Analysis Transcriptome and Phytohormone Changes Associated with the Allelopathic Effects of Ginseng Hairy Roots Induced by Different-Polarity Ginsenoside Components
Abstract
1. Introduction
2. Results
2.1. Composition and Content of Ginsenosides in Different-Polarity Components
2.2. Allelopathic Autotoxicity of Differen-Polarity Ginsenoside Components
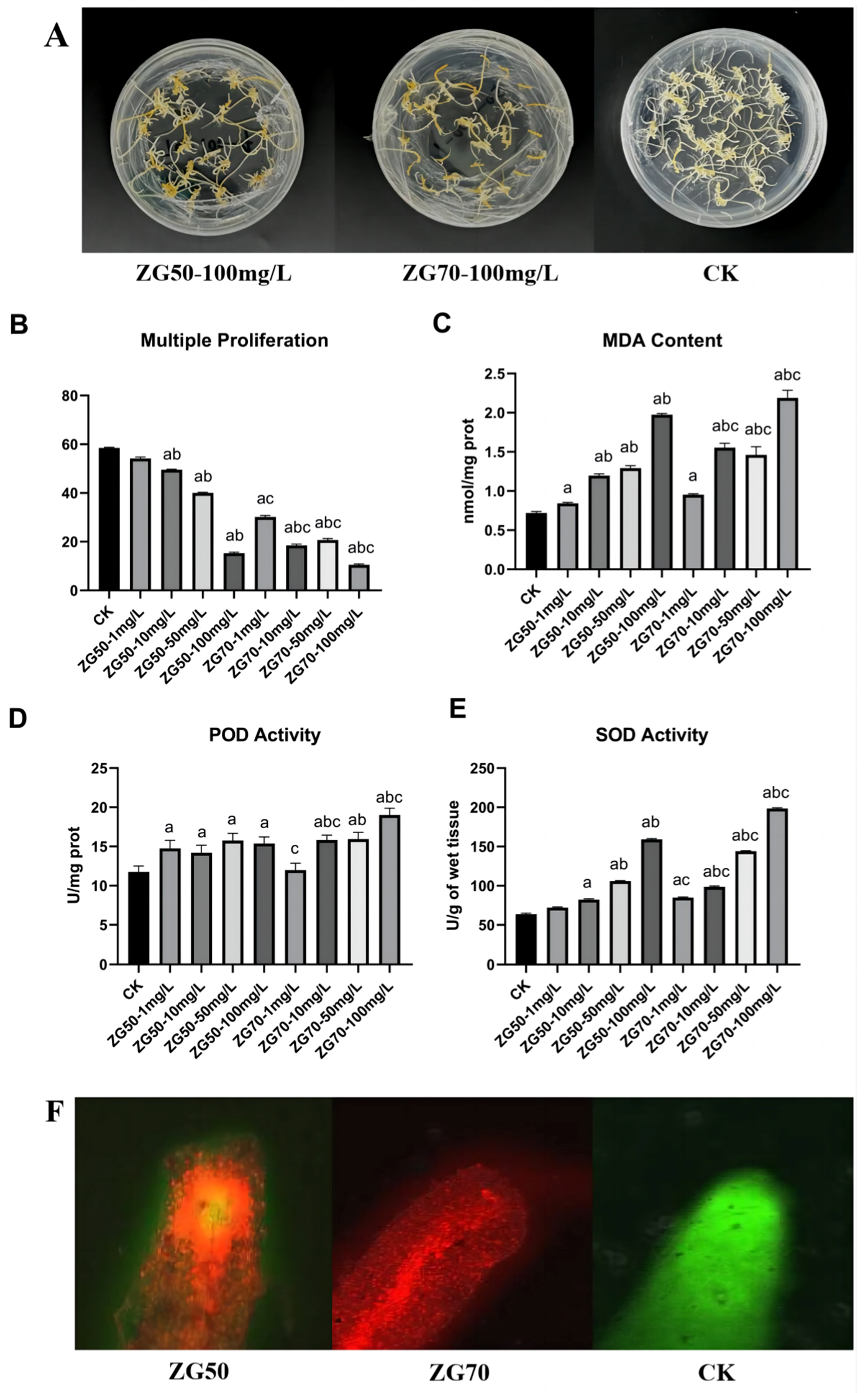
2.2.1. The Influence on Apparent Morphology of Ginseng Hairy Roots
2.2.2. The Influence on Apical Physiological Indexes of Ginseng Hairy Roots
2.2.3. The Influence of Ginseng Hairy Roots on Apical Cell Viability
2.3. Targeted Metabolomics Analysis of the Allelopathic Autotoxicity of Ginsenoside Components with Different Polarities
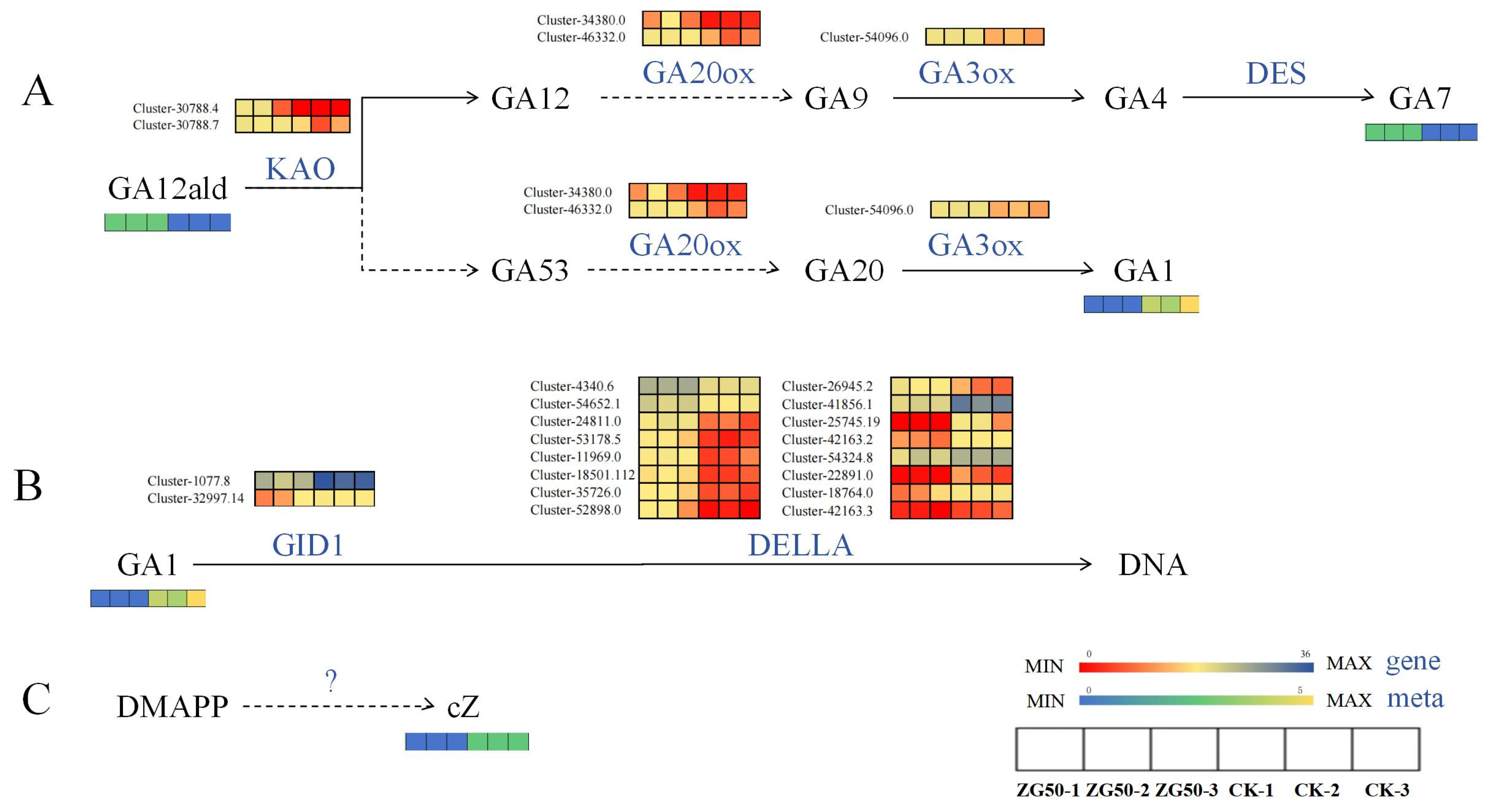
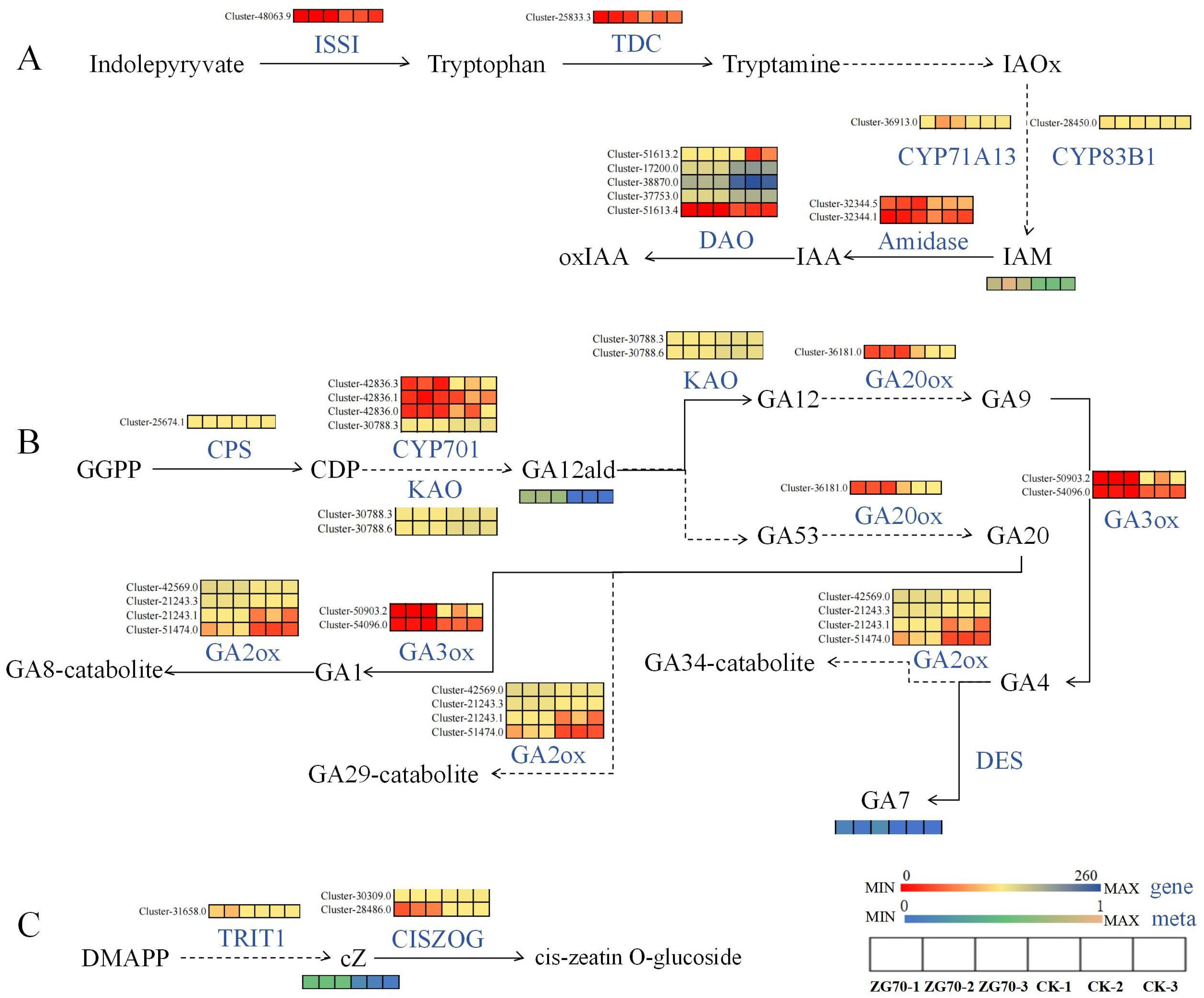
2.3.1. Effects on Endogenous Phytohormone Accumulation and Metabolism in Ginseng Hairy Roots
2.3.2. Metabolic Pathway Analysis of Differential Accumulation of Endogenous Phytohormones in Ginseng Hairy Roots
2.4. Transcriptomics Analysis of the Allelopathic Autotoxicity of Ginsenoside Components with Different Polarities
2.4.1. Transcript Splicing, Gene Annotation, and Differential Expression Gene Screening
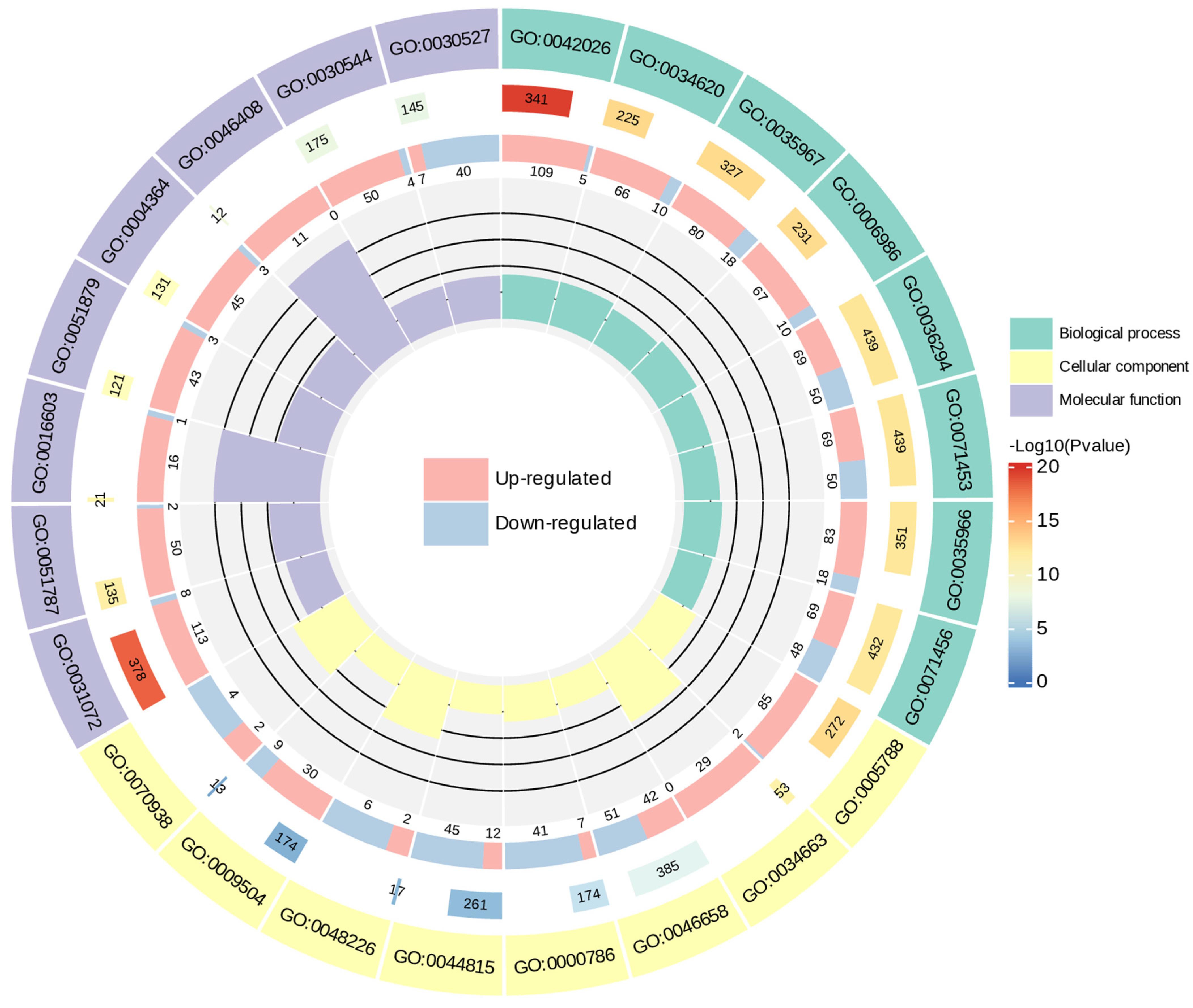
2.4.2. The Functional Annotation and Enrichment of Differential Genes
KEGG Enrichment Analysis
GO Enrichment Analysis
KOG Enrichment Analysis
2.4.3. Validation of the Key Genes by qPCR
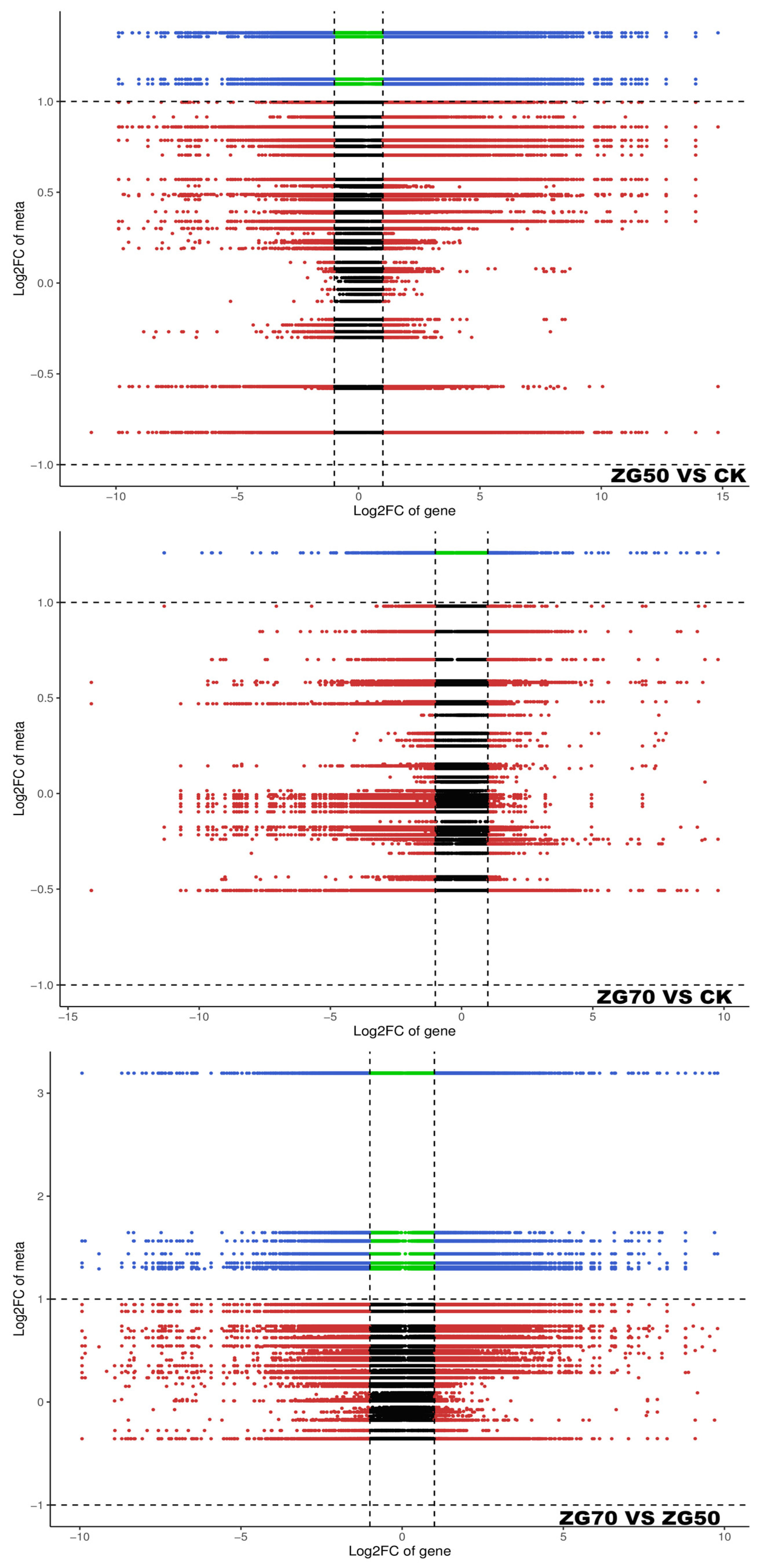
2.5. Integrated Metabolomic and Transcriptomic Analysis of the Correlation between Polarity and Allelopathic Autotoxicity of Ginsenosides
3. Discussion
3.1. Innovative Study on Allelopathic Autotoxicity of Different Polarities of Ginsenoside Components
3.2. Exploring the Possible Mechanism of the Relationship between the Polarity and Allelopathic Autotoxicity of Ginsenoside Components
3.2.1. Reduce the Cell Vitality of Ginseng Roots
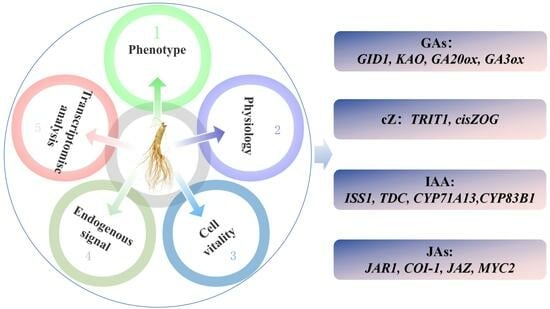
3.2.2. Differential Expression of Key Genes Induced Phytohormones and Their Derivatives
Regulate the Expression of Genes of Key Enzymes in the cZ Synthesis Pathway
Regulate the Expression of Genes of Key Enzymes in the Gibberellin Signal Transduction Pathway
Regulate the Expression of Genes of Key Enzymes in the Pathways of Indole-3-acetic acid (IAA) Synthesis and Oxidation Inactivation
Regulate the Expression of Genes of Key Enzymes in JA Signaling Transduction Pathway
4. Materials and Methods
4.1. Plant Material
4.2. Preparation and Determination of Ginsenoside Components with Different Polarities
4.2.1. Preparation of Ginsenoside Fractions using Medium-pressure Liquid Chromatography
