metalloenzymes: The “Achilles’ Heel” of Viruses and Parasites. Pharmaceuticals 2023, 16, 901. [Google Scholar] [CrossRef]
- Kitagawa, Y.; Liao, Z.; Morikawa, K.; Oda, M. Metal-binding and folding thermodynamics of Escherichia coli ribonuclease HI related to its catalytic function. Biophys. Chem. 2023, 295, 106961. [Google Scholar] [CrossRef]
- Kankanala, J.; Kirby, K.A.; Liu, F.; Miller, L.; Nagy, E.; Wilson, D.J.; Parniak, M.A.; Sarafianos, S.G.; Wang, Z. Design, Synthesis, and Biological Evaluations of Hydroxypyridonecarboxylic Acids as Inhibitors of HIV Reverse Transcriptase Associated RNase H. J. Med. Chem. 2016, 59, 5051–5062. [Google Scholar] [CrossRef]
- Borkow, G.; Fletcher, R.S.; Barnard, J.; Arion, D.; Motakis, D.; Dmitrienko, G.I.; Parniak, M.A. Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry 1997, 36, 3179–3185. [Google Scholar] [CrossRef]
- Messore, A.; Corona, A.; Madia, V.N.; Saccoliti, F.; Tudino, V.; De Leo, A.; Scipione, L.; De Vita, D.; Amendola, G.; Di Maro, S.; et al. Pyrrolyl Pyrazoles as Non-Diketo Acid Inhibitors of the HIV-1 Ribonuclease H Function of Reverse Transcriptase. ACS Med. Chem. Lett. 2020, 11, 798–805. [Google Scholar] [CrossRef]
- Wang, L.; Tang, J.; Huber, A.D.; Casey, M.C.; Kirby, K.A.; Wilson, D.J.; Kankanala, J.; Xie, J.; Parniak, M.A.; Sarafianos, S.G.; et al. 6-Arylthio-3-hydroxypyrimidine-2,4-diones potently inhibited HIV reverse transcriptase-associated RNase H with antiviral activity. Eur. J. Med. Chem. 2018, 156, 652–665. [Google Scholar] [CrossRef]
- Wang, X.; Gao, P.; Menendez-Arias, L.; Liu, X.; Zhan, P. Update on Recent Developments in Small Molecular HIV-1 RNase H Inhibitors (2013–2016): Opportunities and Challenges. Curr. Med. Chem. 2018, 25, 1682–1702. [Google Scholar] [CrossRef]
- Tramontano, E.; Corona, A.; Menéndez-Arias, L. Ribonuclease H, an unexploited target for antiviral intervention against HIV and hepatitis B virus. Antivir. Res. 2019, 171, 104613. [Google Scholar] [CrossRef]
- Cihlar, T.; Ray, A.S. Nucleoside and nucleotide HIV reverse transcriptase inhibitors: 25 years after zidovudine. Antivir. Res. 2010, 85, 39–58. [Google Scholar] [CrossRef] [PubMed]
- de Béthune, M.P. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: A review of the last 20 years (1989–2009). Antivir. Res. 2010, 85, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Beilhartz, G.L.; Götte, M. HIV-1 Ribonuclease H: Structure, Catalytic Mechanism and Inhibitors. Viruses 2010, 2, 900–926. [Google Scholar] [CrossRef] [PubMed]
- Corona, A.; Di Leva, F.S.; Thierry, S.; Pescatori, L.; Cuzzucoli Crucitti, G.; Subra, F.; Delelis, O.; Esposito, F.; Rigogliuso, G.; Costi, R.; et al. Identification of highly conserved residues involved in inhibition of HIV-1 RNase H function by Diketo acid derivatives. Antimicrob. Agents Chemother. 2014, 58, 6101–6110. [Google Scholar] [CrossRef] [PubMed]
- Lansdon, E.B.; Liu, Q.; Leavitt, S.A.; Balakrishnan, M.; Perry, J.K.; Lancaster-Moyer, C.; Kutty, N.; Liu, X.; Squires, N.H.; Watkins, W.J.; et al. Structural and binding analysis of pyrimidinol carboxylic acid and N-hydroxy quinazolinedione HIV-1 RNase H inhibitors. Antimicrob. Agents Chemother. 2011, 55, 2905–2915. [Google Scholar] [CrossRef] [PubMed]
- Costa, G.; Rocca, R.; Corona, A.; Grandi, N.; Moraca, F.; Romeo, I.; Talarico, C.; Gagliardi, M.G.; Ambrosio, F.A.; Ortuso, F.; et al. Novel natural non-nucleoside inhibitors of HIV-1 reverse transcriptase identified by shape- and structure-based virtual screening techniques. Eur. J. Med. Chem. 2019, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Corona, A.; Meleddu, R.; Esposito, F.; Distinto, S.; Bianco, G.; Masaoka, T.; Maccioni, E.; Menéndez-Arias, L.; Alcaro, S.; Le Grice, S.F.; et al. Ribonuclease H/DNA Polymerase HIV-1 Reverse Transcriptase Dual Inhibitor: Mechanistic Studies on the Allosteric Mode of Action of Isatin-Based Compound RMNC6. PLoS ONE 2016, 11, e0147225. [Google Scholar] [CrossRef]
- Corona, A.; Schneider, A.; Schweimer, K.; Rösch, P.; Wöhrl, B.M.; Tramontano, E. Inhibition of foamy virus reverse transcriptase by human immunodeficiency virus type 1 RNase H inhibitors. Antimicrob. Agents Chemother. 2014, 58, 4086–4093. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).