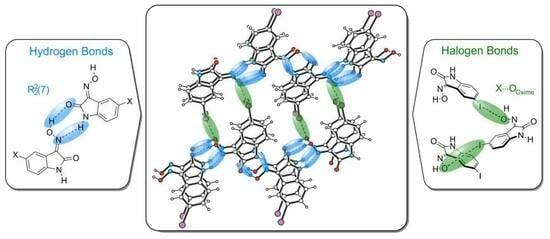
Combination of Hydrogen and Halogen Bonds in the Crystal Structures of 5-Halogeno-1H-isatin-3-oximes: Involvement of the Oxime Functionality in Halogen Bonding
Abstract
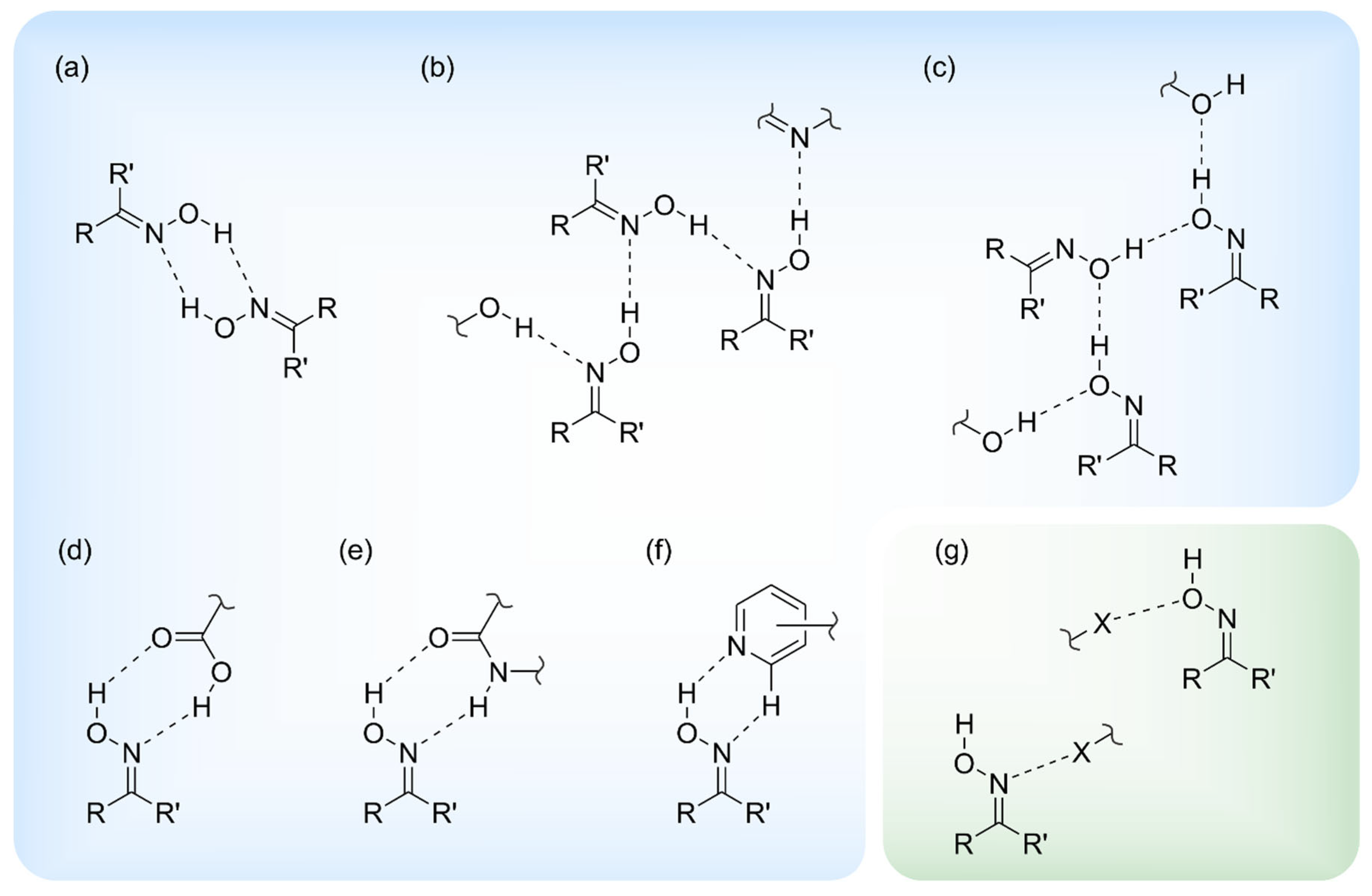
1. Introduction
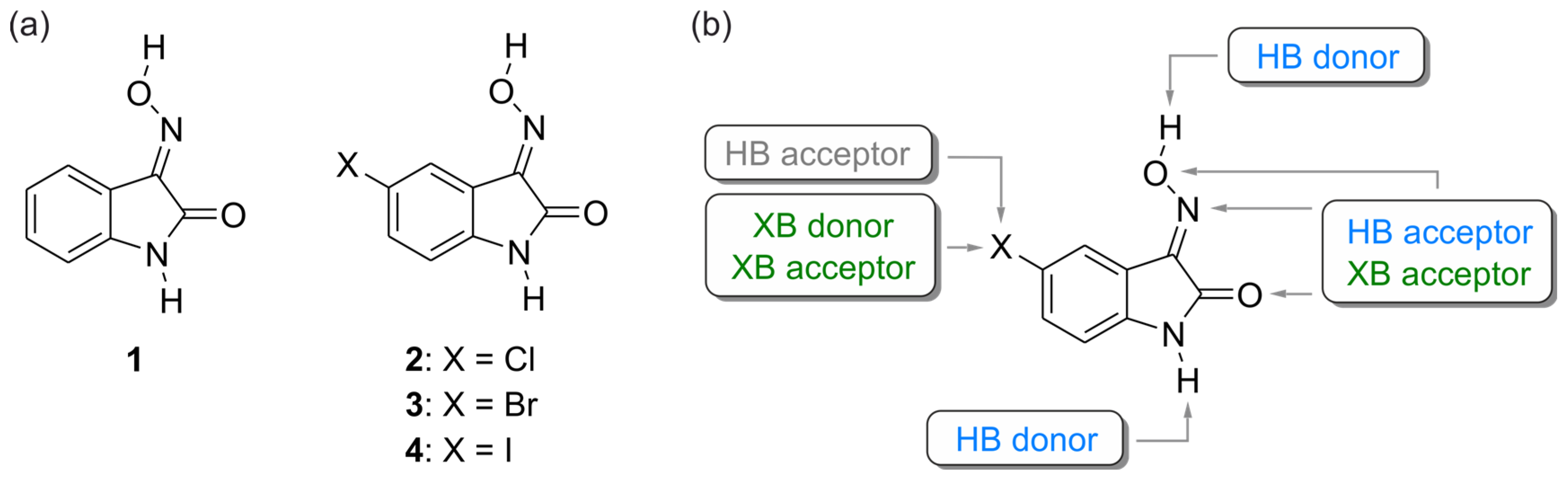