4.2.2. UPLC-QQQ-MS Determination of Ginsenoside Components with Different Polarities
4.3. Allelopathic Autotoxicity Validation
4.3.1. Culture of Ginseng Hairy Roots
4.3.2. Detection of Physiological Indexes of Ginseng Hairy Roots
4.3.3. Detection of Cell Viability of Ginseng Hairy Roots
4.4. Phytohormones’ Metabolomic Profiling and Statistical Analysis
4.5. RNA-seq Test and Statistical Analysis
4.6. Integrated Analysis of Metabolomics and Transcriptomics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, M.; Li, A.X.; Wang, F.Q.; Jiang, J.F.; Wang, Z.B.; You, J.F. Integrative Analysis of Multiple Metabolomes and Transcriptome Revealed Color Expression Mechanism in Red Skin Root syndrome of Panax Ginseng. Ind. Crops Prod. 2022, 177, 114491. [Google Scholar] [CrossRef]
- Wang, X.Y.; Jiang, M.T.; Lou, J.; Zou, Y.D.; Liu, M.Y.; Li, Z.; Guo, D.; Yang, W.Z. Pseudotargeted Metabolomics Approach Enabling the Classification-Induced Ginsenoside Characterization and Differentiation of Ginseng and Its Compound Formulation Products. J. Agric. Food Chem. 2023, 71, 1735–1747. [Google Scholar] [CrossRef]
- Liang, X.L.; Hou, X.; Li, J.Y.; Han, Y.Q.; Zhang, Y.X.; Feng, N.J.; Du, J.D.; Zhang, W.H.; Zheng, D.F.; Fang, S.M. High-resolution DNA Methylome Reveals That Demethylation Enhances Adaptability to Continuous Cropping Comprehensive Stress in Soybean. BMC Plant Biol. 2019, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, X.; Ding, L.; Kong, C.H. Allelopathy and Allelochemicals in Grasslands and Forests. Forests 2023, 14, 562. [Google Scholar] [CrossRef]
- Ladhari, A.; Tufano, L.; DellaGreca, M. Influence of New Effective Allelochemicals on the Distribution of Cleome arabica L. Community in Nature. Nat. Prod. Res. 2018, 34, 773–781. [Google Scholar] [CrossRef]
- Meng, X.R.; Zhang, T.; Chen, C.B.; Li, Q.; Lin, J.W. Regulatory Network of Ginsenoside Biosynthesis under Ro Stress in the Hairy Roots of Panax Ginseng Revealed by RNA Sequencing. Front. Bioeng. Biotechnol. 2022, 10, 1006386. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, L.X.; Guo, T.Z.; Xu, Y.H.; Chen, C.B. Allelopathic Effects of Ginsenoside Rgl on Seed Germination and Seedling Growth of Panax Ginseng. Allelopath. J. 2020, 49, 229–242. [Google Scholar] [CrossRef]
- Shen, Y.L.; Fan, M.L.; Li, G.; Li, Q.; Meng, X.R.; Wang, E.P.; Chen, C.B. Allelopathic Effect of Ginsenoside Rd on Ginseng Seeds and Seedings. Appl. Ecol. Environ. Res. 2022, 21, 59–72. [Google Scholar] [CrossRef]
- Zhang, K.L.; Zhang, S.A.; Ebihara, A.; Zhou, X.Q.; Fan, L.K.; Li, P.F.; Zhang, Z.Q.; Wang, Y.Y.; Shen, Y. The Current Research Progress of Ginseng Species: The Cultivation and Application. Cogent Food Agric. 2023, 9, 2216483. [Google Scholar] [CrossRef]
