Chalcone-Synthase-Encoding RdCHS1 Is Involved in Flavonoid Biosynthesis in Rhododendron delavayi
Abstract
1. Introduction
2. Results
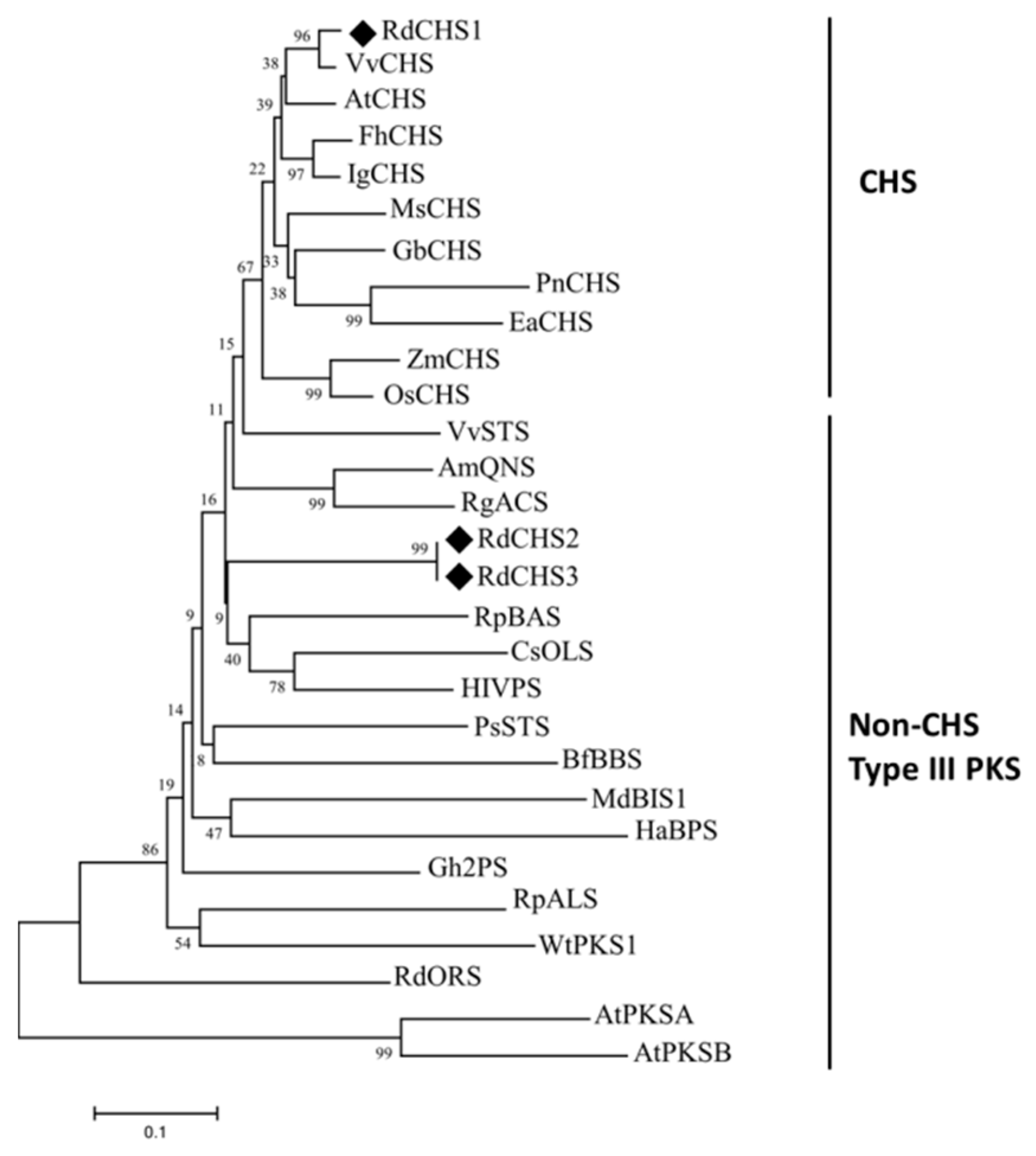
2.1. Cloning and Sequence Analysis of RdCHSs
2.2. RdCHS1 Expression Patterns in Developing Flowers and Different Tissues
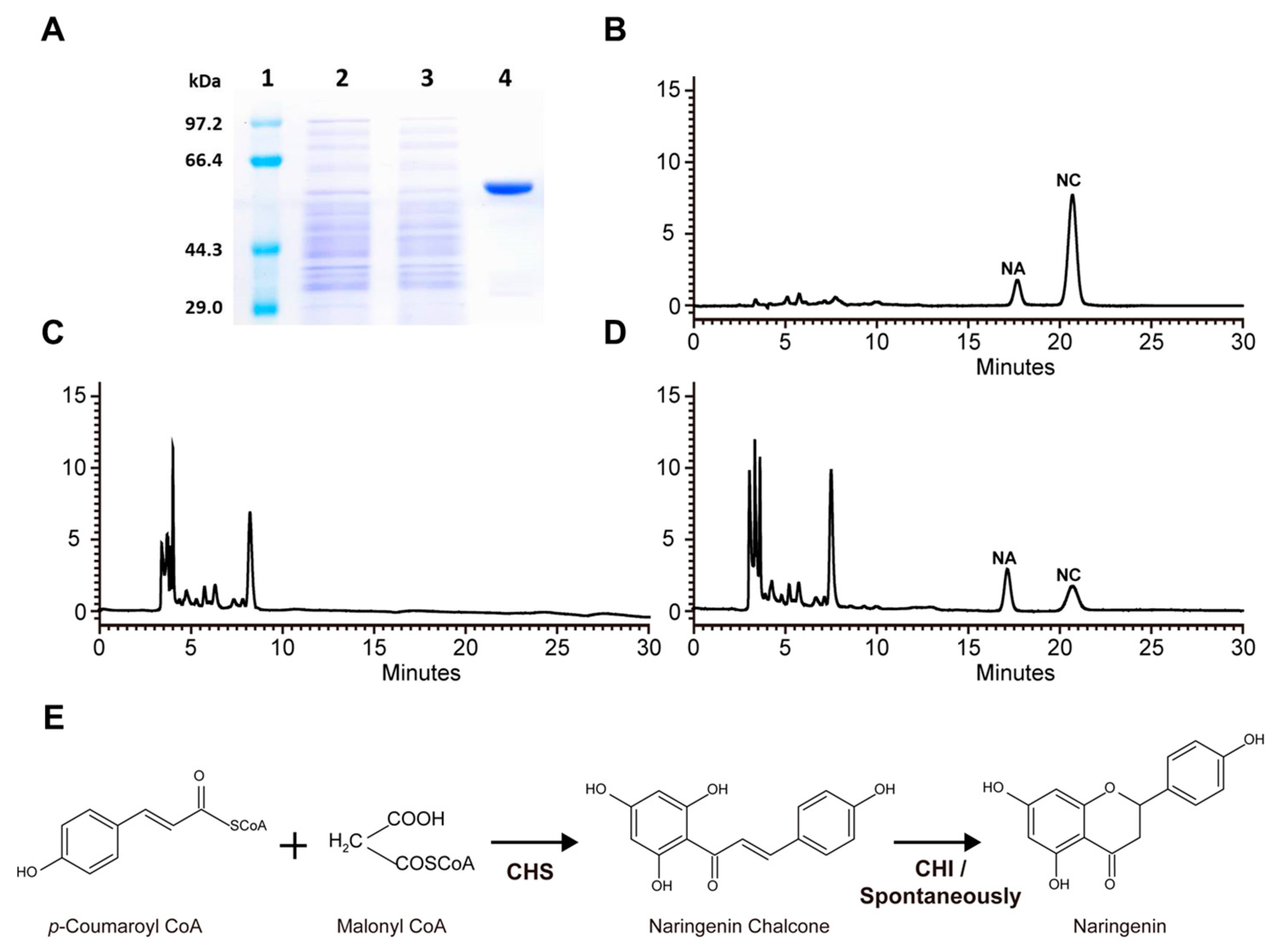
2.3. Biochemical Characterization of RdCHS1
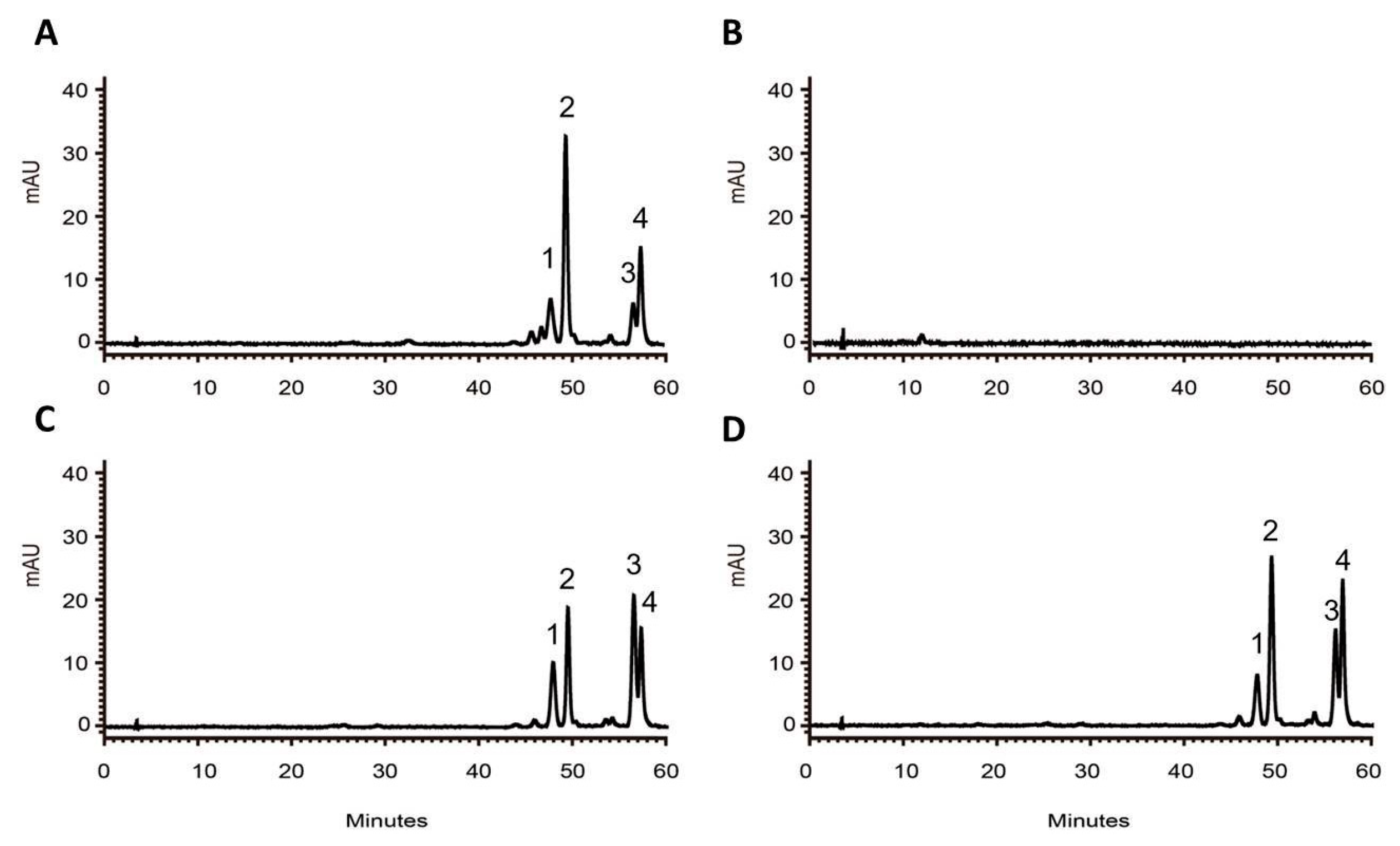
2.4. Complementation of the tt4 Mutant with RdCHS1
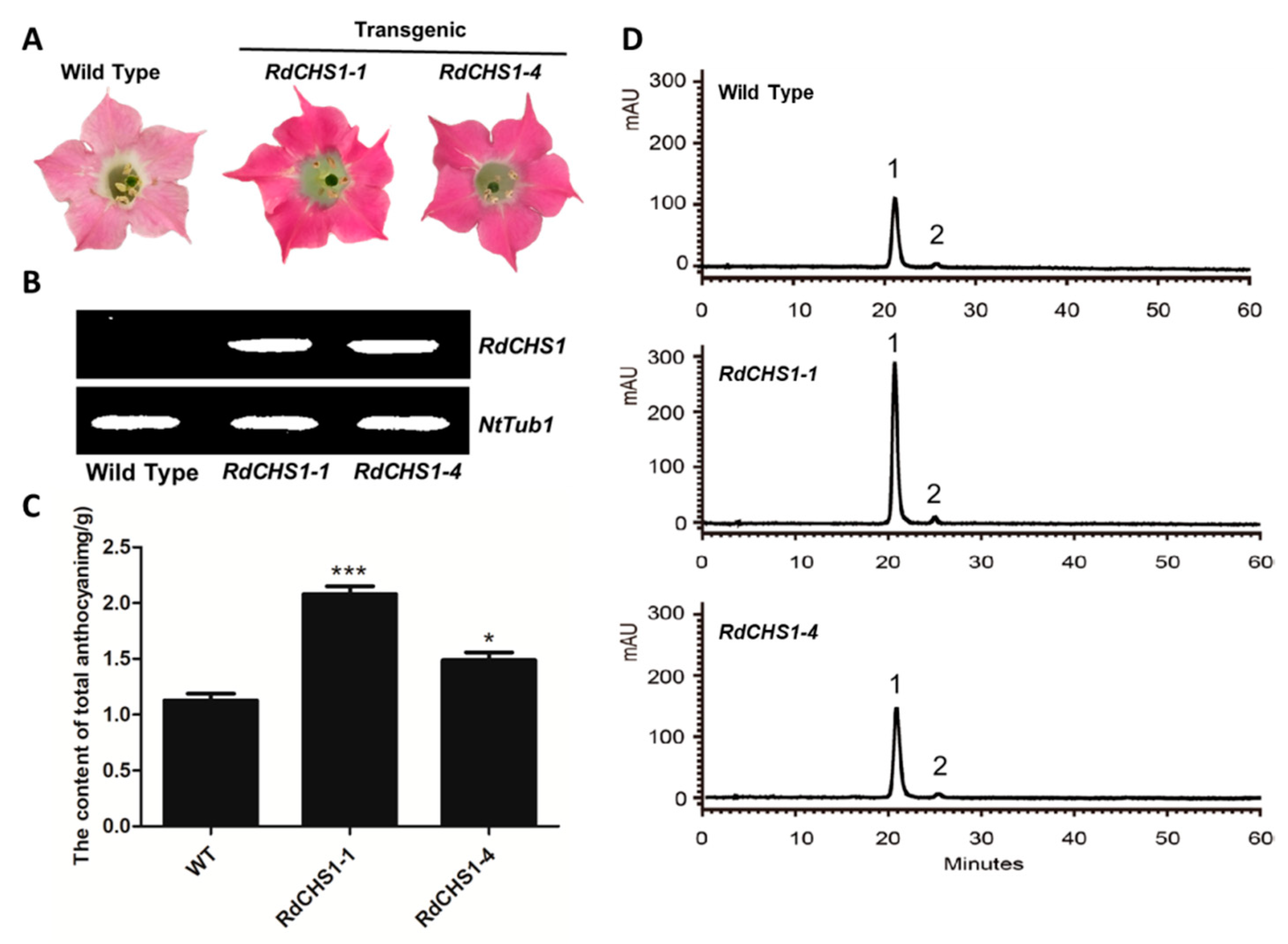
2.5. Overexpression of RdCHS1 in Tobacco
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Chemicals
4.3. Full-Length cDNA Cloning of RdCHS1
4.4. Sequence Alignment and Phylogenetic Analysis
4.5. Gene Expression Analysis
4.6. Soluble Protein Extraction and CHS Enzyme Assay
4.7. Expression Vector Construction and Transformation of Arabidopsis and Tobacco
4.8. Anthocyanin Analysis of Transgenic Arabidopsis Seedlings and Tobacco Flowers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, M.; Rius, S.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar]
- Ma, M.; Zhong, M.; Zhang, Q.; Zhao, W.; Wang, M.; Luo, C. Phylogenetic Implications and Functional Disparity in the Chalcone synthase Gene Family of Common Sea grass Zostera marina. Front. Mar. Sci. 2021, 8, 760902. [Google Scholar] [CrossRef]
- Dong, H.; Li, H.; Xue, Y.; Su, S.; Li, S.; Shan, X.; Liu, H.; Jiang, N.; Wu, X.; Zhang, Z.; et al. E183K Mutation in Chalcone Synthase C2 Causes Protein Aggregation and Maize Colorless. Front. Plant Sci. 2021, 12, 679654. [Google Scholar] [CrossRef]
