Abstract
PETase exhibits a high degradation activity for polyethylene terephthalate (PET) plastic under moderate temperatures. However, the effect of non-active site residues in the second shell of PETase on the catalytic performance remains unclear. Herein, we proposed a crystal structure- and sequence-based strategy to identify the key non-active site residue. D186 in the second shell of PETase was found to be capable of modulating the enzyme activity and stability. The most active PETaseD186N improved both the activity and thermostability with an increase in Tm by 8.89 °C. The PET degradation product concentrations were 1.86 and 3.69 times higher than those obtained with PETaseWT at 30 and 40 °C, respectively. The most stable PETaseD186V showed an increase in Tm of 12.91 °C over PETaseWT. Molecular dynamics (MD) simulations revealed that the D186 mutations could elevate the substrate binding free energy and change substrate binding mode, and/or rigidify the flexible Loop 10, and lock Loop 10 and Helix 6 by hydrogen bonding, leading to the enhanced activity and/or thermostability of PETase variants. This work unraveled the contribution of the key second-shell residue in PETase in influencing the enzyme activity and stability, which would benefit in the rational design of efficient and thermostable PETase.
1. Introduction
Plastics have become an indispensable part of daily life due to their desirable properties, such as lightweight, durability, and low price [1,2,3,4]. However, most of the plastic waste accumulates in the natural environment, exposing serious consequences for the environment and human health [5,6,7]. Polyethylene terephthalate (PET) is a semicrystalline thermoplastic composed of terephthalic acid (TPA) and ethylene glycol (EG), and is widely used in food/beverage packaging and textiles industry [8,9]. In PET, TPA and EG are linked by an ester bond that is highly resistant to biodegradation, leading to the accumulation of large amounts of PET waste in the environment [5,10]. Therefore, it is crucial to explore efficient strategies for the treatment and recycling of PET plastics. Biodegradation methods have recently received significant attention due to their low energy consumption and environmental sustainability [11].
To date, a variety of enzymes that can decompose PET have been identified, including lipases, esterases, and cutinases [12,13]. However, these enzymes generally show poor degradation activity on PET plastics with high crystallinity, and often require high temperatures to achieve the degradation of PET plastics [14,15]. Yoshida et al. isolated a bacterium (Ideonella sakaiensis 201-F6) from PET waste that could use PET as a carbon and energy source at moderate temperatures [16]. This bacterium can secrete a novel enzyme, PET hydrolase (PETase), which can not only degrade PET efficiently at moderate temperatures, but also shows higher activity and specificity for PET with high crystallinity than other hydrolases [4,17]. However, the degradation activity of PETase for highly crystallized PET is still low and rapidly loses enzyme activity at 40 °C, which limits the application in degrading PET waste to resolve the existing environmental crisis [17,18,19].
To overcome these limitations, various protein engineering strategies have been implemented to improve the thermostability of PETase (Table S1). For example, Son et al. applied a rational design strategy to obtain ThermoPETase, which exhibited an 8.8 °C higher Tm value and a 14-fold improvement in PET degradation activity over PETaseWT [17]. The GRAPE strategy was developed by Wu et al. and designed the DuraPETase. The Tm value and the degradation efficiency of DuraPETase against PET films were improved by 31 °C and over 300-fold, respectively [20]. The FAST-PETase, which was designed by a machine learning algorithm, exhibited an increase in Tm of 18.4 °C as compared to PETaseWT and was able to completely degrade 51 different post-consumer PET wastes within 7 days [21]. Moreover, an automated, high-throughput directed evolution platform and a fluorescence-based high-throughput screening assay were designed for the directed evolution of PETase [15,22]. The engineered HotPETase and DepoPETase have both enhanced the robustness of the enzyme, and HotPETase could operate at the glass transition temperature of PET [15,23].
It is interesting to note that most of the mutations that enhanced the thermostability of PETase are the second-shell or distal residues (Table S1 and Figure S1). Actually, the non-active site residues can be classified into different “shells” depending on their locations in the enzyme. The second-shell residues are defined as the residues that interact with the first-shell residues [24,25], while the residues in direct contact with the substrate, cofactor, or product are the first-shell residues [24]. The residues that are located in the second shell or further and are not within van der Waals distance of any part of the substrate, cofactor, or product are distal residues [26,27]. Many works have demonstrated that non-active site residues play an important role in the regulation of enzyme functions such as activity, stability, and selectivity [25,27,28,29,30]. However, there is no research on the molecular mechanism of how the second-shell or distal residues manipulate the catalytic performance of PETase.
Therefore, the present study focused on identifying the key non-active site residue in the second shell of PETase by using a crystal structure- and sequence- (multiple sequence alignment, phylogenetic and conservative analysis) based strategy. The key non-active site residue was then mutated to the other 19 canonical amino acids to investigate how mutations of this residue manipulate the activity and stability of PETase. The structural changes of the variants were characterized by circular dichroism (CD) and fluorescence spectroscopy. Subsequently, MD simulations were performed to explore the molecular mechanisms underlying the effects of these mutations on enzymatic properties.
2. Results and Discussion
2.1. Identification of Key Non-Active Site Residue
The B-factor and root mean square fluctuation (RMSF) values of the β6-β7 strand connecting loop (D186-F191) were found to be significantly higher than the overall structure of PETase (Figure 1a) [17,31]. This connecting loop was one of the most flexible regions in PETase [17], a key region influencing the stability of PETase. In our previous study, we found that the D186 residue in this flexible loop could interact with the I168R and S188E mutations to form an R168-D186-E188 salt bridge network, which significantly improved the thermostability of PETase [32]. It was also reported that the mutation of D186H combined with other residues (E121 and N172) could form a water-mediated hydrogen bond [17,21], or the D186 residue interact with other mutations (I168R and S188Q) to form a hydrogen bond or a salt bridge to improve the stability of PETase [20,31]. The above analysis indicates that D186 is a key residue in the connecting loop influencing PETase stability. To further investigate the role of D186 in PETase, the amino acid sequences of PETase and 71 PETase-like enzymes from phylogenetically distinct organisms were aligned using ClustalX [33], and the phylogenetic tree of PETase-like enzymes was constructed using MEGA 11 software (Figure 1b) as reported previously [4,34]. As shown in Figure 1b, the PETase-like enzymes could be divided into types Ⅰ and Ⅱ, and the type Ⅱ enzymes could be further subdivided into two types (type Ⅱa and type Ⅱb). The type Ⅰ enzymes contained 58 PETase-like enzymes and type Ⅱ contained 14 enzymes, of which PETase belonged to type Ⅱb. Meanwhile, most of the type I and Ⅱa enzymes were reported to exhibit higher thermostability and lower PET degradation activity compared with PETase, such as the cutinase from Thermobifida fusca (TfCut1and TfCut2) [35,36], Saccharomonospora viridis (Cut190) [37], and the lipase from a metagenomic library (PET2), etc. [12,38,39], whereas the type Ⅱb enzyme variant from Burkholderiales bacterium (BurPLDM) showed higher thermostability and PET degradation activity than that of PETase [40]. Thus, the conservation of the D186 residue in different types were then analyzed by the WebLogo sever (Figure 1c) [41]. This site could be amino acid D, S, N, or H in the type Ⅱb enzymes, while it could be amino acid D, Y, or N in type Ⅱa enzymes. In the type Ⅰ enzymes, the amino acid N had the most frequency, followed by H and D. The highly conserved positions determine the general commonality of all homologous enzymes, while the sequence variability and non-strictly conserved position may lead to the functional diversity, substrate, and reaction specificity of enzymes [42,43,44,45]. Based on the above analysis, it is considered that the variability of residue 186 in the different types of PETase-like enzymes was important for changing the enzymatic performance of PETase. To understand how possible mutations at this key non-active site residue in the second shell are likely to manipulate the activity and thermostability of PETase, the D186 residue was mutated to the other 19 canonical amino acids.
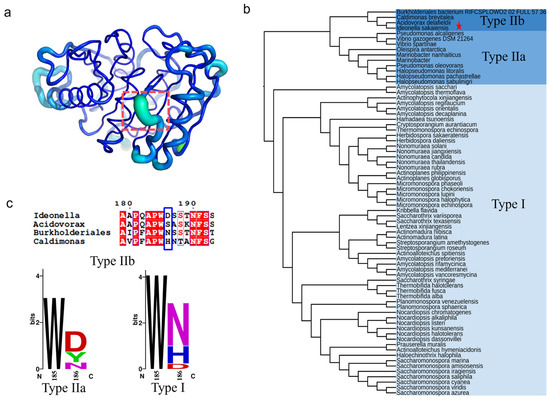
Figure 1.