- Zhang, A.H.; Lei, F.J.; Fu, J.F.; Zhou, R.J.; Zhang, L.X. Influence of Exogenous Ginsenosides on New Forest Soil Microbial Communities. China J. Chin. Mater. Medica 2017, 42, 4756–4761. [Google Scholar]
- Yang, M.; Chuang, Y.C.; Guo, C.W.; Liao, J.J.; Xu, Y.G.; Mei, X.Y.; Liu, Y.X.; Huang, H.C.; He, X.H.; Zhu, S.S. Panax Notoginseng Root Cell Death Caused by the Autotoxic Ginsenoside Rg1 is Due to Over-Accumulation of ROS, as Revealed by Transcriptomic and Cellular Approaches. Front. Plant Sci. 2018, 9, 264. [Google Scholar] [PubMed]
- Meng, X.R.; Huang, X.; Li, Q.; Wang, E.P.; Chen, C.B. Application of UPLC-Orbitrap-HRMS Targeted Metabolomics in Screening of Allelochemicals and Model Plants of Ginseng. J. Plant Physiol. 2023, 285, 153996. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Dai, S.Y.; Wang, B.Y.; Jiang, Y.T.; Ma, Y.Y.; Pan, L.L.; Wu, K.; Huang, X.Q.; Zhang, J.B.; Cai, Z.C.; et al. Autotoxic Ginsenoside Disrupts Soil Fungal Microbiomes by Stimulating Potentially Pathogenic Microbes. Appl. Environ. Microbiol. 2020, 86, e00130-20. [Google Scholar] [CrossRef] [PubMed]
- Nicol, R.W.; Yousef, L.; Traquair, J.A.; Bernards, M.A. Ginsenosides Stimulate the Growth of Soilborne Pathogens of American Ginseng. Phytochemistry 2003, 64, 257–264. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X.D.; Xu, Y.G.; Mei, X.Y.; Jiang, B.B.; Liao, J.J.; Yin, Z.B.; Zheng, J.F.; Zhao, Z.; Fan, L.M.; et al. Autotoxic Ginsenosides in the Rhizosphere Contribute to the Replant Failure of Panax notoginseng. PLoS ONE 2015, 10, e01185552. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.Y.; Zhou, J.; Sun, Y.; Qiao, B.X.; Wan, T.; Guo, R.Q.; Zhang, J.; Shan, D.Q.; Cai, Y.L. Comparative Transcriptome and Metabolome Analyses of Cherry Leaves Spot Disease Caused by Alternaria Alternata. Front. Plant Sci. 2023, 14, 1129515. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P. The Current Status of Research on Gibberellin Biosynthesis. Plant Cell Physiol. 2020, 61, 1832–1849. [Google Scholar] [PubMed]
- Tuderman, L.; Myllylä, R.; Kivirikko, K.I. Mechanism of the Prolyl Hydroxylase Reaction. 1. Role of Co-substrates. Eur. J. Biochem. 1977, 80, 341–348. [Google Scholar] [CrossRef]
- Ward, D.A.; MacMillan, J.; Gong, F.; Phillips, A.L.; Hedden, P. Gibberellin 3-Oxidases in Developing Embryos of the Southern Wild Cucumber, Marah macrocarpus. Phytochemistry 2010, 71, 2010–2018. [Google Scholar] [CrossRef]
- Sarwar, R.; Zhu, K.M.; Jiang, T.; Ding, P.; Gao, Y.; Tan, X.L. DELLAs Directed Gibberellins Responses Orchestrate Crop Development: A Brief Review. Crop Sci. 2023, 63, 1–28. [Google Scholar] [CrossRef]
