In Silico Investigations on the Synergistic Binding Mechanism of Functional Compounds with Beta-Lactoglobulin
Abstract
1. Introduction
2. Results and Discussion
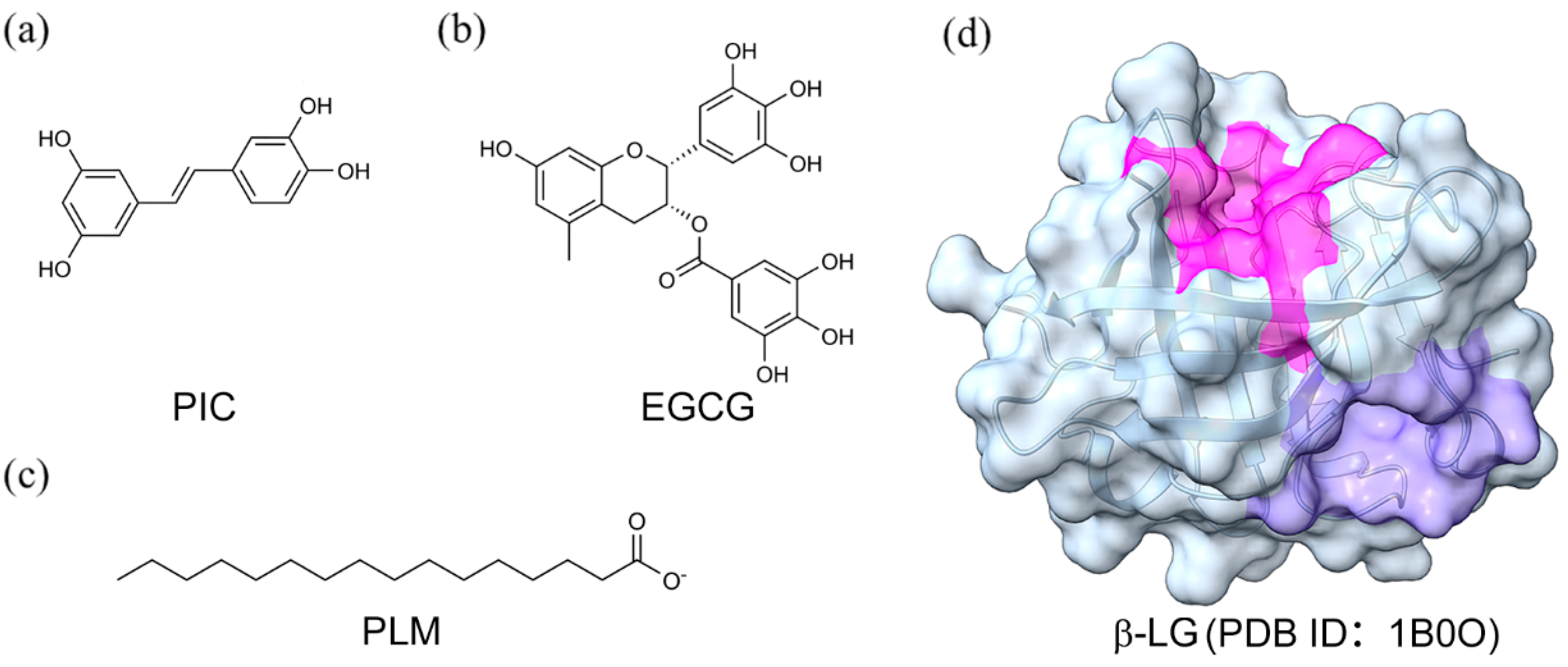
2.1. Binding Modes of PLM, EGCG, and PIC with β-LG
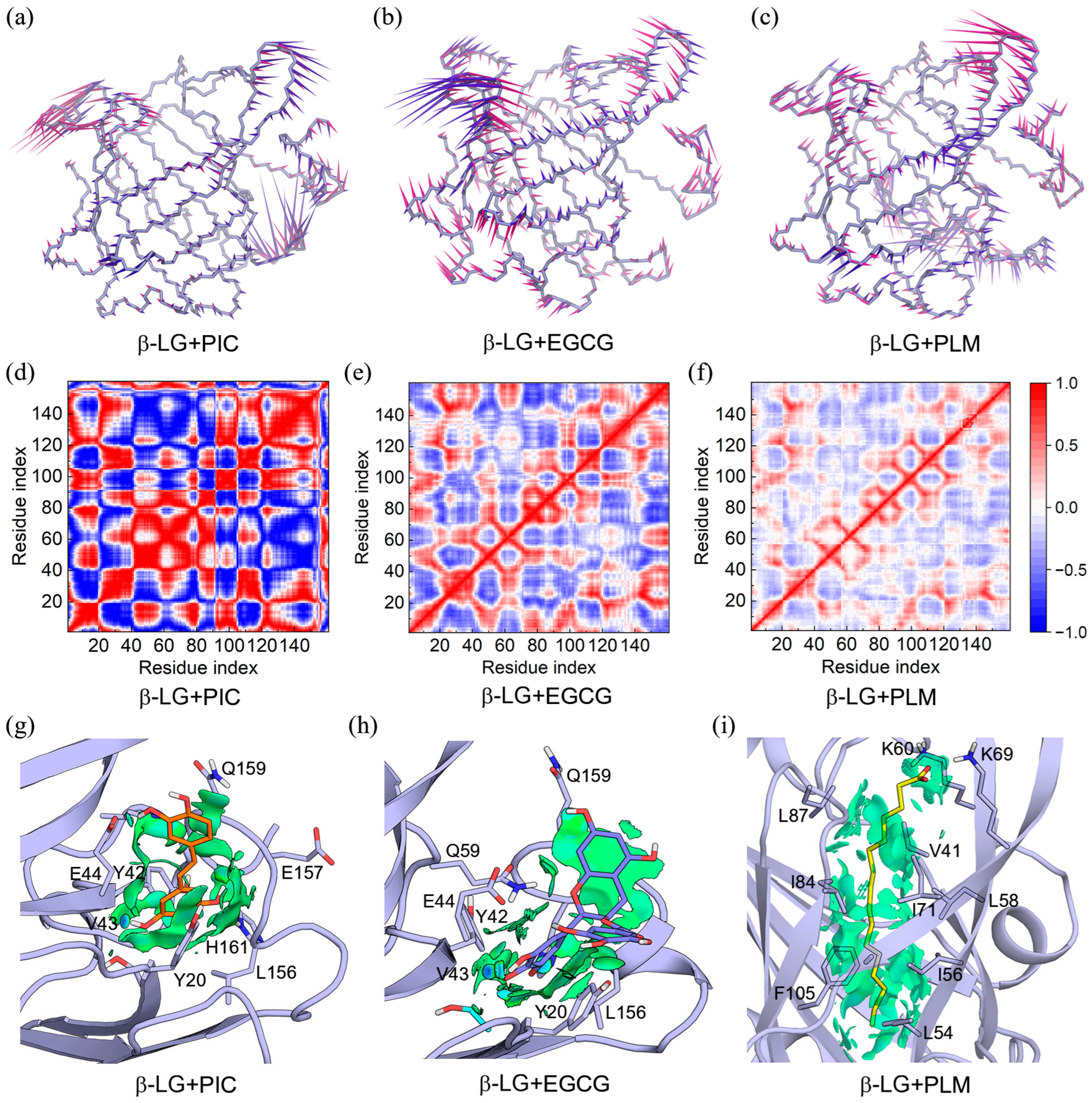
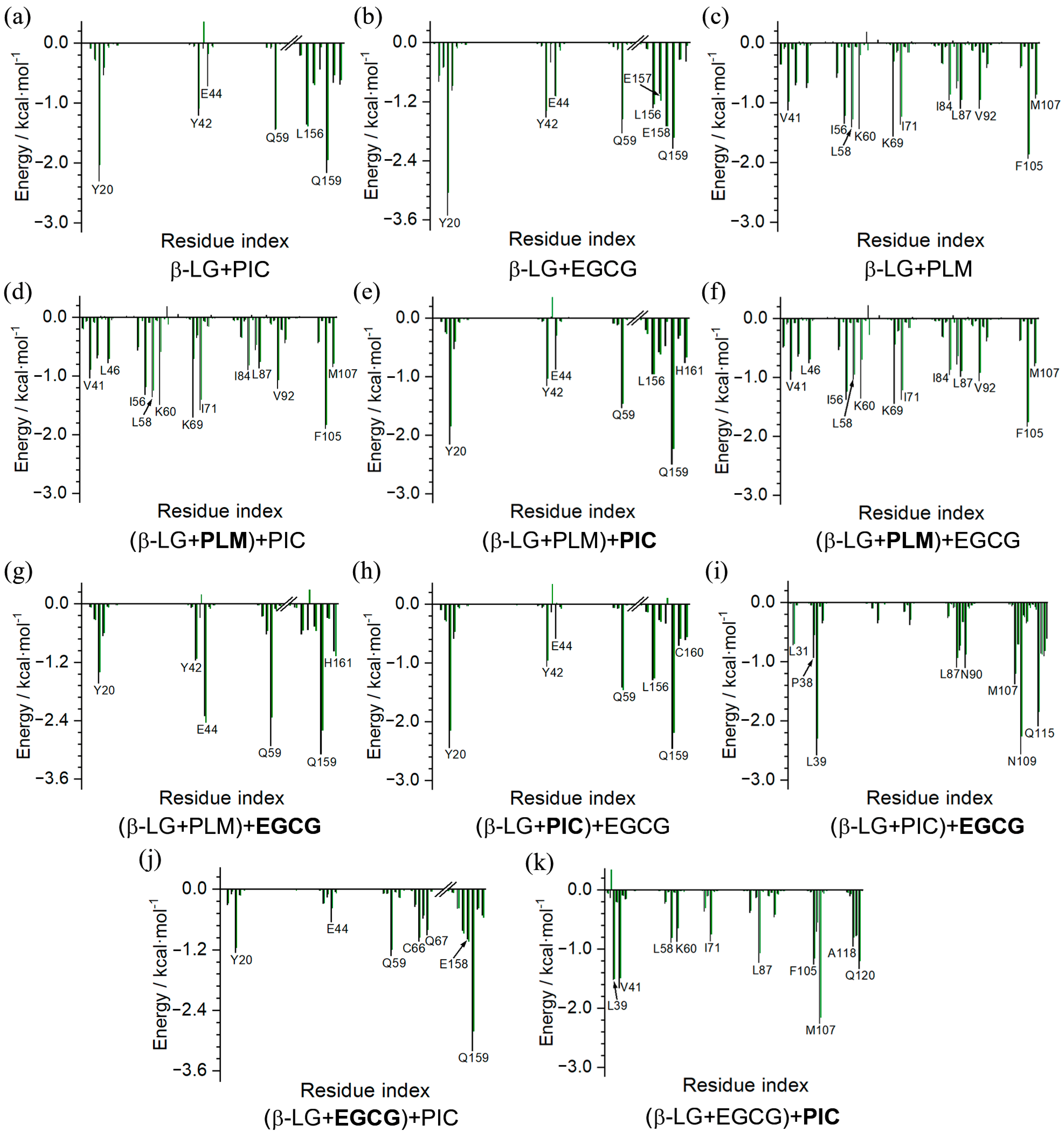
2.2. Binding Characteristics of the β-LG and Compounds of PIC, EGCG, and PLM
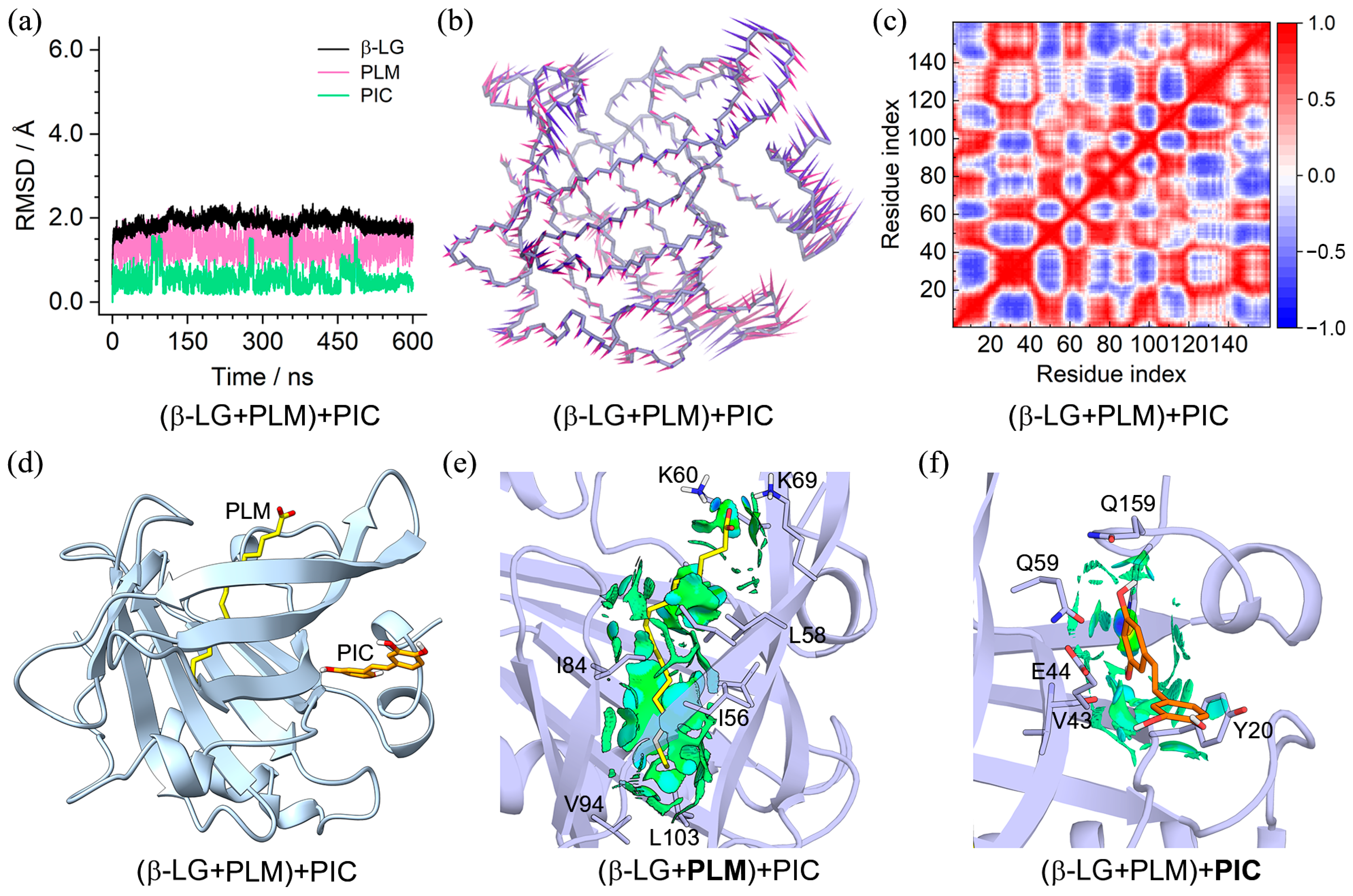
2.3. Characteristics of the (β-LG + PLM) + PIC/EGCG Ternary Complexes
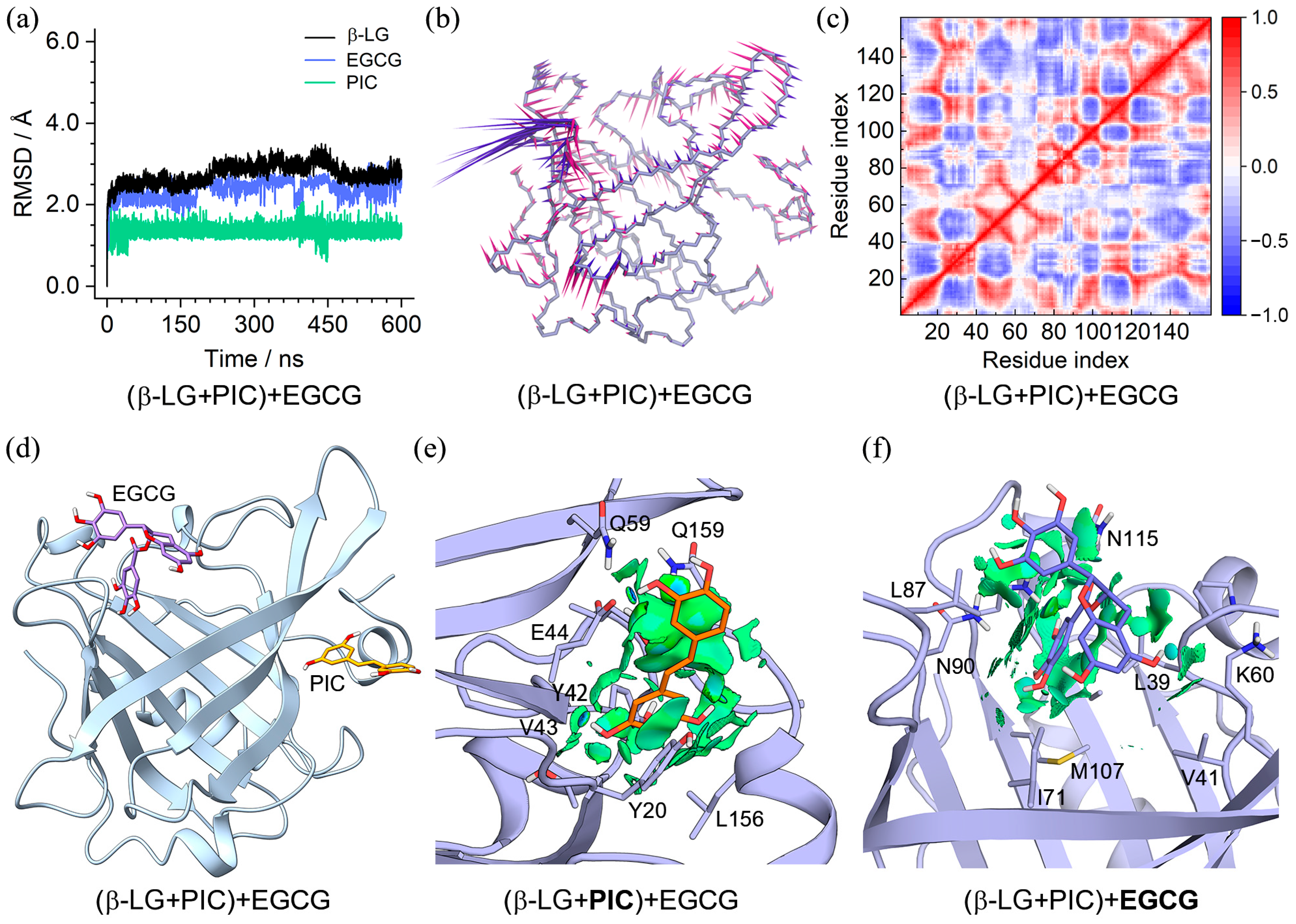
2.4. Binding Characteristics of the (β-LG + PIC/EGCG) + EGCG/PIC Ternary Complexes
2.5. Binding Free Energies between β-LG and the Binding Compounds
3. Methods
3.1. Data
3.2. Molecular Docking
3.3. Molecular Dynamics
3.4. Principal Component Analysis
3.5. Noncovalent Interactions
3.6. Binding Free Energy Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahn, W.S.; Huh, S.W.; Bae, S.M.; Lee, I.P.; Sin, J.I. A major constituent of green tea, EGCG, inhibits the growth of a human cervical cancer cell line, CaSki cells, through apoptosis, G1 arrest, and regulation of gene expression. DNA Cell Biol. 2003, 22, 217–224. [Google Scholar] [CrossRef]
- Roy, A.M.; Baliga, M.S.; Katiyar, S.K. Epigallocatechin-3-gallate induces apoptosis in estrogen receptor–negative human breast carcinoma cells via modulation in protein expression of p53 and Bax and caspase-3 activation. Mol. Cancer Ther. 2005, 4, 81–90. [Google Scholar] [CrossRef]