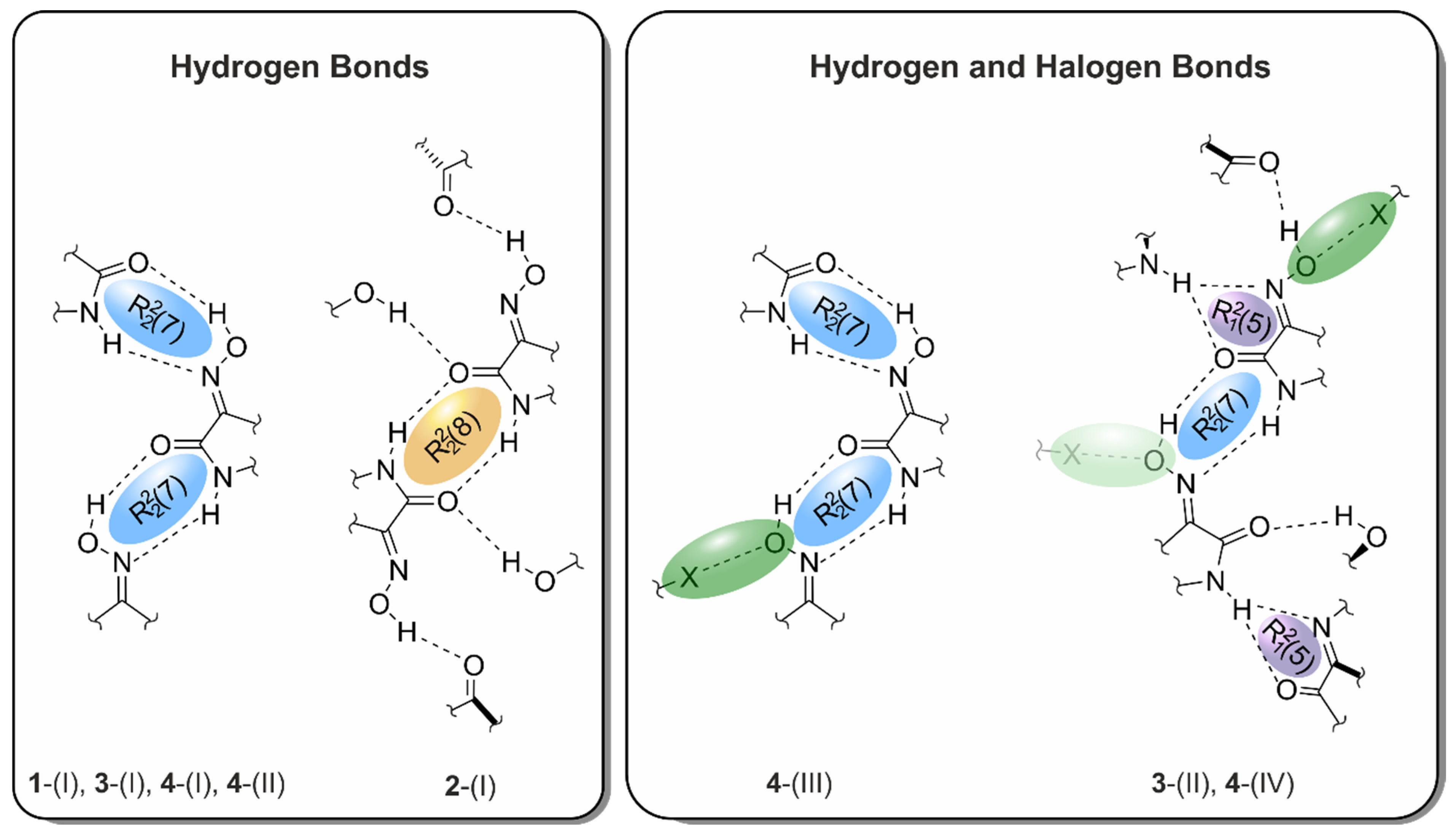
2. Results and Discussion
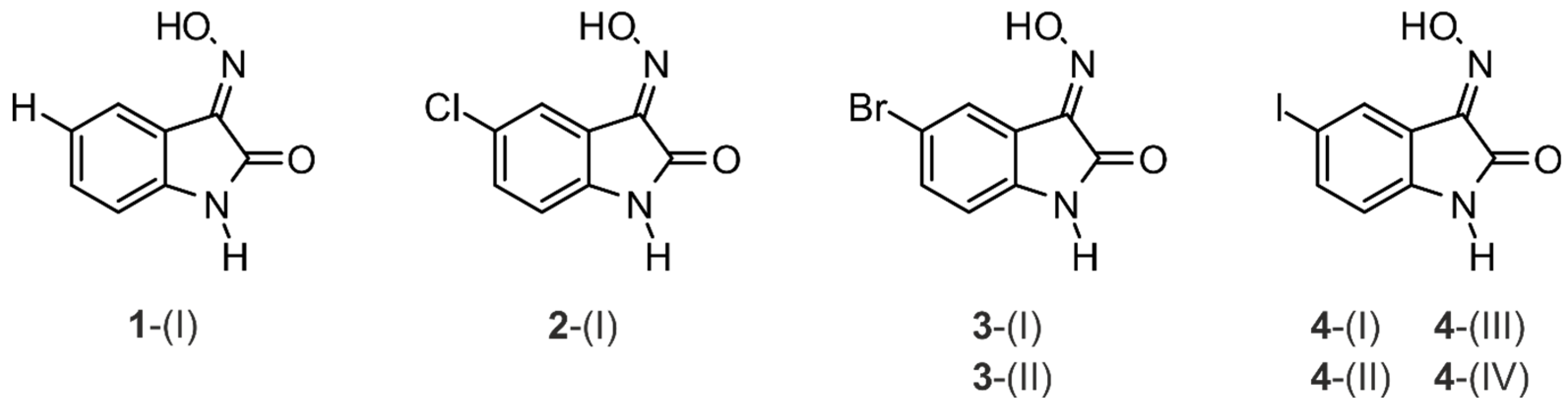