- Lin, B.W.; Gong, C.C.; Song, H.F.; Cui, Y.Y. Effects of anthocyanins on the prevention and treatment of cancer. Br. J. Pharmacol. 2017, 174, 1226–1243. [Google Scholar] [CrossRef]
- Surangi, D.; David, H.; Vasantha, R. Chemopreventive Effect of Dietary Anthocyanins against Gastrointestinal Cancers: A Review of Recent Advances and Perspectives. Int. J. Mol. Sci. 2020, 21, 6555. [Google Scholar] [CrossRef]
- VanderMeer, I.M.; Spelt, C.E.; Mol, J.N.; Stuitje, A.R. Promoter analysis of the chalcone synthase (chsA) gene of Petunia hybrida: A 67 bp promoter region directs flower-specific expression. Plant Mol. Biol. 1990, 15, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Hans, S.; Heinz, S. Structure of the chalcone synthase gene of Antirrhinum majus. Mol. Gen. Genet. 1986, 202, 429434. [Google Scholar]
- Feinbaum, R.; Ausubel, F. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol. Cell. Biol. 1988, 8, 1985–1992. [Google Scholar]
- Colanero, S.; Perata, P.; Gonzali, S. What’s behind purple tomatoes? Insight into the mechanisms of anthocyanin synthesis in tomato fruits. Plant Physiol. 2020, 182, 1841–1853. [Google Scholar] [CrossRef]
- Zhang, X.; Abrahan, C.; Colquhoun, T.A.; Liu, C.J. A Proteolytic Regulator Controlling Chalcone Synthase Stability and Flavonoid Biosynthesis in Arabidopsis. Plant Cell. 2017, 29, 1157–1174. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Ferrer, J.L.; Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 1999, 6, 775–784. [Google Scholar]
- Liu, X.J.; Chuang, Y.N.; Chiou, C.Y.; Chin, D.C.; Shen, F.Q.; Yeh, K.W. Methylation effect on chalcone synthase gene expression determines anthocyanin pigmentation in floral tissues of two on cidium orchid cultivars. Planta 2012, 236, 401–409. [Google Scholar] [CrossRef]