- Sen, T.; Dutta, A.; Chatterjee, A. Epigallocatechin-3-gallate (EGCG) downregulates gelatinase-B (MMP-9) by involvement of FAK/ERK/NFκB and AP-1 in the human breast cancer cell line MDA-MB-231. Anticancer Drugs 2010, 21, 632–644. [Google Scholar] [CrossRef]
- Meeran, S.M.; Patel, S.N.; Chan, T.H. A novel prodrug of epigallocatechin-3-gallate: Differential epigenetic hTERT repression in human breast cancer cells. Cancer Prev. Res. 2011, 4, 1243–1254. [Google Scholar] [CrossRef]
- Li, Y.; Shen, X.; Wang, X.; Li, A.; Wang, P.; Jiang, P.; Zhou, J.; Feng, Q. EGCG regulates the cross-talk between JWA and topoisomerase IIα in non-small-cell lung cancer (NSCLC) cells. Sci. Rep. 2015, 5, 11009. [Google Scholar] [CrossRef]
- Farooqi, A.A.; Mansoor, Q.; Ismail, M.; Bhatti, S. Therapeutic Effect of Epigallocatechin-3-gallate (EGCG) and Silibinin on ATM Dynamics in Prostate Cancer Cell Line LNCaP. World J. Oncol. 2010, 1, 242–246. [Google Scholar] [CrossRef][Green Version]
- Wieder, T.; Prokop, A.; Bagci, B.; Essmann, F.; Bernicke, D.; Schulze-Osthoff, K.; Drken, B.; Schmalz, H.G.; Daniel, P.T.; Henze, G.J. Piceatannol, a hydroxylated analog of the chemopreventive agent resveratrol, is a potent inducer of apoptosis in the lymphoma cell line BJAB and in primary, leukemic lymphoblasts. Leukemia 2001, 15, 1735–1742. [Google Scholar] [CrossRef]