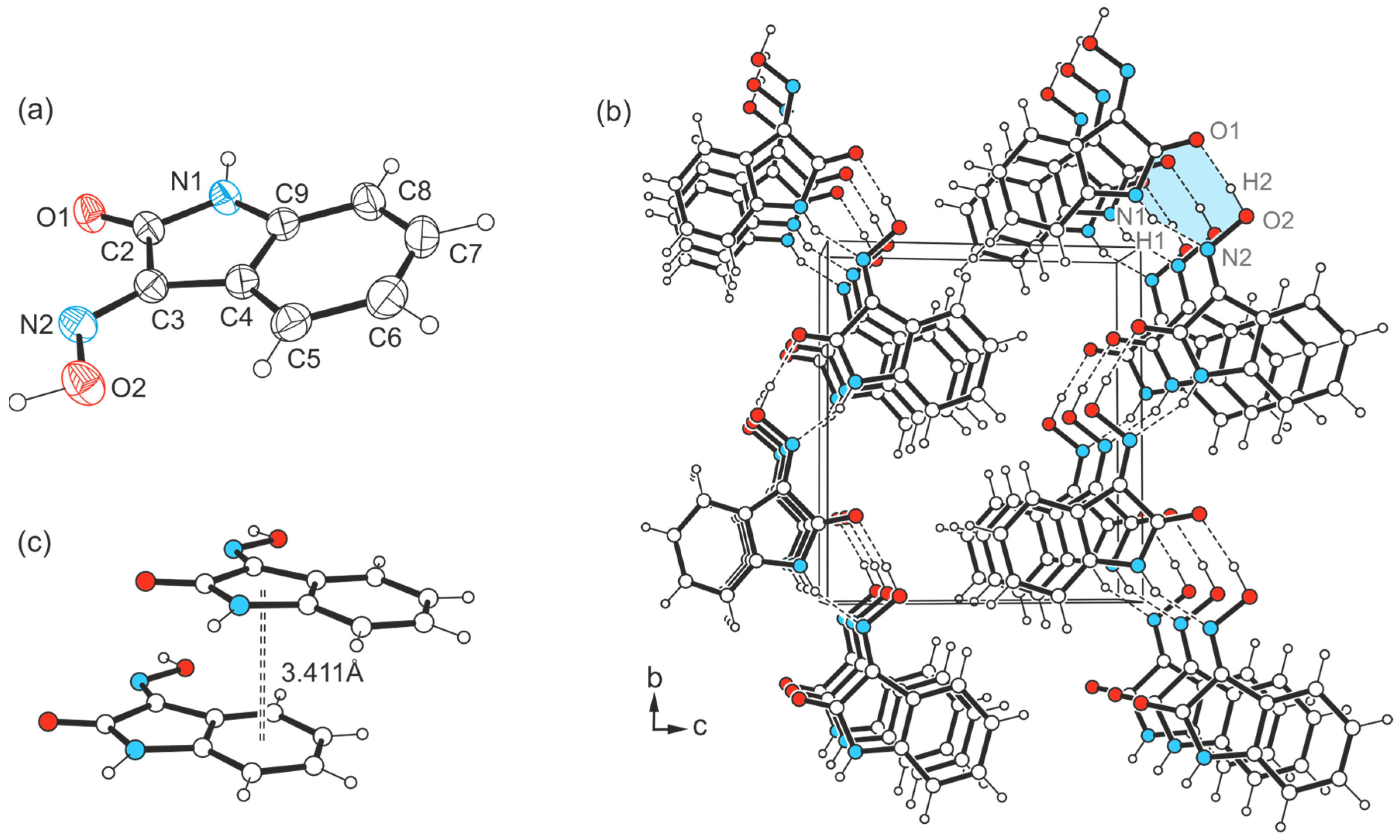
2.1. Crystal Structure of 1H-isatin-3-oxime [Structure 1-(I)]
2.2. Crystal Structure of 5-Chloro-1H-isatin-3-oxime [Structure 2-(I)]