- Dare, A.P.; Tomes, S.; Jones, M.; McGhie, T.K.; Stevenson, D.E.; Johnson, R.A.; Greenwood, D.R.; Hellens, R.P. Phenotypic changes associated with rna interference silencing of chalcone synthase in apple (Malus × domestica). Plant J. 2013, 74, 398–410. [Google Scholar] [CrossRef]
- Harris, N.; Luczo, J.; Robinson, S.; Walker, A. Transcriptional regulation of the three grapevine chalcone synthase genes and their role in flavonoid synthesis in Shiraz. Aust. J. Grape Wine Res. 2013, 19, 221–229. [Google Scholar] [CrossRef]
- Deng, X.; Bashandy, H.; Ainasoja, M.; Kontturi, J.; Pietiäinen, M.; Laitinen, R.A.; Albert, V.A.; Valkonen, J.; Elomaa, P.; Teeri, T.H. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of gerbera hybrida. New Phytol. 2014, 201, 1469–1483. [Google Scholar] [CrossRef]
- Liu, J.; Hao, X.L.; He, X.Q. Characterization of three chalcone synthase-like genes in Dianthus chinensis. Plant Cell. Tiss Org. 2021, 146, 483–492. [Google Scholar] [CrossRef]
- Kuckuck, H. Ober vier neue Serien multipler Allele bei Antirrhinum majus. Z. Induct. Abstamm. Vererbgsl. 1936, 71, 429–440. [Google Scholar]
- Koes, R.E.; Spelt, C.E.; Mol, J.N.M. The chalcone synthase multigene family of Petunia hybrid (V30): Differential, light-regulated expression during flower development and UV light induction. Plant Mol. Biol. 1989, 12, 213–225. [Google Scholar] [CrossRef]
- Koseki, M.; Goto, K.; Masuta, C.; Kanazawa, A. The star-type color pattern in Petunia hybrida “red Star” fowers is induced by the sequence-specifc degradation of the chalcone synthase RNA. Plant Cell Physiol. 2005, 46, 1879–1883. [Google Scholar] [CrossRef]