- Kasahara, H.; Takei, K.; Ueda, N.; Hishiyama, S.; Yamaya, T.; Kamiya, Y.; Yamaguchi, S.; Sakakibara, H. Distinct Isoprenoid Origins of Cis- and Trans-zeatin Biosyntheses in Arabidopsis. J. Biol. Chem. 2004, 279, 14049–14054. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP Isopentenyltransferases and tRNA Isopentenyltransferases in Cytokinin Biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, H.; Hanada, A.; Kuzuyama, T.; Takagi, M.; Kamiya, Y.; Yamaguchi, S. Contribution of the Mevalonate and Methylerythritol Phosphate Pathways to the Biosynthesis of Gibberellins in Arabidopsis. J. Biol. Chem. 2002, 277, 45188–45194. [Google Scholar] [CrossRef] [PubMed]
- Köksal, M.; Potter, K.; Peters, R.J.; Christianson, D.W. 1.55 Å-resolution Structure of Ent-copalyl Diphosphate Synthase and Exploration of General Acid Function by Site-directed Mutagenesis. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J.; Gao, X.L.; Jones, A.D.; Howe, G.A. A Rapid Wound Signal Activates the Systemic Synthesis of Bioactive Jasmonates in Arabidopsis. Plant J. 2009, 59, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Howe, G.A. A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-domain Protein JAZ10 in Arabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, T.; Zhang, J.; Li, C.; Xu, Y.H.; Zheng, H.; Zhou, J.H.; Zha, L.P.; Jiang, C.; Jin, Y.; et al. Ginsenosides Regulate Adventitious Root Formation in Panax Ginseng Via a CLE45-WOX11 Regulatory Module. J. Exp. Bot. 2020, 71, 6396–6407. [Google Scholar] [CrossRef]
- Zhang, A.H.; Lei, F.J.; Fang, S.W.; Jia, M.H.; Zhang, L.X. Effects of Ginsenosides on the Growth and Activity of Antioxidant Enzymes in American Ginseng Seedlings. J. Med. Plants Res. 2011, 5, 3217–3223. [Google Scholar]
- Jones, K.; Kim, D.W.; Park, J.S.; Khang, C.H. Live-cell Fluorescence Imaging to Investigate the Dynamics of Plant Cell Death during Infection by the Rice Blast Fungus Magnaporthe oryzae. BMC Plant Biol. 2016, 16, 69. [Google Scholar] [CrossRef]
- Zhang, S.L.; Zhang, M.M.; Dai, W.; Bi, X.D.; Yang, P.; Zhang, D.J. Evaluation of Berberine’s Algicidal Effects on Toxic Microcystis Aeruginosa Growth using a Double Fluorescein Staining Method. Isr. J. Aquac. Bamidgeh 2014, 66, 940. [Google Scholar] [CrossRef]
- Kojima, M.; Makita, N.; Miyata, K.; Yoshino, M.; Iwase, A.; Ohashi, M.; Surjana, A.; Kudo, T.; Takeda-Kamiya, N.; Toyooka, K.; et al. A Cell Wall-localized Cytokinin/purine Riboside Nucleosidase is involved in Apoplastic Cytokinin Metabolism in Oryza sativa. Proc. Natl. Acad. Sci. USA 2023, 120, e221770812036. [Google Scholar] [CrossRef] [PubMed]