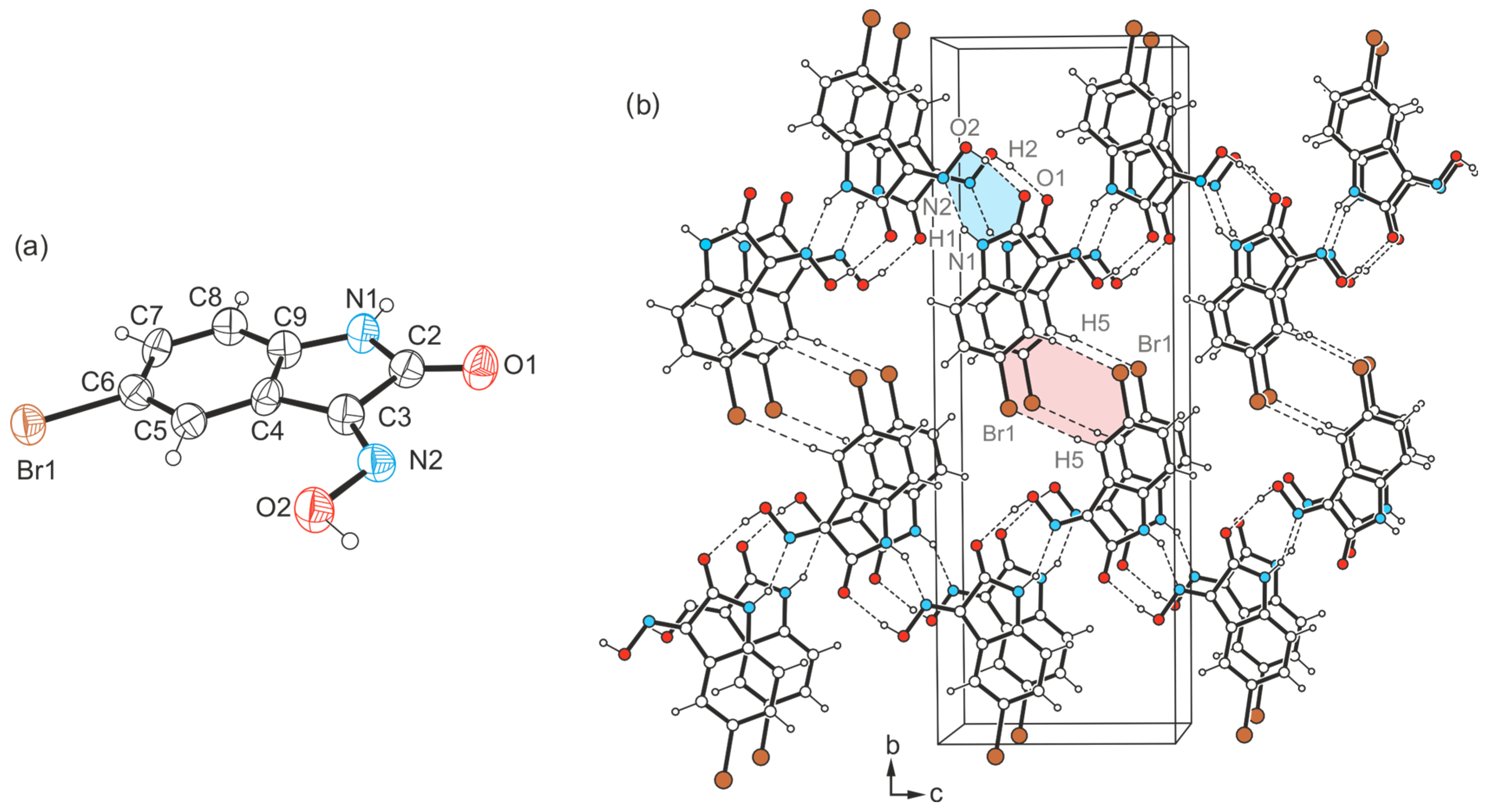
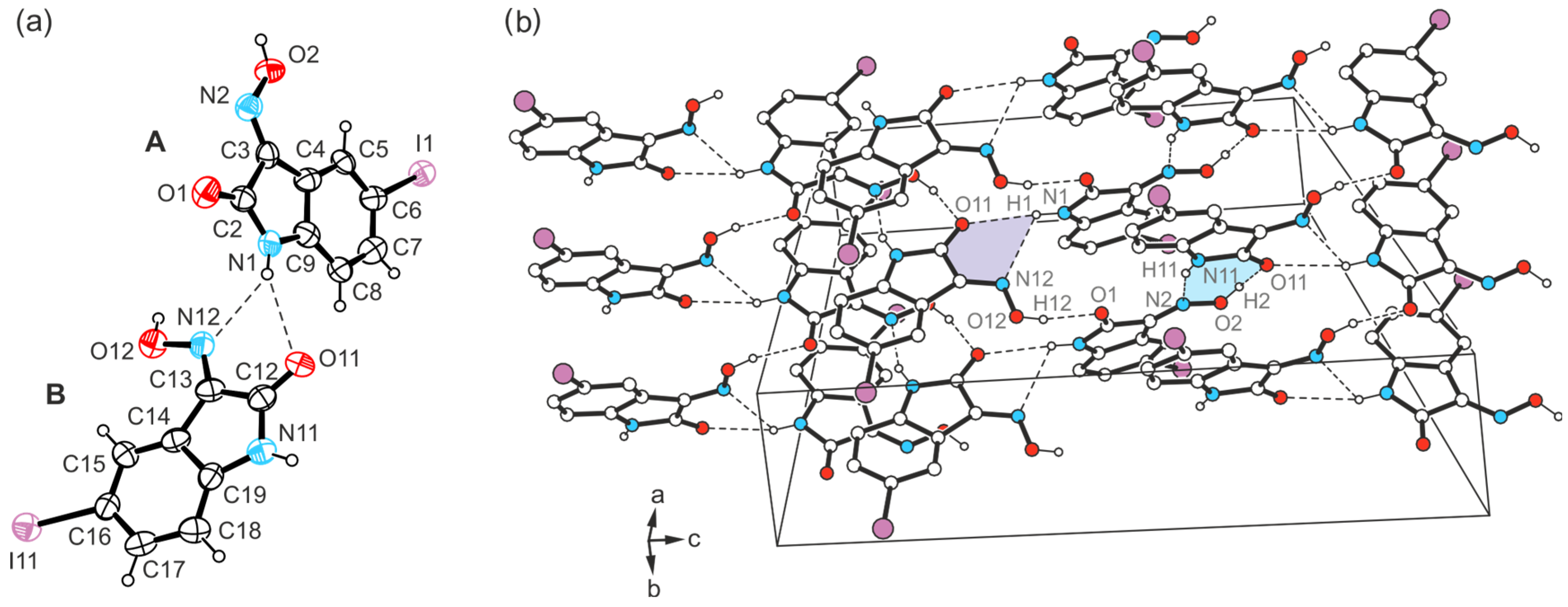
2.3. Crystal Structures of 5-Bromo-1H-isatin-3-oxime (3)