The crystal structure- and sequence-based strategy to identify the key non-active site residue in the second shell of PETase. The structure and B-factors of PETaseWT (a), where the residues D186-F191 were marked by orange dotted box, the highest B-factor region was colored in lime green and the lowest B-factor region was colored in blue. The phylogenetic tree of PETase-like enzymes (b), where the PETase was marked with a pentagram in the type IIb enzymes. The conservation of the D186 residue in different types of PETase-like enzymes (c).
2.2. Catalytic Activity of D186 Variants
In order to investigate the effect of the mutations on the activity of PETase, the hydrolytic activities were tested using BHET as a substrate at 30 and 40 °C because PETase and its variants can hydrolyze BHET to MHET and EG [4,46]. As shown in Figure 2a,b, in all the nonpolar variants, PETaseD186A, PETaseD186V, PETaseD186I, PETaseD186L, PETaseD186M, PETaseD186F, PETaseD186W, PETaseD186P, and PETaseD186G, only PETaseD186F showed enhanced hydrolytic activity as compared to that of PETaseWT, and the others showed reduced activities. In the polar variants, PETaseD186C, PETaseD186Y, PETaseD186S, PETaseD186T, PETaseD186N, and PETaseD186Q, only PETaseD186N showed a 1.03-fold higher activity than PETaseWT at both 30 and 40 °C. In contrast, the activity of PETaseD186Q was significantly reduced to only 21% and 13% of that of PETaseWT at 30 and 40 °C, respectively. When the D186 residue was mutated to charged amino acids (H, K, R and E), PETaseD186H exhibited an increase in activity of 26% and 30% at 30 and 40 °C, respectively, as compared with PETaseWT, while reduced activities were observed for PETaseD186K, PETaseD186R, and PETaseD186E.
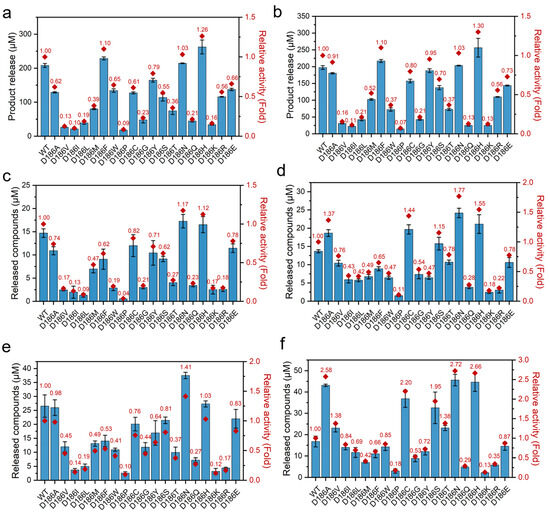
Figure 2.
The activities of PETaseWT and its variants against different substrates: hydrolytic activities of PETaseWT and its variants at 30 °C (a) and 40 °C (b) using BHET as a substrate, where the product release was the concentration of MHET; PET film degradation activities of PETaseWT and its variants at 30 °C and 40 °C for 24 h (c,d) and 72 h (e,f), where the released compounds were the sum of MHET and TPA.
To further evaluate the effect of the mutations on PET degradation, the degradation activity against PET films (crystallinity of 28.34 ± 2.54%, Figure S2) was measured at 30 and 40 °C (Figure 2c–f). As compared with PETaseWT, enhanced degradation activities against PET films were observed at least once at the two temperatures by PETaseD186A, PETaseD186V, PETaseD186C, PETaseD186S, PETaseD186T, PETaseD186N, and PETaseD186H, while the other variants always showed reduced activities. Among these seven variants, the highest activity was obtained by PETaseD186N as compared with PETaseWT at the two temperatures. PETaseD186N exhibited a 1.17- and 1.41-fold increase in activity at 30 °C for 24 h and 72 h, respectively, as compared to the wild type (Figure 2c,e). This variant also showed 1.77- and 2.72-fold higher degradation activity at 40 °C for 24 h and 72 h (Figure 2d,f), respectively. This suggested that the D186N mutant could not only improve the thermostability of PETase, but also increase the PET degradation ability of PETase. However, higher activity was obtained by PETaseD186H for BHET. This difference further demonstrated that the hydrolysis of small molecule BHET by PETase can be different from its degradation activity on rigid PET plastics [4,47,48]. After incubation at 40 °C for 72 h, PETaseD186A, PETaseD186V, PETaseD186C, PETaseD186S, and PETaseD186T also showed 2.58-, 1.38-, 2.20-, 1.95-, and 1.38-fold increased activities (Figure 2f), respectively, as compared to the wild type. The above results suggest that D186 in the second shell plays a critical role in regulating the activity of PETase.
2.3. Thermostability of D186 Variants
In order to evaluate how the mutations influence the stability of PETase, the Tm values of the enzymes were measured by differential scanning fluorimetry (DSF) (Figure 3a). An increase in Tm values was observed with nonpolar variants except for PETaseD186W and PETaseD186P. PETaseD186V had the highest Tm value, which showed a Tm value of 59.49 °C, 12.91 °C higher than that of PETaseWT. The Tm values of most polar variants also showed increases as compared with PETaseWT. In particular, PETaseD186C, PETaseD186S, PETaseD186T, and PETaseD186N showed increases in Tm of 11.83, 9.61, 9.01, and 8.89 °C, respectively. Among the charged amino acid mutations, only PETaseD186H had a markedly improved Tm value of 8.76 °C. These results indicated that the D186 residue could be substituted by some other amino acids to improve the thermostability of PETase, and it could manipulate the thermostability of PETase to a wide extent. Moreover, the PETaseD186N and PETaseD186H variants showed similarly higher Tm values than the wild type (Figure 3a), suggesting that the differences in substrate degradation between the two variants (Figure 2) were mainly attributed to the differences in activity.
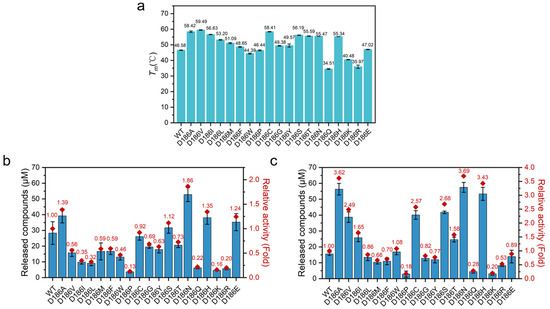
Figure 3.
The Tm and PET film degradation activities of PETaseWT and its variants: Tm values of PETaseWT and its variants (a), PET film degradation activities of PETaseWT and its variants at 30 °C (b) and 40 °C (c) in 6 days, where the released compounds were the sum of MHET and TPA.
To assess the durability of PETase and its variants, the long-term degradation performance of the enzymes was then conducted at 30 and 40 °C for 6 days using PET film as a substrate. As shown in Figure 3b,c, most of the variants with improved stability (increased Tm values) exhibited higher product concentrations at 40 °C than at 30 °C, whereas the variants with poor stability (decreased Tm values) showed lower product concentrations at 40 °C than at 30 °C. This result further indicated that the thermostability of PETase was important for its efficient degradation of PET.
A detailed analysis of the long-term degradation experiments revealed that the degradation activity of PETaseWT could be maintained for 5 days at 30 °C, whereas the degradation activity was completely lost within 1 day at 40 °C (Figure S3). For most of the variants with improved thermostability, the PET degradation product concentration gradually increased at 30 °C for 6 days. Consequently, the PET degradation product concentrations of the PETaseD186N and PETaseD186H were 1.86- and 1.35-fold higher than that of PETaseWT after 6 days, respectively (Figure 3b). At 40 °C, the product concentrations of the PETaseD186N and PETaseD186H rapidly increased in the first 3 days (Figure S3), and the product concentrations were 3.69- and 3.43-fold higher than that of PETaseWT after 6 days, respectively (Figure 3c). For PETaseD186V, the PET product concentration rapidly increased at least in the first 5 days at 40 °C (Figure S3), which was mainly due to its higher stability (Figure 3a), and the product concentration was 2.49-fold higher than that of PETaseWT after 6 days (Figure 3c). In addition, it was worth noting that the product concentration of PETaseD186A was 1.05- and 1.45-fold higher than that of PETaseD186H and PETaseD186V, respectively, at 40 °C for 6 days. This phenomenon was attributed to the higher thermostability of PETaseD186A (Tm = 58.42 °C) than PETaseD186H (Figure 3a) and the significantly higher activity of PETaseD186A than PETaseD186V (Figure 2). Therefore, it could be concluded that the higher product concentrations of PETaseD186N and PETaseD186H than the wild type at both 30 and 40 °C, especially for PETaseD186N, were mainly attributed to the superimposed effects of higher activity and thermostability. Meanwhile, the higher product concentrations of PETaseD186V and PETaseD186A than the wild type were due to their improved thermostability.
Based on the results of the catalytic activity and thermostability of the D186 variants, the most active PETaseD186N, the most stable PETaseD186V, the stable and active PETaseD186H and PETaseD186A, and the less stable and less active PETaseD186Q were selected for the following molecular simulation studies to unravel the molecular mechanisms.