- Baigorri, R.; Conesa, C.M.; del Pozo, J.C.; Garcia-Mina, J.M.; Saez, A.; Silva-Navas, J.; Swarup, R.; Zamarreño, A.M. Role of Cis-zeatin in Root Responses to Phosphate Starvation. New Phytol. 2019, 224, 242. [Google Scholar]
- Wang, W.Q.; Zhang, G.Q.; Wang, W.L.; Wang, Z.G.; Lv, Y.L.; Guo, F.X.; Di, Y.D.; Zhang, J.F.; Wang, Y.H.; Wang, W.; et al. Wheat Cis-zeatin-O-glucosyltransferase cZOGT1 Interacts with the Ca2+-dependent Lipid Binding Protein TaZIP to Regulate Senescence. J. Exp. Bot. 2023, 74, 6619–6630. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.C.; Martin, R.C.; Mok, M.C.; Habben, J.E.; Mok, D.W. A Maize Cytokinin Gene Encoding an O-glucosyltransferase Specific to Cis-zeatin. Proc. Natl. Acad. Sci. USA 2001, 98, 5922–5926. [Google Scholar] [CrossRef] [PubMed]
- Dobra, J.; Motyka, V.; Dobrev, P.; Malbeck, J.; Prasil, I.T.; Haisel, D.; Gaudinova, A.; Havlova, M.; Gubis, J.; Vankova, R. Comparison of Hormonal Responses to Heat, Drought and Combined Stress in Tobacco Plants with Elevated Proline Content. J. Plant Physiol. 2010, 167, 1360–1370. [Google Scholar] [CrossRef] [PubMed]
- Vanková, R.; Kosová, K.; Dobrev, P.; Vítámvás, P.; Trávnícková, A.; Cvikrová, M.; Pesek, B.; Gaudinová, A.; Prerostová, S.; Musilová, J.; et al. Dynamics of Cold Acclimation and Complex Phytohormone Responses in Triticum Monococcum Lines G3116 and DV92 Differing in Vernalization and Frost Tolerance Level. Environ. Exp. Bot. 2014, 101, 12–25. [Google Scholar] [CrossRef]
- Schäfer, M.; Brütting, C.; Meza-Canales, I.D.; Grosskinsky, D.K.; Vankova, R.; Baldwin, I.T.; Meldau, S. The Role of Cis-zeatin-type Cytokinins in Plant Growth Regulation and Mediating Responses to Environmental Interactions. J. Exp. Bot. 2015, 66, 4873–4884. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Yang, H.L.; Shang, C.Q.; Ma, S.; Liu, L.; Cheng, J.L. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef] [PubMed]
- Soeno, K.; Sato, A.; Shimada, Y. Investigation of Auxin Biosynthesis and Action Using Auxin Biosynthesis Inhibitors. Methods Mol. Biol. 2021, 2213, 131–144. [Google Scholar]
- Lv, B.S.; Yan, Z.W.; Tian, H.Y.; Zhang, X.S.; Ding, Z.J. Local Auxin Biosynthesis Mediates Plant Growth and Development. Trends Plant Sci. 2019, 24, 6–9. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin Biosynthesis and Storage Forms. J. Expermental Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef] [PubMed]
- Woodward, C.; Bemis, S.M.; Hill, E.J.; Sawa, S.; Koshiba, T.; Torii, K.U. Interaction of Auxin and ERECTA in Elaborating Arabidopsis Inflorescence Architecture Revealed by the Activation Tagging of A New Member of the YUCCA Family Putative Flavin Monooxygenases. Plant Physiol. 2005, 139, 192–203. [Google Scholar] [CrossRef]