- Siedlecka-Kroplewska, K.; Ślebioda, T.; Kmieć, Z. Induction of autophagy, apoptosis and aquisition of resistance in response to piceatannol toxicity in MOLT-4 human leukemia cells. Toxicol. Vitr. 2019, 59, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; McClements, D.J.; Wei, Z.; Wang, G.; Liu, X.; Liu, F. Delivery of synergistic polyphenol combinations using biopolymer-based systems: Advances in physicochemical properties, stability and bioavailability. Crit. Rev. Food Sci. Nutr. 2019, 60, 2083–2097. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liu, T.; Shi, Y.; Zhao, Y.; Yan, H.; Sun, B.; Wang, Q.; Wang, Z.; Han, J. Comparative study on the interaction of oxyresveratrol and piceatannol with trypsin and lysozyme: Binding ability, activity and stability. Food Funct. 2019, 10, 8182–8194. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cheng, H.; Ni, Y.; Liang, L. Technology, Encapsulation and protection of bioactive components based on ligand-binding property of β-lactoglobulin. Zhongguo Shipin Xuebao 2015, 15, 124–129. [Google Scholar]
- Cheng, H.; Fang, Z.; Liu, T.; Gao, Y.; Liang, L. A study on β-lactoglobulin-triligand-pectin complex particle: Formation, characterization and protection. Food Hydrocoll. 2018, 84, 93–103. [Google Scholar] [CrossRef]
- Ajuwon, K.M.; Spurlock, M.E. Palmitate Activates the NF-κB Transcription Factor and Induces IL-6 and TNFα Expression in 3T3-L1 Adipocytes. J. Nutr. 2005, 135, 1841–1846. [Google Scholar] [CrossRef] [PubMed]
- Eom, D.-W.; Lee, J.H.; Kim, Y.-J.; Hwang, G.S.; Kim, S.-N.; Kwak, J.H.; Cheon, G.J.; Kim, K.H.; Jang, H.-J.; Ham, J.; et al. Synergistic effect of curcumin on epigallocatechin gallate-induced anticancer action in PC3 prostate cancer cells. BMB Reports 2015, 48, 461–466. [Google Scholar] [CrossRef]