2.4. Molecular Mechanism of Enhanced Catalytic Activity by D186 Mutations
To explore the molecular insight into the effect of D186 mutations on the catalytic activity of PETase, the substrate model 2PET was docked into PETaseWT and its variants [46,49], and then MD simulations were performed on enzyme-2PET complexes. From the binding free energy between the substrate and enzymes calculated using molecular mechanics Poisson−Boltzmann surface area (MM-PBSA) method (Figure 4a) [50], it was found that the total binding free energy of PETaseD186N (−93.23 ± 1.03 kJ·mol−1) and PETaseD186H (−90.98 ± 3.35 kJ·mol−1) were higher than that of PETaseWT (−88.23 ± 3.90 kJ·mol−1), whereas the total binding free energy of PETaseD186A (−84.83 ± 2.43 kJ·mol−1), PETaseD186V (−65.27 ± 5.41 kJ·mol−1) and PETaseD186Q (−62.19 ± 6.14 kJ·mol−1) were lower than that of PETaseWT. This indicated that the D186N and D186H mutations promoted the substrate binding to the enzyme and enhanced the catalytic activity, while the D186A, D186V, and D186Q mutations were not beneficial for the substrate binding to the enzyme. Then, the residues involved in the substrate binding were identified by analyzing the contribution of each residue to the binding free energy. According to Figure 4b and Figure S4a, the residues Y87, W159, S160, M161, W185, and I208 had higher binding free energies as compared to other residues in the enzymes, indicating that these six residues were crucial for the binding of the substrate to the enzymes. This result was in good agreement with previous findings that these residues are involved in substrate binding and catalysis, and that substitution of these residues by Ala greatly reduces the catalytic activity of PETase [4,46,47,51]. Among the six residues, the binding free energies of Y87, W159, and I208 in PETaseD186H and PETaseD186N were remarkably higher than those in PETaseWT, which could contribute to the substrate binding and increase the catalytic activity of these two variants. In contrast, the binding free energies of Y87, W159, M161, and I208 in PETaseD186Q, PETaseD186A and PETaseD186V were lower than those in PETaseWT, which were unfavorable for the substrate binding to the variants.
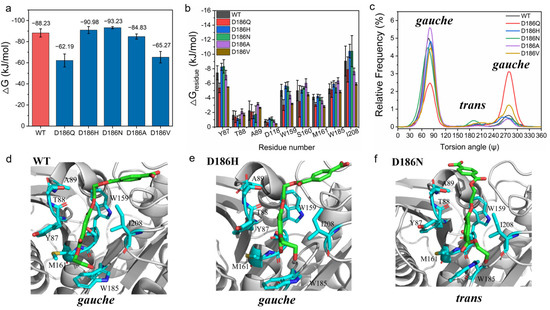
Figure 4.
MD simulations of enzyme-2PET complexes: total binding free energies of PETase and its variants with the 2PET substrate (a); important residues for the 2PET-binding to PETaseWT and its variants (b); the OC-CO torsion angle Ψ in the EG units of 2PET (c); binding modes of the substrate 2PET into the active sites of PETaseWT (d), PETaseD186H (e), and PETaseD186N (f). The results were generated from six independent MD simulation runs.
The relative frequency distribution of the catalytic distance between the hydroxyl oxygen of the catalytic serine (Ser 160) and the carbonyl carbon atom of 2PET was also calculated. As shown in Figure S4b, the catalytic distances in PETaseD186A (3.80 Å), PETaseD186V (3.80 Å), PETaseD186Q (3.80 Å), PETaseD186N (3.75 Å), and PETaseD186H (3.70 Å) were similar to that in PETaseWT (3.70 Å). So, catalytic distance change was not the cause for altering the catalytic activity of the variants. Moreover, the OC-CO torsion angle Ψ in the EG units of the substrate analysis showed that the PET chain could bind to PETaseD186N in the trans conformation with a higher probability, while the PET chain could only bind to other enzymes in the gauche conformation (Figure 4c–f and Figure S4c–f). Recently, it was reported that the ratio of gauche and trans conformations of PET chains was 9:1 in low crystallinity PET films [52], and that a single mutation (S238A) in the loop connecting β8 and α6 could alter the preference of PETase for the conformation of PET chains to increase the degradation activity against PET films [53]. These findings could further explain the higher activity of PETaseD186N as compared to PETaseD186H and other variants.
2.5. Molecular Mechanism of Enhanced Thermostability by D186 Mutations
To explore the molecular insight into the effect of D186 mutations on the thermostability of PETase, the structural changes of PETaseWT and its variants were analyzed. The changes in the secondary structure and tertiary structure of the enzyme variants were examined by CD and fluorescence spectroscopy (Figure S5), respectively. The secondary structure of the variants were slightly different from that of PETaseWT (Figure S5b), but the tertiary structure of the variants were significantly changed from that of PETaseWT (Figure S5c). The emission maxima (λmax) of PETaseD186N, PETaseD186H, PETaseD186A, and PETaseD186V were blue shifted, in which PETaseD186V had the smallest λmax. However, the λmax of PETaseD186Q was red shifted. This result indicated that the overall structures of PETaseD186N, PETaseD186H, PETaseD186A, and PETaseD186V became more compact and the solvent exposure of some tryptophan and tyrosine residues was reduced, which could lead to an improvement in structural stability, whereas the overall structure of PETaseD186Q became looser, resulting in increased solvent exposure of some tryptophan and tyrosine residues, which could reduce its structural stability [46,54].
Subsequently, MD simulations at different temperatures (303, 313, and 403 K) were performed to analyze the overall/local structural stability and flexibility of the enzyme variants. As shown in Figures S6 and S7, the root mean square deviation (RMSD) and radius of gyration (Rg) values of PETaseD186Q were obviously higher than those of PETaseWT at 313 and 403 K during the simulations, indicating its looser structure than the wild type, which was consistent with the results of fluorescence spectroscopy. For PETaseD186H, PETaseD186N, PETaseD186A, and PETaseD186V, the RMSD values were comparable to or lower than that of PETaseWT. Meanwhile, the Rg values of PETaseD186N, PETaseD186A, and PETaseD186V were noticeably lower than that of PETaseWT at 403 K during the last 20 ns (Figure S7c). This suggested that the overall structural rigidity of PETaseD186H, PETaseD186N, PETaseD186A, and PETaseD186V were stronger than that of PETaseWT, which was also consistent with the results of fluorescence spectroscopy (Figure S5c).
According to the average root mean square fluctuation (RMSF) analysis of the Cα atoms of the enzyme variants (Figure 5 and Figure S8), two main regions (residues P181-P197 and residues N205-R222) of fluctuation changed in variants as compared with PETaseWT. The average RMSF values of PETaseWT and its variants in these two regions were remarkably different at 313 and 403 K (Figure 3 and Figure S8b). PETaseD186H, PETaseD186N, PETaseD186A, and PETaseD186V kept lower RMSF values than PETaseWT in these two regions, demonstrating that their structures were more stable than that of PETaseWT. For PETaseD186Q, however, the average RMSF values in these two regions were higher than those of PETaseWT, which should be responsible for its poor thermostability. As shown in Figure 4, residue W185 of region P181-P197 and residue I208 of region N205-R222 were crucial for the binding of the substrate to the enzyme (Figure 4), and the auxiliary catalytic residue D206 located on region N205-R222 [22]. Therefore, reducing the RMSF and increasing the stability of these two regions allow the enzyme to maintain its catalytic activity at high temperatures. The analysis of mobility and structural fluctuation of PETaseWT and its variants by MDLovofit also confirmed the results mentioned above (Figure S9).
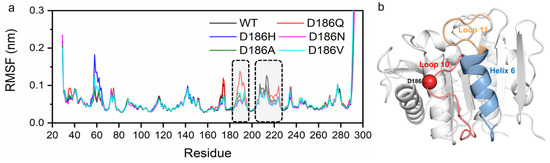
Figure 5.
The RMSF of Cα atoms of PETaseWT and its variants in MD simulations at 313 K, where the regions with significant differences in average RMSF are marked in dotted boxes (a). Corresponding locations of the regions with notable changes of the RMSF in the structure of PETaseWT (b). Loop 10 is colored in red, Helix 6 is colored in blue, Loop 11 is colored in yellow, and the Cα atom of D186 is shown as a red sphere. The RMSF was calculated from six independent MD simulation runs.