- Morita, Y.; Saito, R.; Ban, Y.; Tanikawa, N.; Kuchitsu, K.; Ando, T.; Yoshikawa, M.; Pollak, P.E.; Vogt, T.; Mo, Y.; et al. Chalcone synthase and flavonol accumulation in stigmas and anthers of Petunia hybrida. Plant Physiol. 1993, 102, 925–932. [Google Scholar]
- Pollak, P.; Hansen, K.; Astwood, J.; Taylor, L. Conditional male fertility in maize. Sex. Plant Reprod. 1995, 8, 231–241. [Google Scholar]
- Chang, C.; Bowman, J.L.; DeJohn, A.W.; Lander, E.S.; Meyerowitz, E.M. Restriction fragment length polymorphism linkage map for Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 1988, 85, 6856–6860. [Google Scholar] [CrossRef]
- Buer, C.S.; Djordjevic, M.A. Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J. Exp. Bot. 2009, 60, 751–763. [Google Scholar] [CrossRef]
- Helariutta, Y.; Elomaa, P.; Kotilainen, M.; Griesbach, R.J.; SchrÖder, J.; Teeri, T.H. Chalcone synthase-like genes active during corolla development are diferentially expressed and encode enzymes with diferent catalytic properties in Gerbera hybrida (Aster aceae). Plant Mol. Biol. 1995, 28, 47–60. [Google Scholar] [CrossRef]
- Nakatsuka, A.; Izumi, Y.; Yamagishi, M. Spatial and temporal expression of chalcone synthase and dihydrofavonol 4-reductase genes in the Asiatic hybrid lily. Plant Sci. 2003, 165, 759–767. [Google Scholar] [CrossRef]