- Liu, C.Z.; Lv, N.; Xu, Y.Q. pH-dependent interaction mechanisms between β-lactoglobulin and EGCG: Insights from multi-spectroscopy and molecular dynamics simulation methods. J. Food Hydrocoll. 2022, 133, 108022. [Google Scholar] [CrossRef]
- Liu, T.; Liu, M.; Liu, H.; Ren, Y.; Zhao, Y.; Yan, H.; Wang, Q.; Zhang, N.; Ding, Z.; Wang, Z. Co-encapsulation of (−)-epigallocatechin-3-gallate and piceatannol/oxyresveratrol in β-lactoglobulin: Effect of ligand–protein binding on the antioxidant activity, stability, solubility and cytotoxicity. Food Funct. 2021, 12, 7126–7144. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Subirade, M.; Zhou, P.; Liang, L. A study of multi-ligand beta-lactoglobulin complex formation. Food Chem. 2014, 165, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Kanakis, C.D.; Hasni, I.; Bourassa, P.; Tarantilis, P.A.; Polissiou, M.G.; Tajmir-Riahi, H.-A. Milk β-lactoglobulin complexes with tea polyphenols. Food Chem. 2011, 127, 1046–1055. [Google Scholar] [CrossRef]
- Graves, A.P.; Shivakumar, D.M.; Boyce, S.E.; Jacobson, M.P.; Case, D.A.; Shoichet, B.K. Rescoring Docking Hit Lists for Model Cavity Sites: Predictions and Experimental Testing. J. Mol. Biol. 2008, 377, 914–934. [Google Scholar] [CrossRef]
- Brozell, S.R.; Mukherjee, S.; Balius, T.E.; Roe, D.R.; Case, D.A.; Rizzo, R.C. Evaluation of dock 6 as a pose generation and database enrichment tool. J. Comput. Aid. Mol. Des. 2012, 26, 749–773. [Google Scholar] [CrossRef]