The hydrogen bonding was then analyzed as hydrogen bonds play an important role in protein folding and thermostability [55]. It was reported that H186 could promote the water-mediated hydrogen bond formation between E121 and N172 in PETaseS121E/D186H/R280A [17], or H186 could form a water-mediated hydrogen-bonding network with E121 and N172 in PETaseS121E/D186H/R280A/R224Q/N233K [21]. The residues P181-P197, N205-S213, and S214-R222 correspond to Loop 10, Loop 11, and Helix 6 in PETase, respectively (Figure 5b). So, the interactions between residue 186 and surrounding residues were analyzed at first. For PETaseWT, two hydrogen bonds were formed between D186 and S187 (D186OD1…S187N and D186OD2…S187N) with the length of 3.2 Å and 3.3 Å at 313 K, respectively (Figure 6a). However, the distance/angle between the N atom of S187 and the OD1/OD2 atoms of D186 clearly increased/reduced in the last 20 ns at 403 K (Figures S10e and S11e), showing that the two hydrogen bonds were unstable in PETaseWT [56]. The D186Q mutation could only form a 3.5 Å hydrogen bond with S187 at 313 K and this unstable hydrogen bond rapidly cleaved at 403 K (Figure 6b, Figures S12e and S13e), indicating that this mutation enhanced the flexibility of Loop 10 and decreased the stability of PETase. In contrast, the D186H and D186N mutations introduced three stable hydrogen bonds in Loop 10, which could result in higher Tm values of PETaseD186H and PETaseD186N than PETaseWT as revealed in the experiment (Figure 3a). The three stable hydrogen bonds in PETaseD186H included a hydrogen bond (H186ND1…S187N, 3.0 Å), a carbon–hydrogen bond (H186CD2…S188O, 3.3 Å), and a π–donor hydrogen bond (H186…S188N, 3.8 Å) (Figure 6c, Figures S14 and S15). For PETaseD186N, the three stable hydrogen bonds included N186OD1…S187N (2.9 Å), N186ND2…S188O (3.2 Å) and N186ND2…S188N (3.4 Å) (Figure 6d, Figures S16 and S17). Therefore, the Tm value of PETaseD186H was similar to that of PETaseD186N (Figure 3a). In addition, the occupancy rates of the conventional hydrogen bonds between residue 186 and surrounding residues were also analyzed [57]. According to Tables S3–S5, the occupancy rates of the D186OD1…S187N and D186OD2…S187N in PETaseWT were similar to those of Q186OE1…S187N and Q186NE2…S188O in PETaseD186Q, whereas the occupancy rates of H186ND1…S187N in PETaseD186H and N186ND2…S188O and N186ND2…S188N in PETaseD186N were always higher than those of PETaseWT and PETaseD186Q. Especially at 403 K, the occupancy rates of the conventional hydrogen bonds in PETaseWT and PETaseD186Q were significantly lower than those of PETaseD186H and PETaseD186N. These results further indicated that these hydrogen bonds were more stable in PETaseD186H and PETaseD186N than in PETaseWT and PETaseD186Q.
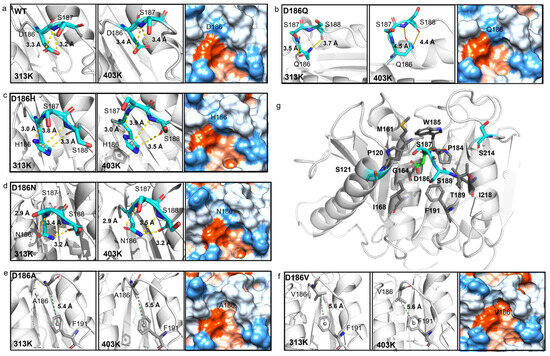
Figure 6.
Molecular interactions between residue 186 and the surrounding residues in PETaseWT and its variants. PETaseWT (a), PETaseD186Q (b), PETaseD186H (c), PETaseD186N (d), PETaseD186A (e), PETaseD186V (f), and collision of polarity between D186 and the surrounding residues (5 Å) (g). The residue 186 and surrounding residues are shown as sticks and colored according to the elements (C: cyan/grey, N: blue, O: red, S: yellow). The protein surfaces are colored according to hydrophilicity, where the most hydrophilic are colored blue and the most hydrophobic are colored orange. Each typical structure was obtained by RMSD-based clustering analysis (cutoff = 0.075 nm) of six independent MD simulation runs.
In the structure of PETaseWT, there are nine hydrophobic (P120, M161, G164, I168, P184, W185, T189, F191, and I218) and four neutral (S121, S187, S188, and S214) residues within 5Å of the residue D186, which may cause D186 to conflict with the surrounding residues (Figure 6a,g). This phenomenon has been described by Son et al. as a “collision of polarity” [17]. By substituting the negatively charged residue D to the hydrophobic residue A/V, stable π−alkyl hydrophobic interactions formed between A186/V186 and F191 (Figure 6e,f, Figures S18 and S19), and the hydrophobic interactions between Loop 10 and its surrounding hydrophobic residues enhanced (Figure 6e,f). Furthermore, the D186A and D186V mutations changed the region from hydrophilic to hydrophobic, leading to the release of this collision of polarity (Figure 6a,e,f) and the reduction in the flexibility of Loop 10 in PETaseD186A and PETaseD186V (Figure 5a, Figures S8 and S9). Among the 20 amino acids, Val is considerably more hydrophobic than Ala, so the hydrophobic interactions were stronger in PETaseD186V than those in PETaseD186A, which could lead to a higher Tm of PETaseD186V than that of PETaseD186A (Figure 3a).
Next, the hydrogen bonds between Loop 10 and the two neighboring helixes, Helix 5 (residues S160-N173) and Helix 6, were analyzed in detail. Between Loop 10 and Helix 5 there were only two hydrogen bonds, W185O…S160O and Q182O…S160N. The occupancy rates of the two hydrogen bonds were comparable as in PETaseWT and its variants, while only the occupancy rate of W185O…S160O (76.4 ± 3.1%) in PETaseD186Q was obviously lower than PETaseWT (87.3 ± 2.1%) at 403 K (Table S6).
According to the typical structures obtained by RMSD-based clustering analysis (cutoff = 0.075 nm) (Figure 7a), the hydrogen bonds P184O…S214OG, F191N…S221OG, and S192OG…S221O formed between Loop 10 and Helix 6 in PETaseD186H, PETaseD186N, PETaseD186A, and PETaseD186V. For PETaseWT and PETaseD186Q, only a hydrogen bond F191N…S221OG was observed in Loop 10 interacting with Helix 6. The occupancy rates of the hydrogen bonds F191N…S221OG and S192OG…S221O in PETaseWT and its variants were similar at temperatures below 403 K (Figure 7b and Table S7). However, the occupancy rates of P184O…S214OG in PETaseWT and PETaseD186Q were always lower than those of the other four variants (Figure 7b,c and Table S7). At 313 K, the occupancy rates of the P184O…S214OG in PETaseD186H (22.2 ± 3.8%) and PETaseD186N (21.9 ± 4.7%) were similar, which were lower than that of PETaseD186A (30.3 ± 5.4%) and PETaseD186V (38.0 ± 4.2%) (Figure 7b). This indicated that the more robust the hydrogen bond P184O…S214OG was, the higher the thermostability of the variants would be (Figure 3a). Furthermore, the occupancy rates of the three hydrogen bonds in PETaseWT and PETaseD186Q were obviously lower than those of the other four variants, and PETaseD186Q had the lowest occupancy rates of the three hydrogen bonds at 403 K (Figure 7c). These results showed that the hydrogen bonds P184O…S214OG, F191N…S221OG, and S192OG…S221O in PETaseD186H, PETaseD186N, PETaseD186A, and PETaseD186V were more robust than those in PETaseWT, whereas these hydrogen bonds in PETaseD186Q were more unstable than those in PETaseWT. Therefore, locking the flexible Loop 10 and Helix 6 with more robust hydrogen bonding increased the interactions between these two regions and reduced the flexibility of the amino acid backbone, which led to the improved thermostability of the four PETase variants. In addition, the auxiliary catalytic residue D206 is located on the flexible Loop 11, and Helix 6 connects to one end of Loop 11; thus, the stabilizing effect of Helix 6 was transferred to Loop 11, leading to the flexibility decrease in the loop (Figure 5 and Figure S8b). The flexibility decrease (or rigidity increase) of Loop 11 could stabilize the auxiliary catalytic residue D206, and further enhance the thermostability of the variants [22].
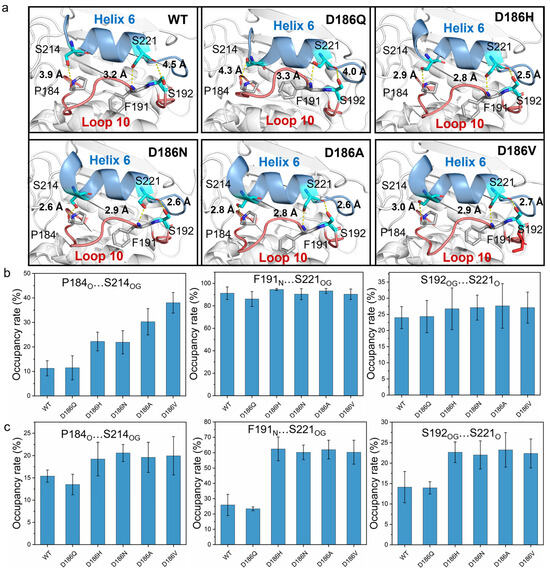
Figure 7.