- Suzuki, K.; Suzuki, T.; Nakatsuka, T.; Dohra, H.; Yamagishi, M.; Matsuyama, K.; Matsuura, H. RNA-seq-based evaluation of bicolor tepal pigmentation in Asiatic hybrid lilies (Lilium spp.). BMC Genom. 2016, 17, 611–629. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, P.W.; Cai, Y.F.; Ma, L.L.; Li, S.F.; Xie, W.L. The draft genome assembly of Rhododendron delavayi Franch var. delavayi. GigaScience 2017, 6, 1–11. [Google Scholar] [CrossRef]
- Sun, W.; Sun, S.; Xu, H.; Wang, Y.; Chen, Y.; Xu, X.; Yi, Y.; Ju, Z. Characterization of Two Key Flavonoid 3-O-Glycosyltransferases Involved in the Formation of Flower Color in Rhododendron Delavayi. Front. Plant Sci. 2022, 13, 863482. [Google Scholar] [CrossRef]
- Okada, Y.; Sano, Y.; Kaneko, T.; Abe, I.; Noguchi, H.; Ito, K. Enzymatic reactions by five chalcone synthase homologs from Hop (Humulus lupulus L.). Biosci. Biotechnol. Biochem. 2004, 68, 1142–1145. [Google Scholar] [CrossRef]
- Ohno, S.; Hosokawa, M.; Kojima, M.; Kitamura, Y.; Hoshino, A.; Tatsuzawa, F.; Doi, M.; Yazawa, S. Simultaneous post-transcriptional gene silencing of two diferent chalcone synthase genes resulting in pure white flowers in the octoploid dahlia. Planta 2011, 234, 945–958. [Google Scholar] [CrossRef]
- Yang, J.; Gu, H.; Yang, Z. Likelihood analysis of the chalcone synthase genes suggests the role of positive selection in morning glories (Ipomoea) J. Mol. Evol. 2004, 58, 54–63. [Google Scholar] [CrossRef][Green Version]
- Koes, R.E.; Spelt, C.E.; Vandenelzen, P.J.M.; Mol, J.N.M. Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene 1989, 81, 245–257. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, Q.; Shen, W.; ElMohtar, C.A.; Zhao, X.; Gmitter, F.G. Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biol. 2018, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef]