- Novák, O.; Hényková, E.; Sairanen, I.; Kowalczyk, M.; Pospísil, T.; Ljung, K. Tissue-specific Profiling of the Arabidopsis Thaliana Auxin Metabolome. Plant J. 2012, 72, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, S.; Hishiyama, S.; Jikumaru, Y.; Hanada, A.; Nishimura, T.; Koshiba, T.; Zhao, Y.; Kamiya, Y.; Kasahara, H. Biochemical Analyses of Indole-3-acetaldoximedependent Auxin Biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5430–5435. [Google Scholar] [CrossRef] [PubMed]
- Chamarro, J.; Ostin, A.; Sandberg, G. Metabolism of Indole-3-acetic Acid by Orange (Citrus sinensis) Flavedo Tissue during Fruit Development. Phytochemistry 2001, 57, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, S.; Neu, D.; Weiler, E.W. Molecular Cloning and Characterization of An Amidase from Arabidopsis Thaliana Capable of Converting Indole-3-acetamide into the Plant Growth Hormone, Indole-3-acetic Acid. Phytochemistry 2003, 62, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.P.; Hu, Y.; Ge, C.N.; Zhao, Y.D.; Kasahara, H.; et al. The Main Oxidative Inactivation Pathway of the Plant Hormone Auxin. Nat. Commun. 2021, 12, 6752. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.G.; Zhang, Y.H.; Liu, X.; Zhang, X.; Liu, S.C.; Yu, X.W.; Ren, Y.L.; Zheng, X.M.; Zhou, K.N.; Jiang, L. A Role for A Dioxygenase in Auxin Metabolism and Reproductive Development in Rice. Dev. Cell 2013, 27, 113–122. [Google Scholar] [CrossRef]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, Action, and Interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef]
- Li, M.Y.; Yu, G.H.; Cao, C.L.; Liu, P. Metabolism, Signaling, and Transport of Jasmonates. Plant Commun. 2021, 2, 100231. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Yao, N.; Jing, L.; Zheng, H.; Chen, M.; Shen, Y. Research Progress of Jasmonate-responsive Transcription Factors in Regulating Plant Secondary Metabolism. China J. Chin. Mater. Medica 2018, 43, 897–903. [Google Scholar]
- Sato, T.; Miyanoiri, Y.; Takeda, M.; Naoe, Y.; Mitani, R.; Hirano, K.; Takehara, S.; Kainosho, M.; Matsuoka, M.; Ueguchi-Tanaka, M.; et al. Expression and Purification of A GRAS Domain of SLR1, the Rice DELLA Protein. Protein Expr. Purifiction 2014, 95, 248–258. [Google Scholar] [CrossRef]
- Hauvermale, A.L.; Cárdenas, J.J.; Bednarek, S.Y.; Steber, C.M. GA Signaling Expands: The Plant UBX Domain-containing Protein 1 Is A Binding Partner for the GA Receptor. Plant Physiol. 2022, 190, 2651–2670. [Google Scholar] [CrossRef] [PubMed]
- Hynes, L.W.; Peng, J.R.; Richards, D.E.; Harberd, N.P. Transgenic Expression of the Arabidopsis DELLA Proteins GAI and Gai confers Altered Gibberellin Response in Tobacco. Transgenic Res. 2003, 12, 707–714. [Google Scholar] [CrossRef]