Molecular interactions between Loop 10 and Helix 6 in PETaseWT and its variants. The hydrogen bonds between Loop 10 and Helix 6 in PETaseWT and its variants determined by MD simulations (a). The occupancy rate of the hydrogen bonding analysis at 313 K (b) and 403 K (c), where only hydrogen bonds with occupancy rates > 10% were considered. Each typical structure was obtained by RMSD-based clustering analysis (cutoff = 0.075 nm) and the average occupancy rates were calculated from six independent MD simulation runs.
3. Materials and Methods
3.1. General Information
Bis-2(hydroxyethyl) terephthalate (BHET) and terephthalic acid (TPA) of analytical grade were purchased from Aladdin (Shanghai, China). Mono(2-hydroxyethyl) terephthalate (MHET) of analytical grade was purchased from MOLBASE (Shanghai, China). PET films (transparent, 0.25 mm thickness) were obtained from Goodfellow (Cambridge, England). All other chemicals and reagents of at least reagent grade were purchased from commercial sources and used without further purification unless otherwise stated. TransStart® FastPfu DNA polymerase, DMT enzyme, pEASY®-Basic Seamless Cloning and Assembly Kit were purchased from TransGen Biotech (Beijing, China). Escherichia coli (E. coli) BL21 (DE3) and expression vector pET-22b (+) were obtained from Solarbio (Beijing, China). The primer synthesis and DNA sequencing analysis were conducted by GENEWIZ and Tsingke (Tianjin, China).
3.2. Site-Directed Mutagenesis, Protein Expression, and Purification
The gene encoding PETase from Ideonella sakaiensis 201-F6 (GenBank accession number: GAP38373.1) was commercially synthesized by GENEWIZ (Beijing, China), and then subcloned into pET-22b (+) vector between Nde I and Xho I restriction sites. The final construct vector pET-22b: PETase was transformed into E. coli BL21 (DE3).
The site-directed mutagenesis was constructed by PCR using a recombinant plasmid pET22b (+) containing the PETase gene as a template. The PCR procedure was as follows: 95 °C for 2 min, (95 °C for 20 s, 56 °C for 20 s, 72 °C for 3.5 min) with 35 cycles and extension at 72 °C for 5 min. The PCR reaction mixture contained 32 μL distilled water, 10 μL 5× buffer, 4 μL 2.5 mM dNTPs, 1 μL each forward and reverse primer (10 μM), 1 μL recombinant plasmid, and 1 μL TransStart®FastPfu DNA polymerase (TransGen Biotech, Beijing, China). The PCR products were digested with 1 μL DMT enzyme at 37 °C for 1 h and then separately transformed into E. coli BL21 (DE3) cells for protein expression. Residue D186 was substituted by the other 19 canonical amino acids by this method (Table S2), and the sequences were verified by DNA sequencing.
A single colony of wild type or variants was picked and incubated in a 5 mL LB medium containing 100 μg·mL−1 ampicillin at 37 °C and 220 rpm for 12 h. Then, 2 mL of the preculture cells were inoculated into 200 mL of LB medium containing 100 μg mL−1 ampicillin at 37 °C and 220 rpm. When the optical density at 600 nm was between 0.8 and 1.0, 0.5 mM isopropyl-β-D-1-thiogalactoside (IPTG) was added to the LB medium. The cultures were then incubated for another 24 h at 16 °C and 160 rpm for protein overexpression. The cells were harvested by centrifugation (5000× g, 30 min) at 4 °C, and the cell pellet was resuspended in lysis buffer (50 mM Na2HPO4-HCl, 100 mM NaCl, 20 mM imidazole, pH 7.0).
The resuspended cells were disrupted by ultrasonication in an ice-water bath for 30 min. Insoluble cell debris was removed by centrifugation (12,000× g, 30 min, 4 °C) and filtrated through a 0.45 μm syringe filter. The supernatant was loaded onto a 5 mL Ni-NTA FF column (GE Healthcare, Solingen, Germany), and the affinity column was pre-equilibrated with lysis buffer. Non-specific adsorbed proteins were removed by washing with lysis buffer, and the target enzyme was eluted with elution buffer (50 mM Na2HPO4-HCl, 100 mM NaCl, 300 mM imidazole, pH 7.0). The protein fractions were then transferred into desalting buffer (50 mM Na2HPO4-HCl, 100 mM NaCl, pH 7.0) by size-exclusion chromatography on an AKTA Basic system with a Superdex200 Increase 10_300 GL column (GE Healthcare, Germany). All purification steps were conducted at 4 °C. The purified enzyme was checked by SDS-PAGE, and the concentration of the enzyme was quantified by BCA Protein Assay Kit (Solarbio, Beijing, China).
3.3. Enzyme Activity Assay for BHET and PET Film Degradation
The activities of the wild-type PETase and its variants were first determined at 30 and 40 °C by monitoring the hydrolysis of BHET to MHET and EG using a high-performance liquid chromatography (HPLC) system (1100 Series HPLC, Agilent, Santa Clara, CA, USA), as reported previously [46,58]. The reaction mixture contained 910 μL reaction buffer (80 mM Na2HPO4−HCl, 40 mM NaCl, pH 7.0), 80 μL BHET substrate (2.5 g·L−1), and 10 μL enzyme solution (5 μM). The enzyme reaction was terminated by adding an equal volume of 160 mM phosphoric acid solution (20% (v/v) DMSO, pH 2.5) and heating at 85 °C for 10 min. Then, the reaction mixture was centrifuged at 13,000× g for 10 min, and the supernatant was filtered through a 0.22 μm membrane for HPLC analysis. Enzyme activity was defined as the concentration (μM) of MHET released by enzyme (50 nM) catalyzed degradation of the BHET substrate (200 mg·L−1) for 30 min, as previously reported [46,58].
The PET film was used as a substrate to determine the degradation activity of PET by the wild-type PETase and its variants. The PET film has a crystallinity of 28.34 ± 2.54% (Figure S1) and was cut into a circular form with a diameter of 6 mm. The PET film was soaked with 12.5 μL of enzyme (5 μM) in 300 μL of glycine-NaOH buffer (50 mM, pH 9.0) at 30 and 40 °C for 1 to 6 days. After removing the PET film from the reaction mixture, the reaction was terminated by adding an equal volume of 160 mM phosphoric acid solution (20% (v/v) DMSO, pH 2.5) and heating at 85 °C for 10 min. Then, the reaction mixture was centrifuged at 13,000× g for 10 min. The supernatant was filtered through a 0.22 μm membrane and analyzed by HPLC.
3.4. Assay of Enzyme Thermostability
To analyze the thermostability of the wild-type PETase and its variants, the melting temperature (Tm) was determined by differential scanning fluorimetry (DSF). The concentration of purified protein was diluted to 5 μM with desalting buffer (50 mM Na2HPO4-HCl, 100 mM NaCl, pH 7.0). The SYPRO Orange dye 5000× (Sigma-Aldrich, Shanghai, China) was diluted to 100× with pure water. The diluted protein solution (20 μL) was mixed with 5 μL of diluted SYPRO Orange dye and loaded onto a 96-well PCR plate (Roche, Shanghai, China). DSF experiments were performed using a Light Cyder480 real-time PCR system (Roche, Santa Clara, CA, USA). The 96-well PCR plate was heated from 25 to 100 °C at a rate of 1.8 °C·min−1. The excitation and emission wavelengths were set at 465 nm and 580 nm, respectively. A single fluorescence measurement was taken every 0.03 s. The apparent Tm was determined from the first derivative curve.
3.5. CD and Fluorescence Spectroscopy
CD spectroscopy was used to assess the second structure of the wild-type PETase and its variants. The concentration of protein was diluted to 0.143 mg·mL−1 with desalting buffer. CD studies were then carried out using the J-810 CD spectrometer (JASCO, Tokyo, Japan) in a 1.0 mm quartz cell at 25 °C. The spectra were recorded at a far-UV wavelength from 190 to 260 nm with a bandwidth of 1 nm at a speed of 100 nm·min−1. The spectra of the desalting buffer were subtracted as background.
Fluorescence spectroscopy was used to determine the conformational change in the wild-type PETase and its variants. The concentration of the sample was maintained at 3 μM. The intrinsic fluorescence spectra were performed on a luminescence spectrometer (PerkinElmer, Shelton, CT, USA) with a 1 cm quartz cell at 25 °C. The excitation wavelength was set at 280 nm, and the emission spectra were recorded from 285 nm to 450 nm. The slit widths of excitation and emission were both set to 5.0 nm, and the scanning speed of emission spectra was set to 200 nm·min−1 [59].