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003, 20, 79–110. [Google Scholar] [CrossRef] [PubMed]
- Abe, I.; Morita, H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat. Prod. Rep. 2010, 27, 809–838. [Google Scholar] [CrossRef]
- Schroder, J. A family of plant-specific polyketide synthases: Facts and predictions. Trends Plant Sci. 1997, 2, 373–378. [Google Scholar] [CrossRef]
- Morita, H.; Wong, C.P.; Abe, I. How structural subtleties lead to molecular diversity for the type III polyketide synthases. J. Biol. Chem. 2019, 294, 15121–15136. [Google Scholar] [CrossRef]
- Park, H.L.; Yoo, Y.; Bhoo, S.H.; Lee, T.H.; Lee, S.W.; Cho, M.H. Two Chalcone Synthase Isozymes Participate Redundantly in UV-Induced Sakuranetin Synthesis in Rice. Int. J. Mol. Sci. 2020, 21, 3777. [Google Scholar] [CrossRef]
- Oakley, T.H.; Østman, B.; Wilson, A.C. Repression and loss of gene expression outpaces activation and gainin recently duplicated fly genes. Proc. Natl. Acad. Sci. USA 2006, 103, 11637–11641. [Google Scholar] [CrossRef]
- Gu, X. Statistical framework for phylogenomic analysis of gene family expression profiles. Genetics 2004, 167, 531–542. [Google Scholar] [CrossRef]
- Jiang, C.; Schommer, C.K.; Kim, S.Y.; Suh, D.Y. Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens. Phytochemistry 2006, 67, 2531–2540. [Google Scholar] [CrossRef]
- Abe, I.; Takahashi, Y.; Noguchi, H. Enzymatic formation of an unnatural C6-C5 aromatic polyketideby plant type III polyketide synthases. Org. Let. 2002, 4, 3623–3626. [Google Scholar] [CrossRef]