- Wu, S.Y.; Perez, M.D.; Puyol, P.; Sawyer, L. β-lactoglobulin binds palmitate within its central cavity. J. Biol. Chem. 1999, 274, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, G.; Meng, T.; Qi, L.; Yan, H.; Wang, Z.G. Molecular insights into the selective binding mechanism targeting parallel human telomeric G-quadruplex. J. Mol. Graph. Model. 2022, 110, 108058. [Google Scholar] [CrossRef] [PubMed]
- Cieplak, P.; Cornell, W.D.; Bayly, C.; Kollman, P.A. Application of the multimolecule and multiconformational RESP methodology to biopolymers: Charge derivation for DNA, RNA, and proteins. J. Comput. Chem. 2010, 16, 1357–1377. [Google Scholar] [CrossRef]
- Díaz-Gómez, D.G.; Galindo-Murillo, R.; Cortés-Guzmán, F. The Role of the DNA Backbone in Minor-Groove Ligand Binding. Chem. Phys. Chem. 2017, 18, 1909–1915. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 3, 198–210. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef]
- Braun, E.; Moosavi, S.M.; Smit, B. Anomalous effects of velocity rescaling algorithms: The flying ice cube effect revisited. J. Chem. Theory Comput. 2018, 14, 5262–5272. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- He, X.; Man, V.H.; Yang, W.; Li, T.-S.; Wang, J. A fast and high-quality charge model for the next generation general AMBER force field. J. Chem. Phys. 2020, 153, 114502. [Google Scholar] [CrossRef]