- Wen, C.K.; Chang, C. Arabidopsis RGL1 Encodes a Negative Regulator of Gibberellin Responses. Plant Cell 2002, 14, 87–100. [Google Scholar] [CrossRef]
- Lee, S.; Cheng, H.; King, K.E.; Wang, W.; He, Y.; Hussain, A.; Lo, J.; Harberd, N.P.; Peng, J. Gibberellin Regulates Arabidopsis Seed Germination Via RGL2, A GAI/RGA-like Gene Whose Expression Is Up-regulated Following Imbibition. Genes Dev. 2002, 16, 646–658. [Google Scholar] [CrossRef]
- Tyler, L.; Thomas, S.G.; Hu, J.H.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.P. Della Proteins and Gibberellin-regulated Seed Germination and Floral Development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef]
- Li, B.; Zhao, Y.T.; Yue, Z.C.; Lei, J.L.; Hu, Q.Z.; Tao, P. Identification of DELLA Gene Family in Head Cabbage and Analysis of mRNA Transport in the Heterograft. Yi Chuan Hered. 2023, 45, 156–164. [Google Scholar]
- Zhang, N.; Huang, X.; Guo, Y.L.; Yue, H.; Chen, C.B.; Liu, S.Y. Evaluation of Storage Period of Fresh Ginseng for Quality Improvement of Dried and Red Processed Varieties. J. Ginseng Res. 2022, 46, 290–295. [Google Scholar] [CrossRef]
- Schupp, D.G.; Erlandsen, S.L. A New Method to Determine Giardia Cyst Viability: Correlation of Fluorescein Diacetate and Propidium Iodide Staining with Animal Infectivity. Appl. Environ. Microbiol. 1987, 53, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Li, X.; Niu, L.; Jameson, P.E.; Zhou, W. Transcription-associated Metabolomic Adjustments in Maize Occur During Combined Drought and Cold Stress. Plant Physiol. 2021, 186, 677–695. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wei, L.; Huang, S.; Yang, C.; Wang, Y.; Yuan, H.; Xu, Q.; Zhang, W.; Wang, M.; Zeng, X.; et al. Drought Resistance in Qingke Involves a Reprogramming of the Phenylpropanoid Pathway and UDP-Glucosyltransferase Regulation of Abiotic Stress Tolerance Targeting Flavonoid Biosynthesis. J. Agric. Food Chem. 2021, 69, 3992–4005. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: An Ultra-fast All-in-one FASTQ Preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]





| Group | Raw Reads | Clean Reads | Clean Base (G) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| CK-1 | 44,419,584 | 43,914,206 | 6.59 | 0.03 | 97.49 | 92.81 | 43.15 |
| CK-2 | 48,091,324 | 46,950,398 | 7.04 | 0.03 | 97.96 | 94.06 | 43.22 |
| CK-3 | 47,199,614 | 46,069,272 | 6.91 | 0.03 | 97.92 | 93.95 | 43.11 |
| ZG50-1 | 44,735,532 | 43,812,496 | 6.57 | 0.03 | 97.52 | 92.92 | 43.26 |
| ZG50-2 | 46,572,572 | 45,450,844 | 6.82 | 0.03 | 97.48 | 92.83 | 43.12 |
| ZG50-3 | 43,992,620 | 43,192,292 | 6.48 | 0.03 | 97.53 | 92.93 | 43.05 |