- Shirley, B.W.; Kubasek, W.L.; Storz, G.; Bruggemann, E.; Koornneef, M.; Ausubel, F.M.; Goodman, H.M. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995, 8, 659–671. [Google Scholar] [CrossRef]
- Dong, X.; Braun, E.L.; Grotewold, E. Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol. 2001, 127, 46–57. [Google Scholar] [CrossRef]
- Sun, W.; Shen, H.; Xu, H.; Tang, X.; Tang, M.; Ju, Z.; Yi, Y. Chalcone Isomerase a Key Enzyme for Anthocyanin Biosynthesis in Ophiorrhiza japonica. Front. Plant Sci. 2019, 10, 865. [Google Scholar] [CrossRef]
- Chen, X.; Liu, W.; Huang, X.; Fu, H.; Wang, Q.; Wang, Y. Arg-type dihydroflavonol 4-reductase genes from the fern Dryopteris erythrosora play important roles in the biosynthesis of anthocyanins. PLoS ONE 2020, 15, e0232090. [Google Scholar] [CrossRef]
- Schijlen, E.G.; RicdeVos, C.H.; vanTunen, A.J.; Bovy, A.G. Modification of flavonoid biosynthesis in crop plants. Phytochemistry 2004, 65, 2631–2648. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Irani, N.G.; Koo, A.J.; Bohorquez-Restrepo, A.; Howe, G.A.; Grotewold, E.A. chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J. 2013, 74, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lou, Q.; Ma, J. Cloning and functional characterization of dihydroflavonol 4-reductase gene involved in anthocyanidin biosynthesis of grape hyacinth. Int. J. Mol. Sci. 2019, 20, 4743. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Meng, X.; Liang, L.; Jiang, W.; Huang, Y.; He, J. Molecular and Biochemical Analysis of Chalcone Synthase from Freesia hybrid in Flavonoid Biosynthetic Pathway. PLoS ONE 2015, 10, e0119054. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusionproteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 4, 2019–2025. [Google Scholar] [CrossRef]