- Wang, Z.; Li, G.; Tian, Z.; Lou, X.; Huang, Y.; Wang, L.; Li, J.; Hou, T.; Liu, J.-P. Insight derived from molecular dynamics simulation into the selectivity mechanism targeting c-MYC G-quadruplex. J. Phys. Chem. B 2020, 124, 9773–9784. [Google Scholar] [CrossRef]
- Amadei, A.; Linssen, A.B.M.; Berendsen, H.J.C. Essential dynamics of proteins. Proteins 1993, 17, 412–425. [Google Scholar] [CrossRef]
- Mongan, J. Interactive essential dynamics. J. Comput. Aid. Mol. Des. 2004, 118, 433–436. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K.K. VMD—Visual molecular dynamics. J. Mol. Graph. 1995, 14, 33–38. [Google Scholar] [CrossRef]
- Contreras-García, J.; Johnson, E.R.; Keinan, S.; Chaudret, R.; Piquemal, J.-P.; Beratan, D.N.; Yang, W. NCIPLOT: A program for plotting noncovalent interaction regions. J. Chem. Theory Comput. 2011, 7, 625–632. [Google Scholar] [CrossRef]
- Islam, B.; Stadlbauer, P.; Neidle, S.; Haider, S.; Sponer, J. Can we execute reliable MM-PBSA free energy computations of relative stabilities of different guanine quadruplex folds? J. Phys. Chem. B 2016, 120, 2899–2912. [Google Scholar] [CrossRef]
- Hou, T.J.; Wang, J.M.; Li, Y.Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. How to obtain statistically converged MM/GBSA results. J. Comput. Chem. 2010, 31, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Onufriev, A.; Bashford, D.; Case, D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 2004, 55, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Sun, H.; Wang, J.; Zhu, F.; Liu, H.; Wang, Z.; Lei, T.; Li, Y.