2.3.1. Polymorph 3-(I)
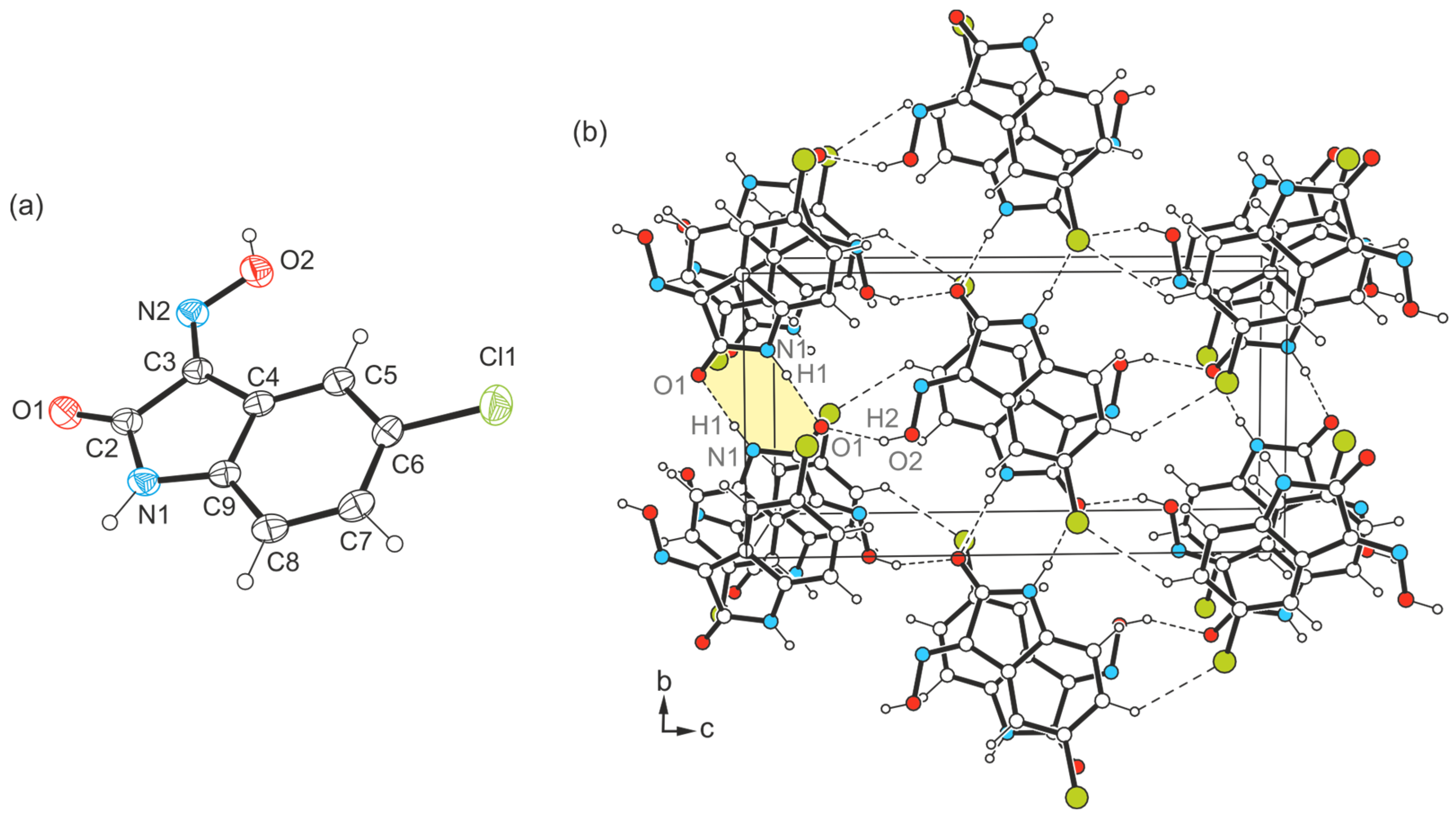
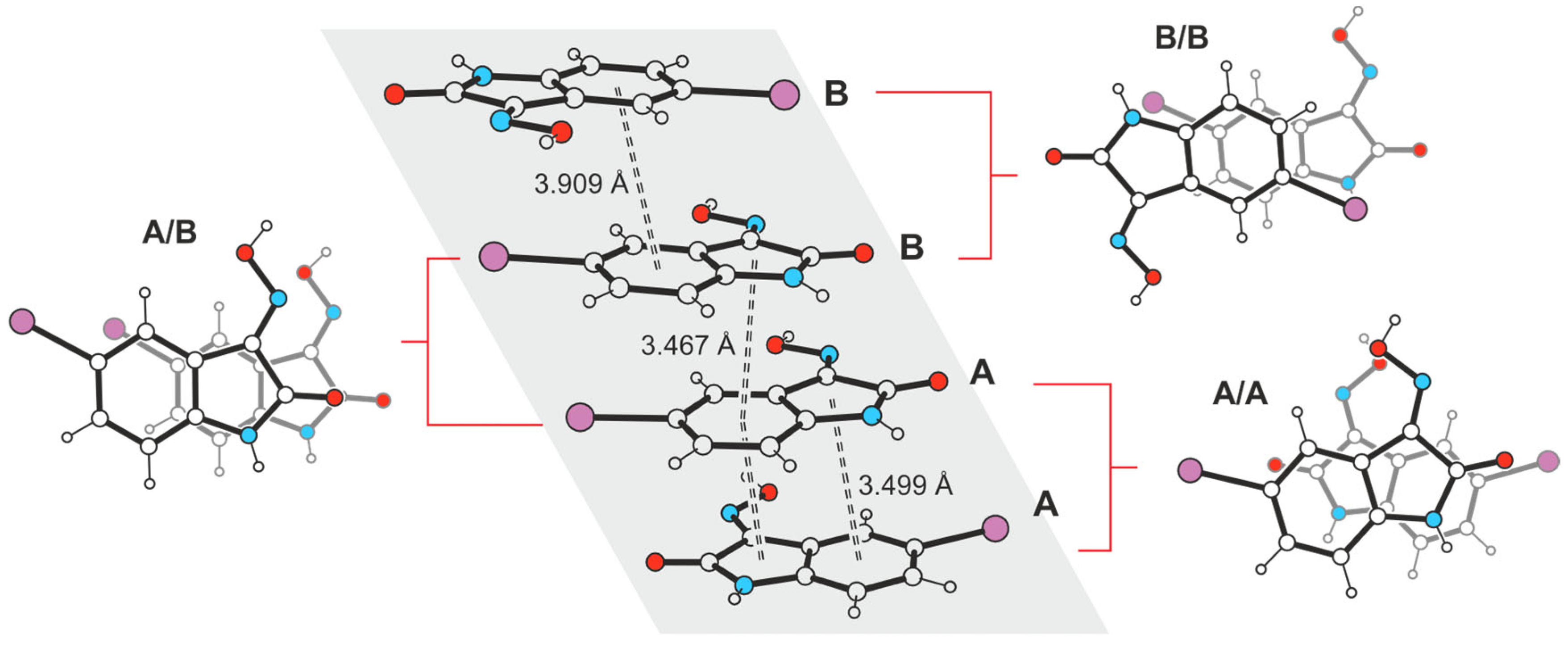
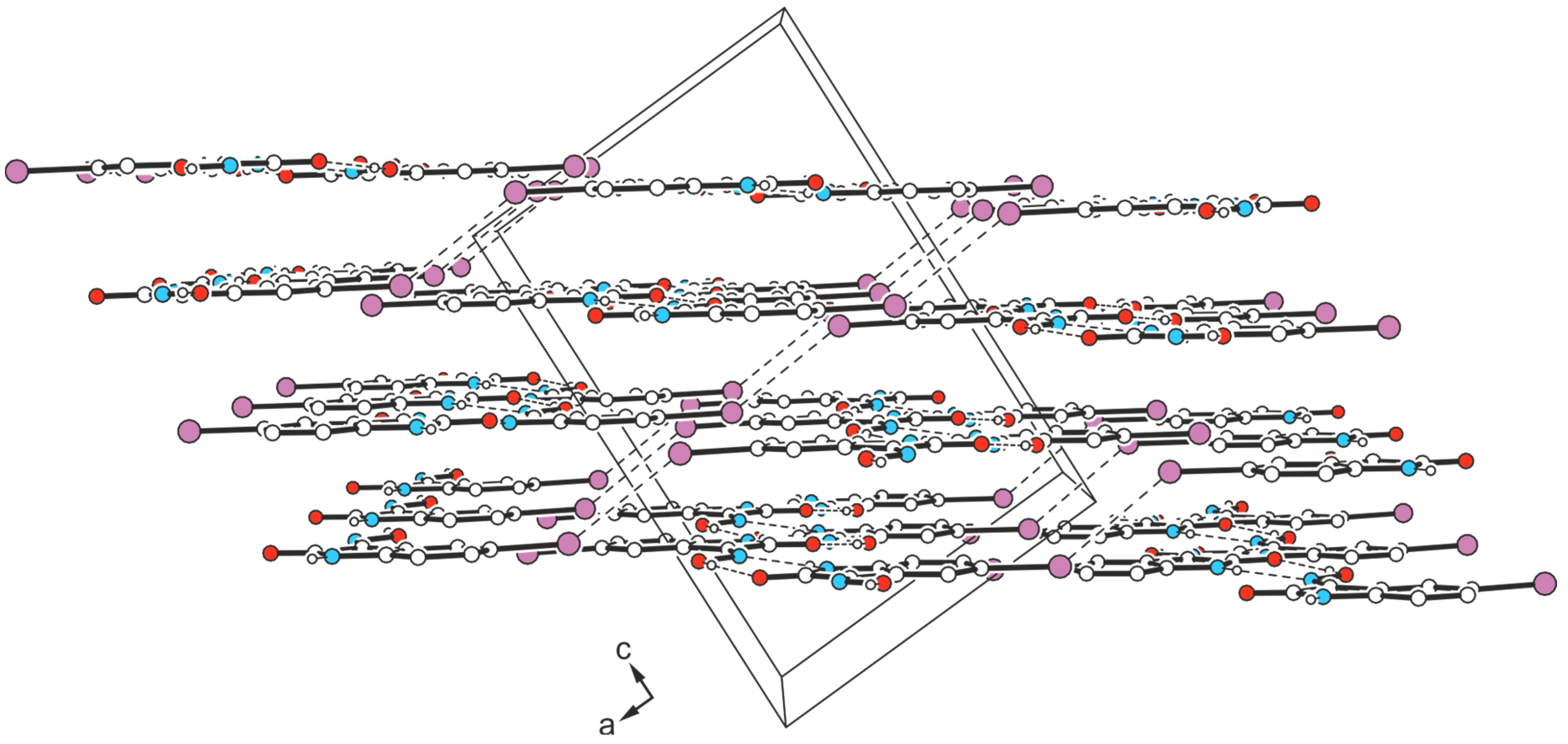