- Fanali, C.; Dugo, L.; D’Orazio, G.; Lirangi, M.; Dacha, M.; Dugo, P. Analysis of anthocyanins in commercial fruit juices by using nano-liquid chromatography-electrospray-mass spectrometry and high-performance liquid chromatography with UV-visdetector. J. Sep. Sci. 2011, 34, 150–159. [Google Scholar] [CrossRef]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef]
- Bloora, S.J.; Abrahams, S. The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 2002, 59, 343–346. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zhao, X.; Zhang, Y.; Chen, Y.; Zhang, X.; Yi, Y.; Ju, Z.; Sun, W. Chalcone-Synthase-Encoding RdCHS1 Is Involved in Flavonoid Biosynthesis in Rhododendron delavayi. Molecules 2024, 29, 1822. https://doi.org/10.3390/molecules29081822
Huang J, Zhao X, Zhang Y, Chen Y, Zhang X, Yi Y, Ju Z, Sun W. Chalcone-Synthase-Encoding RdCHS1 Is Involved in Flavonoid Biosynthesis in Rhododendron delavayi. Molecules. 2024; 29(8):1822. https://doi.org/10.3390/molecules29081822
Chicago/Turabian StyleHuang, Ju, Xin Zhao, Yan Zhang, Yao Chen, Ximin Zhang, Yin Yi, Zhigang Ju, and Wei Sun. 2024. "Chalcone-Synthase-Encoding RdCHS1 Is Involved in Flavonoid Biosynthesis in Rhododendron delavayi" Molecules 29, no. 8: 1822. https://doi.org/10.3390/molecules29081822
APA StyleHuang, J., Zhao, X., Zhang, Y., Chen, Y., Zhang, X., Yi, Y., Ju, Z., & Sun, W. (2024). Chalcone-Synthase-Encoding RdCHS1 Is Involved in Flavonoid Biosynthesis in Rhododendron delavayi. Molecules, 29(8), 1822. https://doi.org/10.3390/molecules29081822