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 8. Predicting binding free energies and poses of protein–RNA complexes. RNA 2018, 24, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.-Q.; Chen, S.-B.; Tan, J.-H.; Luo, H.-B.; Li, D.; Gu, L.-Q.; Huang, Z.-S. New insights from molecular dynamic simulation studies of the multiple binding modes of a ligand with G-quadruplex DNA. J. Comput. Aid. Mol. Des. 2012, 26, 1355–1368. [Google Scholar] [CrossRef] [PubMed]
- Weiser, J.; Shenkin, P.S.; Still, W.C. Approximate solvent-accessible surface areas from tetrahedrally directed neighbor densities. J. Biopolym. 1999, 50, 373–380. [Google Scholar] [CrossRef]




| Receptor | Ligand | Energy Component | |||||
|---|---|---|---|---|---|---|---|
| Eele | EvdW | GGB | GSA | –TΔS | Gbind | ||
| β-LG | PIC | −13.66 ± 1.47 | −24.28 ± 3.28 | 13.81 ± 0.94 | −3.68 ± 0.12 | 17.50 ± 11.76 | −10.32 |
| β-LG | EGCG | −13.74 ± 3.23 | −35.50 ± 3.03 | 15.76 ± 2.75 | −4.91 ± 0.23 | 21.18 ± 10.27 | −17.20 |
| β-LG | PLM | 12.36 ± 7.19 | −36.38 ± 2.76 | −12.82 ± 5.77 | −6.23 ± 0.31 | 23.78 ± 11.07 | −19.30 |
| β-LG | PLM | 19.47 ± 7.58 | −37.04 ± 0.05 | −18.87 ± 6.15 | −6.16 ± 0.17 | 23.43 ± 10.49 | −19.17 |
| PIC | −12.84 ± 1.54 | −23.33 ± 3.10 | 13.01 ± 0.94 | −3.75 ± 0.09 | 14.09 ± 10.99 | −12.83 | |
| β-LG | PLM | 15.4 ± 8.84 | −35.85 ± 2.84 | −15.54 ± 7.15 | −6.26 ± 0.21 | 20.38 ± 11.04 | −21.88 |
| EGCG | −17.07 ± 5.37 | −36.93 ± 4.78 | 19.1 ± 4.25 | −5.22 ± 0.20 | 24.41 ± 11.44 | −15.71 | |