| ZG70-1 | 41,996,536 | 41,172,528 | 6.18 | 0.03 | 97.51 | 92.87 | 43.11 |
| ZG70-2 | 43,724,370 | 42,959,844 | 6.44 | 0.03 | 97.56 | 92.95 | 43.17 |
| ZG70-3 | 44,232,050 | 43,265,614 | 6.49 | 0.03 | 97.6 | 93.08 | 43.1 |
| Class | KEGG Map | Description | Count Meta | Index Meta | Count Gene |
|---|---|---|---|---|---|
| ZG50 vs. CK | ko01100 | Metabolic pathways | 3 | cZ; GA12-ald; GA1 | 798 |
| ZG50 vs. CK | ko00904 | Diterpenoid biosynthesis | 3 | GA12-ald; GA7; GA1 | 6 |
| ZG50 vs. CK | ko04075 | Phytohormone signal transduction | 1 | GA1 | 94 |
| ZG50 vs. CK | ko00908 | Zeatin biosynthesis | 1 | cZ | 5 |
| ZG50 vs. CK | ko01110 | Biosynthesis of secondary metabolites | 4 | cZ; GA12-ald; GA7; GA1 | 390 |
| ZG70 vs. CK | ko01110 | Biosynthesis of secondary metabolites | 3 | cZ; GA12-ald; GA7 | 691 |
| ZG70 vs. CK | ko01100 | Metabolic pathways | 3 | IAM; cZ; GA12-ald | 1156 |
| ZG70 vs. CK | ko00904 | Diterpenoid biosynthesis | 2 | GA12-ald; GA7 | 15 |
| ZG70 vs. CK | ko00908 | Zeatin biosynthesis | 1 | cZ | 14 |
| ZG70 vs. CK | ko00380 | Tryptophan metabolism | 1 | IAM | 28 |
| ZG70 vs. ZG50 | ko04075 | Phytohormone signal transduction | 2 | GA1; JA-ILE | 305 |
| ZG70 vs. ZG50 | ko01110 | Biosynthesis of secondary metabolites | 2 | cZ; GA1 | 1136 |
| ZG70 vs. ZG50 | ko01100 | Metabolic pathways | 2 | cZ; GA1 | 2070 |
| ZG70 vs. ZG50 | ko00904 | Diterpenoid biosynthesis | 1 | GA1 | 22 |
| ZG70 vs. ZG50 | ko00908 | Zeatin biosynthesis | 1 | cZ | 19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Li, Q.; Huang, X.; Chen, C. Analysis Transcriptome and Phytohormone Changes Associated with the Allelopathic Effects of Ginseng Hairy Roots Induced by Different-Polarity Ginsenoside Components. Molecules 2024, 29, 1877. https://doi.org/10.3390/molecules29081877
Zhou T, Li Q, Huang X, Chen C. Analysis Transcriptome and Phytohormone Changes Associated with the Allelopathic Effects of Ginseng Hairy Roots Induced by Different-Polarity Ginsenoside Components. Molecules. 2024; 29(8):1877. https://doi.org/10.3390/molecules29081877
Chicago/Turabian StyleZhou, Tingting, Qiong Li, Xin Huang, and Changbao Chen. 2024. "Analysis Transcriptome and Phytohormone Changes Associated with the Allelopathic Effects of Ginseng Hairy Roots Induced by Different-Polarity Ginsenoside Components" Molecules 29, no. 8: 1877. https://doi.org/10.3390/molecules29081877
APA StyleZhou, T., Li, Q., Huang, X., & Chen, C. (2024). Analysis Transcriptome and Phytohormone Changes Associated with the Allelopathic Effects of Ginseng Hairy Roots Induced by Different-Polarity Ginsenoside Components. Molecules, 29(8), 1877. https://doi.org/10.3390/molecules29081877