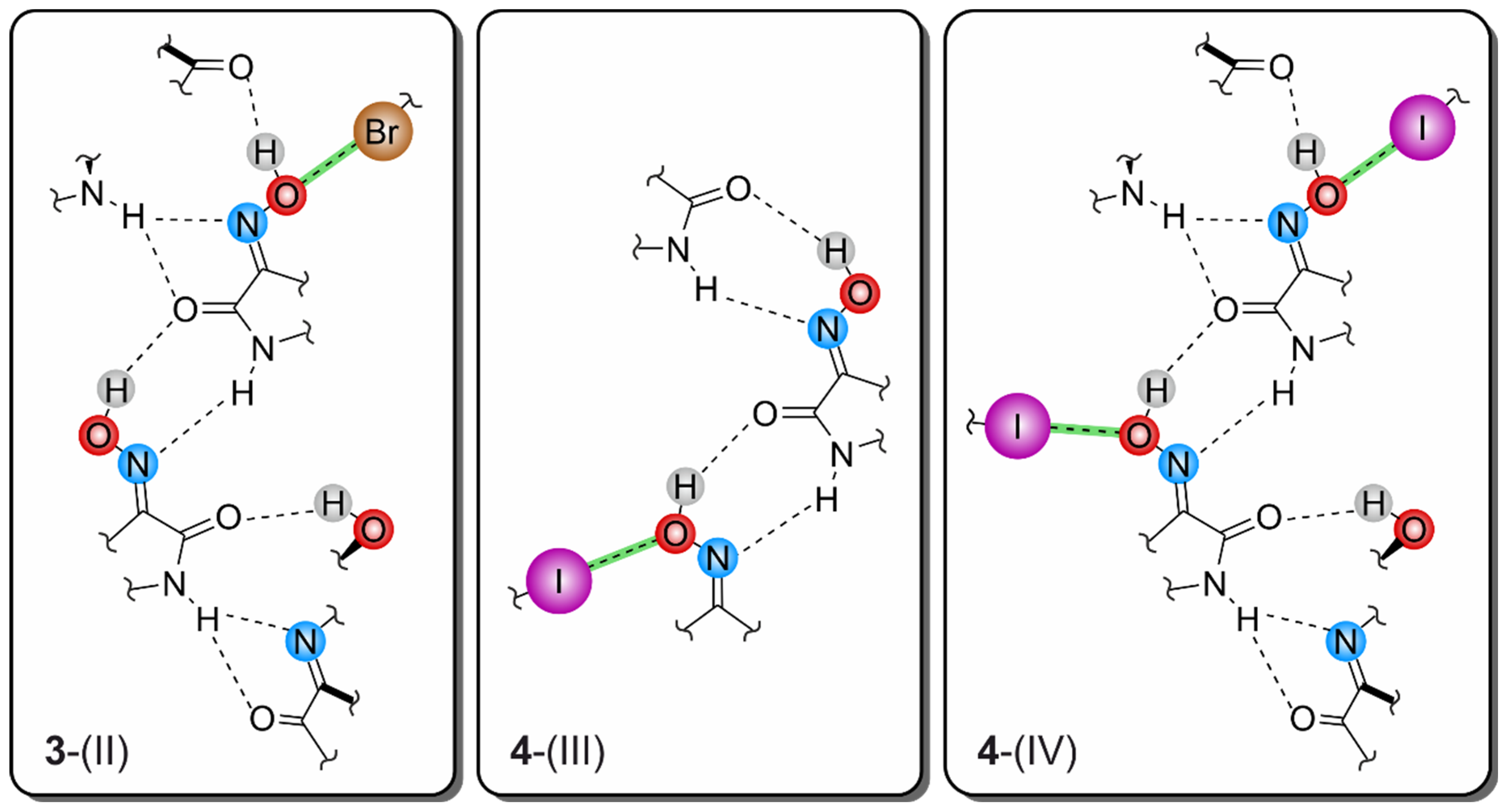
2.3.2. Polymorph 3-(II)
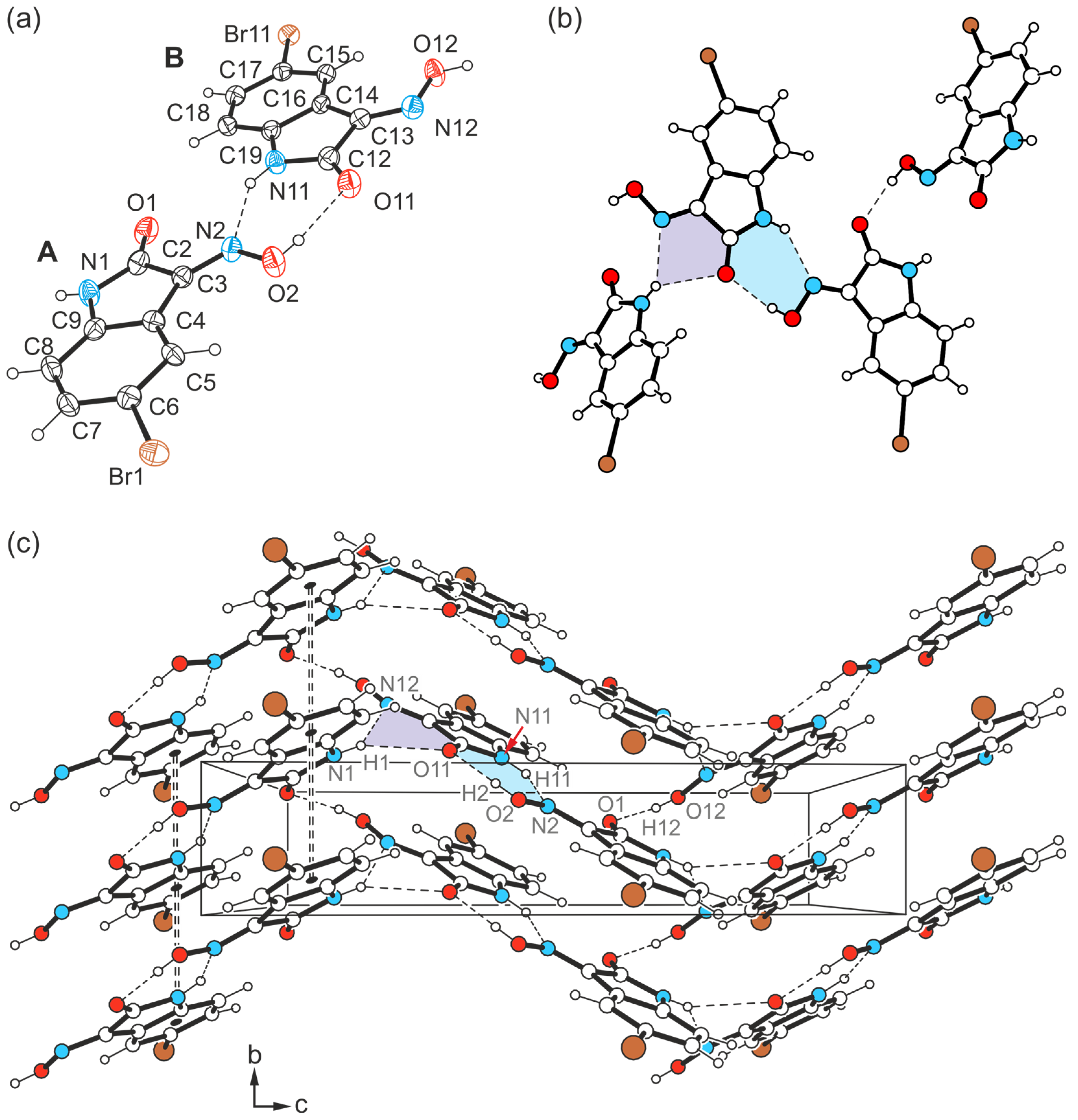
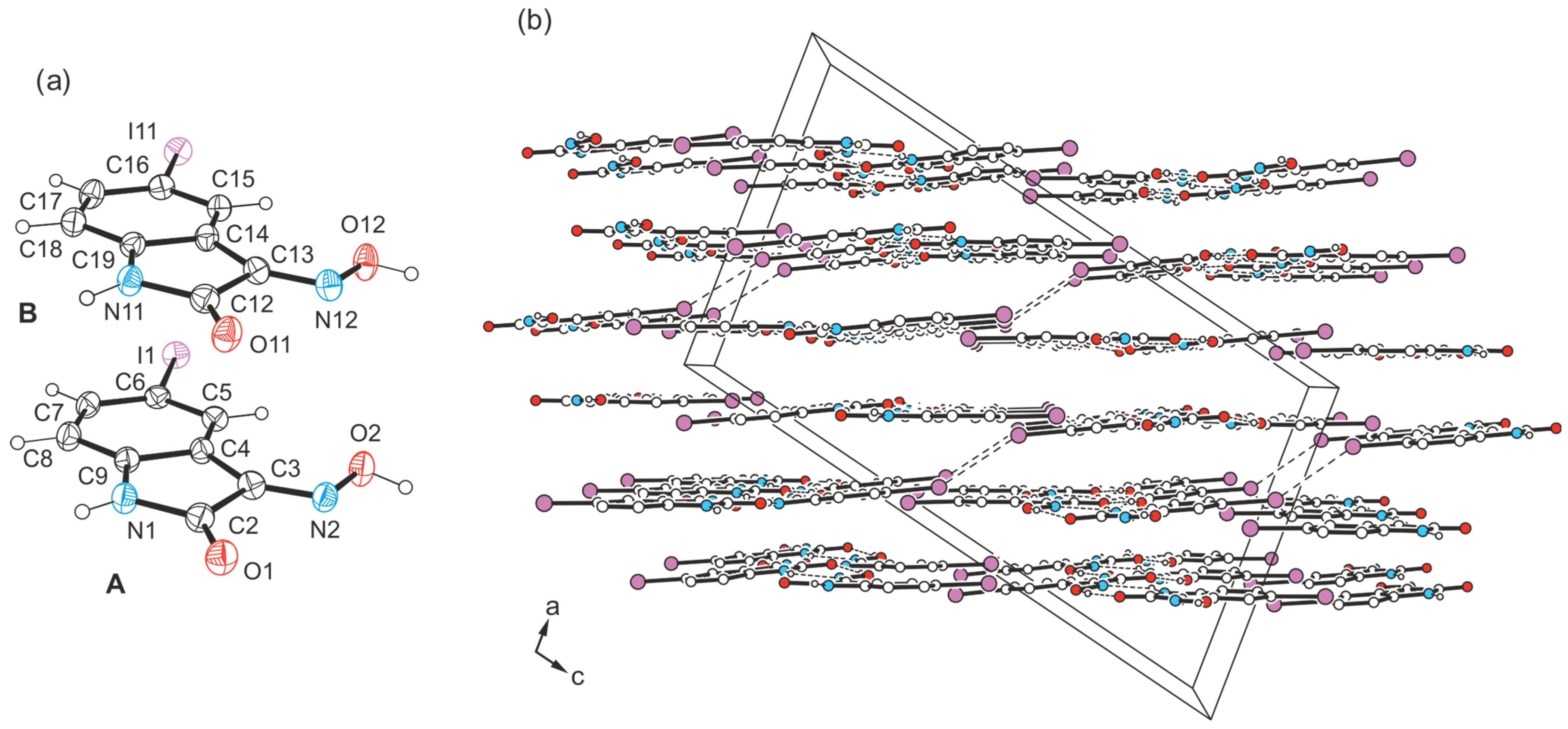