3.6. Molecular Docking and Molecular Dynamics (MD) Simulations
The three-dimensional structure of the wild-type PETase was obtained from the Protein Data Bank (PDB ID: 6EQE) [60]. The structure of variants (D186Q, D186H, D186N, D186A, D186V) was constructed by the mutate module of Pymol software (https://pymol.org/ (accessed on 1 October 2023), with the structure of the wild-type PETase as a template. The MD simulations of wild type and variants were performed using GROMACS 5.1.4 software with the AMBER99SB-ILDN force field [61,62,63]. The enzymes were placed in the center of a cubic box at a distance of 10 Å from the edge of the box. The six systems were then solvated with the TIP3P water model and Cl- ions were added to keep all the systems neutral. All the systems were minimized for 50,000 steps until the maximum force reached 1000 kJ·mol−1·nm−1 using the steepest descent algorithm. Then, the systems were heated from 0 K to 303 K through v-rescal under NVT ensemble for 100 ps with a time-step of 2 fs. Subsequently, the systems were equilibrated for 100 ps under NPT ensemble at the constant temperature of 303 K and pressure of 1.0 atm using the Parrinello–Rahman thermostat. After this, the restraints for the protein were removed and six independent 50 ns MD simulations were performed for each system under NPT ensemble. The MD simulations for the wild-type PETase and its variants at 313 K and 403 K were performed using the same protocol as that at 303 K.
The typical structures obtained from the above simulations were used for subsequent docking. The model substrate, consisting of two repeating units of ethylene terephthalate (2PET) [46,49], was docked into the wild-type PETase and its variants by Autodock [64]. The MD simulations for the enzyme-substrate complexes were performed at 303 K for 50 ns, using the same protocol as that described above. Six independent simulations were performed for each system.
The linear constraint solver (LINCS) algorithm was used to constrain the bonds. The particle-mesh Ewald (PME) algorithm was used to calculate the electrostatic interactions. The cutoffs of the neighboring atom list, Lennard-Jones (LJ) potential and Coulomb potential energies were all set as 12 Å. The trajectories analysis were performed using the Gromacs package and VMD 1.9.3 software [65].
3.7. Measurement of PET Crystallinity
The percentage of crystallinity (Xc) for PET film was analyzed by the differential scanning calorimetry (DSC) instrument (Q20, TA instrument, New Castle, DE, USA) with the methods reported previously [17,46,66]. Approximately 2–3 mg of the PET sample was cooled to 0 °C and equilibrated for 1 min. Then, the sample was heated to 300 °C (10 °C·min−1) and maintained at 300 °C for 1 min. The sample was then cooled from 300 °C to 0 °C (10 °C·min−1). The percentage of crystallinity was calculated using the equation:
where is the value of melting enthalpy (J·g−1), is the value of cold crystallization (J·g−1), and is the melting enthalpy of 100% crystalline PET (140.1 J·g−1).
3.8. HPLC Analysis
The supernatant of reaction solution (20 μL) was analyzed by an Agilent 1100 series LC system (Agilent, USA) equipped with an Ultimate XB-C18 column (4.6 × 250 mm, 5 μm, Welch Materials, Shanghai, China) at 25 °C. The mobile phase consisted of 70% (v/v) distilled water, 20% (v/v) acetonitrile, and 10% (v/v) formic acid. The flow rate was set at 1.0 mL·min−1, and the detection wavelength for aromatic products (BHET, MHET, and TPA) was set at 254 nm.
4. Conclusions
In the present study, a non-active key amino acid residue in the second shell, D186, was identified using a crystal structure- and sequence-based strategy. The effects of this residue on the catalytic activity and thermostability of PETase were investigated by mutating D186 to the other 19 canonical amino acids. Among the variants, the Tm values of PETaseD186V and PETaseD186A increased by 12.91 and 11.84 °C, respectively, and their degradation efficiency of PET were 2.49 and 3.62 times higher than that of PETaseWT at 40 °C. PETaseD186N and PETaseD186H showed both improved thermostability and activity in PET degradation. PETaseD186N had the highest degradation efficiency against PET, being 1.86 and 3.69 times higher than the wild type at 30 and 40 °C, respectively. These results indicated that this key non-active amino acid was important for modulating the catalytic activity and thermostability of PETase. The structural analysis and MD simulations revealed that the D186 mutations (1) altered the tertiary structure rather than the secondary structure of the enzyme molecule, (2) elevated the binding free energy of the substrate to the enzyme and changed the substrate binding mode, resulting in the increase in catalytic activity of the enzyme, and/or (3) reduced the flexibility of Loop 10 mainly by hydrogen bonding and hydrophobic interactions, and locked flexible Loop 10 and Helix 6 by robust hydrogen bonding, leading to the enhanced enzyme thermostability. It is anticipated that this finding can be applied as a rational design strategy to improve thermostability of PET hydrolases and other enzymes, and it suggests that other non-active site residues in the second shell may be targeted as hot spots to further improve the catalytic activity and thermostability of PETase.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29061338/s1, Table S1: Engineered PETases for improved thermostability [15,17,20,21,22,31,67,68,69]. Table S2: The sequences of primers for site-directed mutation. Table S3: The occupancy rates of the conventional hydrogen bonds between residue 186 and surrounding residues at 303 K. Table S4: The occupancy rates of the conventional hydrogen bonds between residue 186 and surrounding residues at 313 K. Table S5: The occupancy rates of the conventional hydrogen bonds between residue 186 and surrounding residues at 403 K. Table S6: The occupancy rates of the hydrogen bonds between Loop 10 and Helix 5. Table S7: The occupancy rates of the hydrogen bonds between Loop 10 and Helix 6. Figure S1: Spatial distribution of point mutations that influence the thermostability of PETase. Figure S2: The differential scanning calorimetry (DSC) spectrum of PET film. Figure S3: PET film degradation activities of PETaseWT and its variants at 30 °C and 40 °C for 6 days. Figure S4: Molecular dynamics (MD) simulations of enzyme-2PET complexes. Figure S5: The crystal structure of PETase, the circular dichroism spectra and the intrinsic fluorescence spectra of PETaseWT and its variants at 25 °C. Figure S6: Root mean square deviation (RMSD) of the Cα atoms of PETaseWT and its variants in the MD simulations. Figure S7: Radius of gyration (Rg) of the Cα atoms of PETaseWT and its variants in the MD simulations. Figure S8: Root mean square fluctuations (RMSF) of the Cα atoms of PETaseWT and its variants in the MD simulations. Figure S9: The mobility and structural fluctuations of the Cα atoms of PETaseWT and its variants obtained with MDLovofit. Figure S10: The distance between the atom OD1 or OD2 of D186 and the atom N of S187, and the relative frequency of these two distances in PETaseWT during the MD simulations. Figure S11: The angle between the acceptor atom (D) of D186OD1 or D186OD2, the hydrogen atom (H), and the donor atom (A) of S187N, and the relative frequency of these two angles in PETaseWT during the MD simulations [56]. Figure S12: The distance between the atom OE1 of Q186 and the atom N of S187, the distance between the atom NE2 of Q186 and the atom O of S188, and the relative frequency of these two distances in PETaseD186Q during the MD simulations. Figure S13: The angle between the acceptor atom (A) of Q186OE1, the hydrogen atom (H), and the donor atom (D) of S187N, the angle between the acceptor atom (A) of S186O, the hydrogen atom (H), and the donor atom (D) of S186NE2, and the relative frequency of these two angles in PETaseD186Q during the MD simulations [56]. Figure S14: The distance between the atom ND1 of H186 and the atom N of S187, the distance between the atom CD2 of H186 and the atom O of S188, the distance between the centroid of imidazole ring of H186 and the N atom of S188, and the relative frequency of these three distances in PETaseD186H during the MD simulations. Figure S15: The angle between the acceptor atom (A) of H186ND1, the hydrogen atom (H), and the donor atom (D) of S187N, the angle between the acceptor atom (A) of S186O, the hydrogen atom (H), and the donor atom (D) of H186CD2, and the relative frequency of these two angles in PETaseD186H during the MD simulations [56]. Figure S16: The distance between the atom OD1 of N186 and the atom N of S187, the distance between the atom ND2 of N186 and the atom O of S188, the distance between the atom ND2 of N186 and the atom N of S188, and the relative frequency of these two distances in PETaseD186N during the MD simulations. Figure S17: The angle between the acceptor atom (A) of N186OD1, the hydrogen atom (H), and the donor atom (D) of S187N, the angles between the donor atom (D) of N186ND2, the hydrogen atom (H), and the acceptor atom (A) of S188O or S188N, and the relative frequency of these three angles in PETaseD186N during the MD simulations [56]. Figure S18: The distance between the centroid of alkyl of A186 and π ring of F191, and the relative frequency of this distance in PETaseD186A during the MD simulations. Figure S19: The distance between the centroid of alkyl of V186 and π ring of F191, and the relative frequency of this distance in PETaseD186V during the MD simulations.