| β-LG | PIC | −12.85 ± 1.58 | −22.91 ± 3.23 | 12.85 ± 0.99 | −3.63 ± 0.11 | 16.72 ± 10.55 | −10.00 |
| EGCG | −3.95 ± 2.52 | −37.32 ± 5.03 | 7.83 ± 2.14 | −5.40 ± 0.40 | 21.81 ± 11.98 | −17.03 | |
| β-LG | EGCG | −9.45 ± 4.74 | −26.87 ± 6.59 | 11.73 ± 3.97 | −4.09 ± 0.48 | 22.31 ± 10.25 | −6.37 |
| PIC | −3.71 ± 1.22 | −32.83 ± 2.45 | 6.78 ± 0.81 | −4.73 ± 0.12 | 17.31 ± 9.25 | −17.18 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, T.; Wang, Z.; Zhang, H.; Zhao, Z.; Huang, W.; Xu, L.; Liu, M.; Li, J.; Yan, H. In Silico Investigations on the Synergistic Binding Mechanism of Functional Compounds with Beta-Lactoglobulin. Molecules 2024, 29, 956. https://doi.org/10.3390/molecules29050956
Meng T, Wang Z, Zhang H, Zhao Z, Huang W, Xu L, Liu M, Li J, Yan H. In Silico Investigations on the Synergistic Binding Mechanism of Functional Compounds with Beta-Lactoglobulin. Molecules. 2024; 29(5):956. https://doi.org/10.3390/molecules29050956
Chicago/Turabian StyleMeng, Tong, Zhiguo Wang, Hao Zhang, Zhen Zhao, Wanlin Huang, Liucheng Xu, Min Liu, Jun Li, and Hui Yan. 2024. "In Silico Investigations on the Synergistic Binding Mechanism of Functional Compounds with Beta-Lactoglobulin" Molecules 29, no. 5: 956. https://doi.org/10.3390/molecules29050956
APA StyleMeng, T., Wang, Z., Zhang, H., Zhao, Z., Huang, W., Xu, L., Liu, M., Li, J., & Yan, H. (2024). In Silico Investigations on the Synergistic Binding Mechanism of Functional Compounds with Beta-Lactoglobulin. Molecules, 29(5), 956. https://doi.org/10.3390/molecules29050956