Author Contributions
Conceptualization, Y.S.; methodology, Z.Q. and L.Z.; validation, Z.Q.; formal analysis, Z.Q.; investigation, Z.Q. and L.Z.; data curation, Y.S.; writing—original draft preparation, Z.Q. and L.Z.; writing—review and editing, Y.S.; supervision, Y.S.; funding acquisition, Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China, grant number 2018YFA0900702.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available on request from the authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Moog, D.; Schmitt, J.; Senger, J.; Zarzycki, J.; Rexer, K.H.; Linne, U.; Erb, T.J.; Maier, U.G. Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microb. Cell Fact. 2020, 18, 171. [Google Scholar] [CrossRef] [PubMed]
- Sagong, H.Y.; Seo, H.; Kim, T.; Son, H.F.; Joo, S.; Lee, S.H.; Kim, S.; Woo, J.S.; Hwang, S.Y.; Kim, K.J. Decomposition of the PET Film by MHETase Using Exo-PETase Function. ACS Catal. 2020, 10, 4805–4812. [Google Scholar] [CrossRef]
- Kumar, A.G.; Anjana, K.; Hinduja, M.; Sujitha, K.; Dharani, G. Review on plastic wastes in marine environment-Biodegradation and biotechnological solutions. Mar. Pollut. Bull. 2020, 150, 110733. [Google Scholar] [CrossRef]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.J. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef]
- Gibb, B.C. Plastics are forever. Nat. Chem. 2019, 11, 394–395. [Google Scholar] [CrossRef]
- Magalhaes, R.P.; Fernandes, H.S.; Sousa, S.F. The critical role of Asp206 stabilizing residues on the catalytic mechanism of the Ideonella sakaiensis PETase. Catal. Sci. Technol. 2022, 12, 3474–3483. [Google Scholar] [CrossRef]
- Webb, H.K.; Arnott, J.; Crawford, R.J.; Ivanova, E.P. Plastic Degradation and Its Environmental Implications with Special Reference to Poly(ethylene terephthalate). Polymers 2013, 5, 1–18. [Google Scholar] [CrossRef]
- Gong, J.X.; Kong, T.T.; Li, Y.Q.; Li, Q.J.; Li, Z.; Zhang, J.F. Biodegradation of Microplastic Derived from Poly(ethylene terephthalate) with Bacterial Whole-Cell Biocatalysts. Polymers 2018, 10, 1326. [Google Scholar] [CrossRef]
- Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K. Biodegradation of PET: Current Status and Application Aspects. ACS Catal. 2019, 9, 4089–4105. [Google Scholar] [CrossRef]
- Jerves, C.; Neves, R.P.P.; Ramos, M.J.; da Silva, S.; Fernandes, P.A. Reaction Mechanism of the PET Degrading Enzyme PETase Studied with DFT/MM Molecular Dynamics Simulations. ACS Catal. 2021, 11, 11626–11638. [Google Scholar] [CrossRef]
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.Z.; Hazen, T.; Streit, W.R. New Insights into the Function and Global Distribution of Polyethylene Terephthalate (PET)-Degrading Bacteria and Enzymes in Marine and Terrestrial Metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17. [Google Scholar] [CrossRef]
- Muller, R.J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.D. Enzymatic degradation of poly(ethylene terephthalate): Rapid hydrolyse using a hydrolase from T-fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Liu, B.; He, L.H.; Wang, L.P.; Li, T.; Li, C.C.; Liu, H.Y.; Luo, Y.Z.; Bao, R. Protein Crystallography and Site-Direct Mutagenesis Analysis of the Poly(ethylene terephthalate) Hydrolase PETase from Ideonella sakaiensis. ChemBioChem 2018, 19, 1471–1475. [Google Scholar] [CrossRef]
- Bell, E.L.; Smithson, R.; Kilbride, S.; Foster, J.; Hardy, F.J.; Ramachandran, S.; Tedstone, A.A.; Haigh, S.J.; Garforth, A.A.; Day, P.J.R.; et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 2022, 5, 673–681. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.-Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Rational Protein Engineering of Thermo-Stable PETase from Ideonella sakaiensis for Highly Efficient PET Degradation. ACS Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- Son, H.F.; Joo, S.; Seo, H.; Sagong, H.Y.; Lee, S.H.; Hong, H.; Kim, K.J. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzym. Microb. Technol. 2020, 141, 109656. [Google Scholar] [CrossRef]
- Feng, S.S.; Yue, Y.; Zheng, M.N.; Li, Y.W.; Zhang, Q.Z.; Wang, W.X. IsPETase- and IsMHETase-Catalyzed Cascade Degradation Mechanism toward Polyethylene Terephthalate. ACS Sustain. Chem. Eng. 2021, 9, 9823–9832. [Google Scholar] [CrossRef]
- Cui, Y.L.; Chen, Y.C.; Liu, X.Y.; Dong, S.J.; Tian, Y.E.; Qiao, Y.X.; Mitra, R.; Han, J.; Li, C.L.; Han, X.; et al. Computational Redesign of a PETase for Plastic Biodegradation under Ambient Condition by the GRAPE Strategy. ACS Catal. 2021, 11, 1340–1350. [Google Scholar] [CrossRef]
- Lu, H.Y.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.Z.; Kim, W.T.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y.; et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.X.; Liu, P.; Tan, Z.J.; Zhao, W.; Gao, J.F.; Gu, Q.; Ma, H.W.; Liu, H.F.; Zhu, L.L. Complete Depolymerization of PET Wastes by an Evolved PET Hydrolase from Directed Evolution. Angew. Chem. Int. Edit 2023, 62, e202218390. [Google Scholar] [CrossRef]
- Zhang, Y.F. A relay for improving the catalytic efficiency and thermostability of PET hydrolases. Chem. Catal. 2022, 2, 2420–2422. [Google Scholar] [CrossRef]
- Anishchenko, I.; Ovchinnikov, S.; Kamisetty, H.; Baker, D. Origins of coevolution between residues distant in protein 3D structures. Proc. Natl. Acad. Sci. USA 2017, 114, 9122–9127. [Google Scholar] [CrossRef]
- Truong, D.P.; Rousseau, S.; Machala, B.W.; Huddleston, J.P.; Zhu, M.Z.; Hull, K.G.; Romo, D.; Raushel, F.M.; Sacchettini, J.C.; Glasner, M.E. Second-Shell Amino Acid R266 Helps Determine N-Succinylamino Acid Racemase Reaction Specificity in Promiscuous N-Succinylamino Acid Racemase/o-Succinylbenzoate Synthase Enzymes. Biochemistry 2021, 60, 3829–3840. [Google Scholar] [CrossRef]
- Lee, J.; Goodey, N.M. Catalytic Contributions from Remote Regions of Enzyme Structure. Chem. Rev. 2011, 111, 7595–7624. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xu, Y.; Nie, Y. Role of distal sites in enzyme engineering. Biotechnol. Adv. 2023, 63, 108094. [Google Scholar] [CrossRef]
- Bi, J.; Chen, S.; Zhao, X.; Nie, Y.; Xu, Y. Computation-aided engineering of starch-debranching pullulanase from Bacillus thermoleovorans for enhanced thermostability. Appl. Microbiol. Biotechnol. 2020, 104, 7551–7562. [Google Scholar] [CrossRef]
- Parasuram, R.; Coulther, T.A.; Hollander, J.M.; Keston-Smith, E.; Ondrechen, M.J.; Beuning, P.J. Prediction of Active Site and Distal Residues in E. coli DNA Polymerase III alpha Polymerase Activity. Biochemistry 2018, 57, 1063–1072. [Google Scholar] [CrossRef]
- Ma, E.J.; Siirola, E.; Moore, C.; Kummer, A.; Stoeckli, M.; Faller, M.; Bouquet, C.; Eggimann, F.; Ligibel, M.; Huynh, D.; et al. Machine-Directed Evolution of an Imine Reductase for Activity and Stereoselectivity. ACS Catal. 2021, 11, 12433–12445. [Google Scholar] [CrossRef]
- Yin, Q.D.; You, S.P.; Zhang, J.X.; Qi, W.; Su, R.X. Enhancement of the polyethylene terephthalate and mono-(2-hydroxyethyl) terephthalate degradation activity of Ideonella sakaiensis PETase by an electrostatic interaction-based strategy. Bioresour. Technol. 2022, 364, 128026. [Google Scholar] [CrossRef]
- Qu, Z.; Chen, K.; Zhang, L.; Sun, Y. Computation-Based Design of Salt Bridges in PETase for Enhanced Thermostability and Performance for PET Degradation. ChemBioChem 2023, 21, e202300373. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Wei, R.; Breite, D.; Song, C.; Gräsing, D.; Ploss, T.; Hille, P.; Schwerdtfeger, R.; Matysik, J.; Schulze, A.; Zimmermann, W. Biocatalytic Degradation Efficiency of Postconsumer Polyethylene Terephthalate Packaging Determined by Their Polymer Microstructures. Adv. Sci. 2019, 6, 1900491. [Google Scholar] [CrossRef] [PubMed]
- Espino-Rammer, L.; Ribitsch, D.; Przylucka, A.; Marold, A.; Greimel, K.J.; Acero, E.H.; Guebitz, G.M.; Kubicek, C.P.; Druzhinina, I.S. Two Novel Class II Hydrophobins from spp. Stimulate Enzymatic Hydrolysis of Poly(Ethylene Terephthalate) when Expressed as Fusion Proteins. Appl. Environ. Microbiol. 2013, 79, 4230–4238. [Google Scholar] [CrossRef] [PubMed]
- Inaba, S.; Kamiya, N.; Bekker, G.J.; Kawai, F.; Oda, M. Folding thermodynamics of PET-hydrolyzing enzyme Cut190 depending on Ca concentration. J. Therm. Anal. Calorim. 2019, 135, 2655–2663. [Google Scholar] [CrossRef]
- Bollinger, A.; Thies, S.; Knieps-Grünhagen, E.; Gertzen, C.; Kobus, S.; Höppner, A.; Ferrer, M.; Gohlke, H.; Smits, S.H.J.; Jaeger, K.E. A Novel Polyester Hydrolase from the Marine Bacterium-Structural and Functional Insights. Front. Microb. 2020, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Meilleur, C.; Hupé, J.F.; Juteau, P.; Shareck, F. Isolation and characterization of a new alkali-thermostable lipase cloned from a metagenomic library. J. Ind. Microbiol. Biotechnol. 2009, 36, 853–861. [Google Scholar] [CrossRef]
- Chen, C.C.; Han, X.; Li, X.; Jiang, P.C.; Niu, D.; Ma, L.X.; Liu, W.D.; Li, S.Y.; Qu, Y.Y.; Hu, H.B.; et al. General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis. Nat. Catal. 2021, 4, 425–430. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Fesko, K.; Suplatov, D.; Svedas, V. Bioinformatic analysis of the fold type I PLP-dependent enzymes reveals determinants of reaction specificity in L-threonine aldolase from. FEBS Open Bio 2018, 8, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- Suplatov, D.; Shalaeva, D.; Kirilin, E.; Arzhanik, V.; Svedas, V. Bioinformatic analysis of protein families for identification of variable amino acid residues responsible for functional diversity. J. Biomol. Struct. Dyn. 2014, 32, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Suplatov, D.; Voevodin, V.; Svedas, V. Robust enzyme design: Bioinformatic tools for improved protein stability. Biotechnol. J. 2015, 10, 344–355. [Google Scholar] [CrossRef]
- Pleiss, J. Systematic Analysis of Large Enzyme Families: Identification of Specificity- and Selectivity-Determining Hotspots. ChemCatChem 2014, 6, 944–950. [Google Scholar] [CrossRef]
- Chen, K.; Hu, Y.; Dong, X.Y.; Sun, Y. Molecular Insights into the Enhanced Performance of EKylated PETase Toward PET Degradation. ACS Catal. 2021, 11, 7358–7370. [Google Scholar] [CrossRef]
- Han, X.; Liu, W.; Huang, J.W.; Ma, J.; Zheng, Y.; Ko, T.P.; Xu, L.; Cheng, Y.S.; Chen, C.C.; Guo, R.T. Structural insight into catalytic mechanism of PET hydrolase. Nat. Commun. 2017, 8, 2106. [Google Scholar] [CrossRef]
- Eiamthong, B.; Meesawat, P.; Wongsatit, T.; Jitdee, J.; Sangsri, R.; Patchsung, M.; Aphicho, K.; Suraritdechachai, S.; Huguenin-Dezot, N.; Tang, S.; et al. Discovery and Genetic Code Expansion of a Polyethylene Terephthalate (PET) Hydrolase from the Human Saliva Metagenome for the Degradation and Bio-Functionalization of PET. Angew. Chem. Int. Edit 2022, 61, e202203061. [Google Scholar] [CrossRef]
- Fecker, T.; Galaz-Davison, P.; Engelberger, F.; Narui, Y.; Sotomayor, M.; Parra, L.P.; Ramirez-Sarmiento, C.A. Active Site Flexibility as a Hallmark for Efficient PET Degradation by I-sakaiensis PETase. Biophys. J. 2018, 114, 1302–1312. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A.; Consort, O.S.D.D. g_mmpbsa-A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Chen, K.; Dong, X.Y.; Sun, Y. Sequentially co-immobilized PET and MHET hydrolases via Spy chemistry in calcium phosphate nanocrystals present high-performance PET degradation. J. Hazard. Mater. 2022, 438, 129517. [Google Scholar] [CrossRef]
- Wei, R.; Song, C.; Gräsing, D.; Schneider, T.; Bielytskyi, P.; Böttcher, D.; Matysik, J.; Bornscheuer, U.T.; Zimmermann, W. Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Nat. Commun. 2019, 10, 5581. [Google Scholar] [CrossRef]
- Guo, B.Y.; Vanga, S.R.; Lopez-Lorenzo, X.; Saenz-Mendez, P.; Ericsson, S.R.; Fang, Y.; Ye, X.C.; Schriever, K.; Backstrom, E.; Biundo, A.; et al. Conformational Selection in Biocatalytic Plastic Degradation by PETase. ACS Catal. 2022, 12, 3397–3409. [Google Scholar] [CrossRef]
- Prasad, A.R.; Luduena, R.F.; Horowitz, P.M. Detection of energy transfer between tryptophan residues in the tubulin molecule and bound bis(8-anilinonaphthalene-1-sulfonate), an inhibitor of microtubule assembly, that binds to a flexible region on tubulin. Biochemistry 1986, 25, 3536–3540. [Google Scholar] [CrossRef]
- Lopez-Chavez, E.; Perez-Hernandez, G.; Aparicio, F.; Alas, S.J. On the Thermal Stability of O-6-Methylguanine-DNA Methyltransferase from Archaeon Pyrococcus kodakaraensis by Molecular Dynamics Simulations. J. Chem. Inf. Model 2020, 60, 2138–2154. [Google Scholar] [CrossRef]
- Baker, E.N.; Hubbard, R.E. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 1984, 44, 97–179. [Google Scholar] [CrossRef] [PubMed]
- Pierce, A.C.; Sandretto, K.L.; Bemis, G.W. Kinase inhibitors and the case for CH...O hydrogen bonds in protein-ligand binding. Proteins 2002, 49, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Quan, M.Q.; Dong, X.Y.; Shi, Q.H.; Sun, Y. Low modification of PETase enhances its activity toward degrading PET: Effect of conjugate monomer property. Biochem. Eng. J. 2021, 175, 108151. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Y.; Sun, Y.; Dong, X. Complicated effects of a zwitterionic polymer containing dimethyl chains on the structures, activities and stabilities of different enzymes. Biochem. Eng. J. 2021, 165, 107813. [Google Scholar] [CrossRef]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Dominick, G.; Duman, R.; El Omari, K.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef] [PubMed]
- Kutzner, C.; Páll, S.; Fechner, M.; Esztermann, A.; de Groot, B.L.; Grubmüller, H. Best bang for your buck: GPU nodes for GROMACS biomolecular simulations. J. Comput. Chem. 2015, 36, 1990–2008. [Google Scholar] [CrossRef]
- Kutzner, C.; Páll, S.; Fechner, M.; Esztermann, A.; de Groot, B.L.; Grubmüller, H. More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 2019, 40, 2418–2431. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.J.; Zhang, L.; Sun, Y. Rational Design of Disulfide Bridges in BbPETase(CD) for Enhancing the Enzymatic Performance in PET Degradation. Molecules 2023, 28, 3528. [Google Scholar] [CrossRef]
- Meng, X.X.; Yang, L.X.; Liu, H.Q.; Li, Q.B.; Xu, G.S.; Zhang, Y.; Guan, F.F.; Zhang, Y.H.; Zhang, W.; Wu, N.F.; et al. Protein engineering of stable IsPETase for PET plastic degradation by Premuse. Int. J. Biol. Macromol. 2021, 180, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Zhong-Johnson, E.Z.L.; Voigt, C.A.; Sinskey, A.J. An absorbance method for analysis of enzymatic degradation kinetics of poly(ethylene terephthalate) films. Sci. Rep. 2021, 11, 2045–2322. [Google Scholar] [CrossRef]
- Enhancement of the degradation capacity of IsPETase for PET plastic degradation by protein engineering. Sci. Total Environ. 2022, 834, 154947. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).