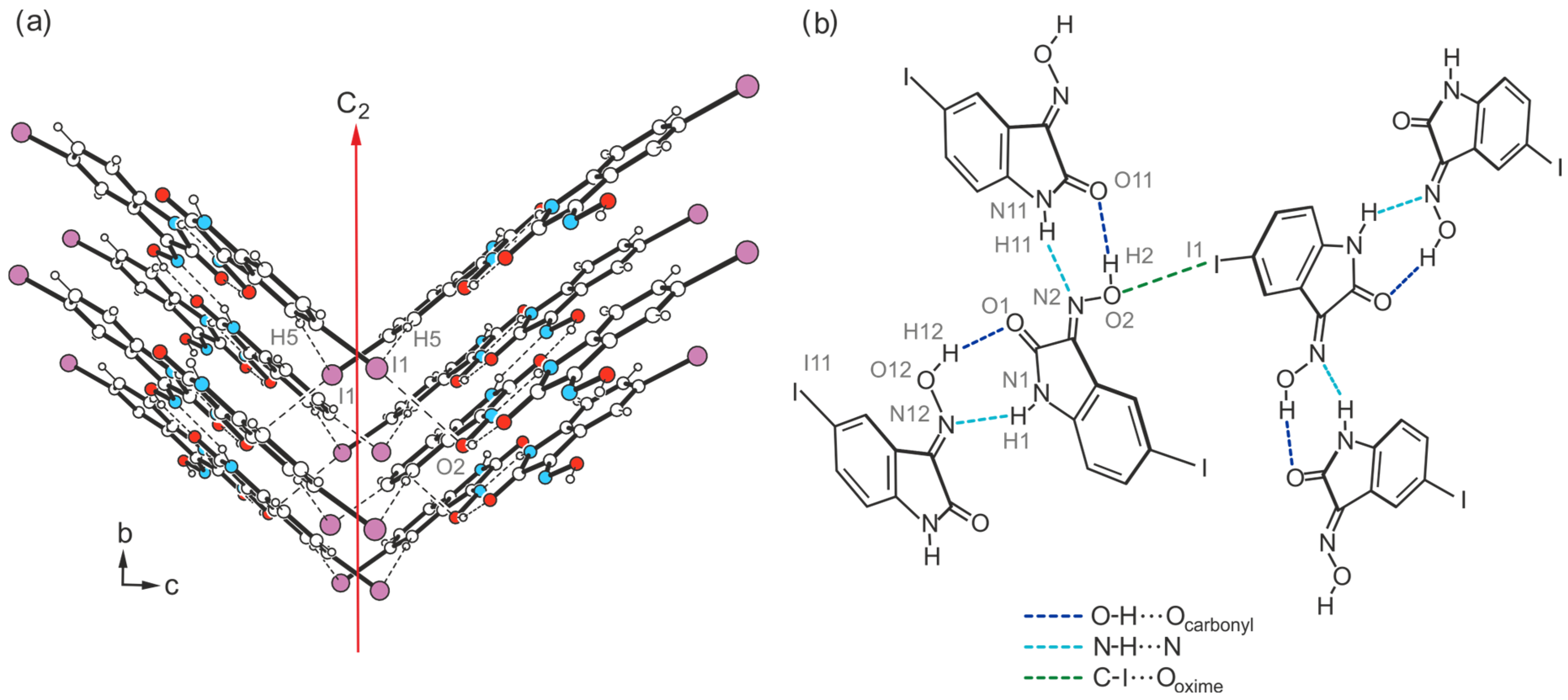
2.4. Crystal Structures of 5-Iodo-1H-isatin-3-oxime (4)
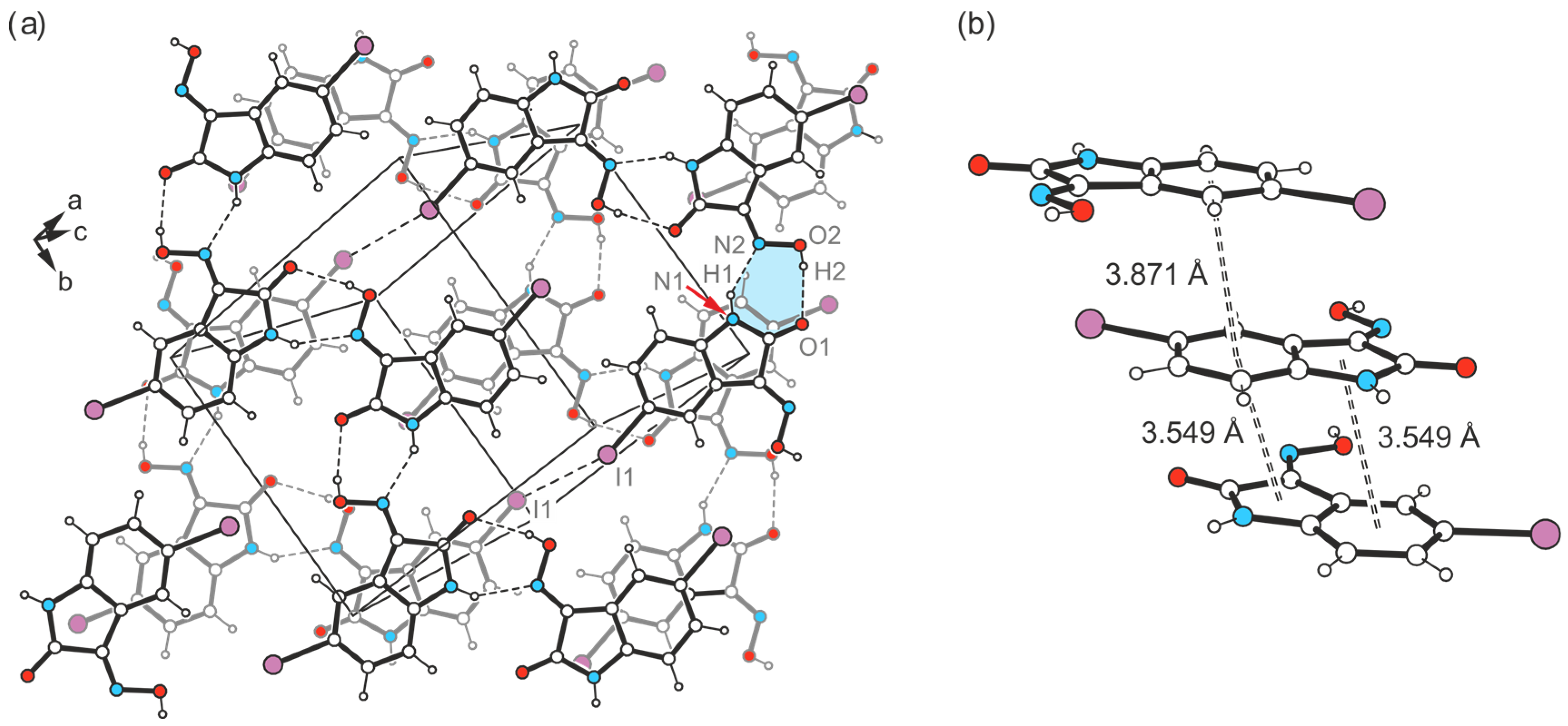
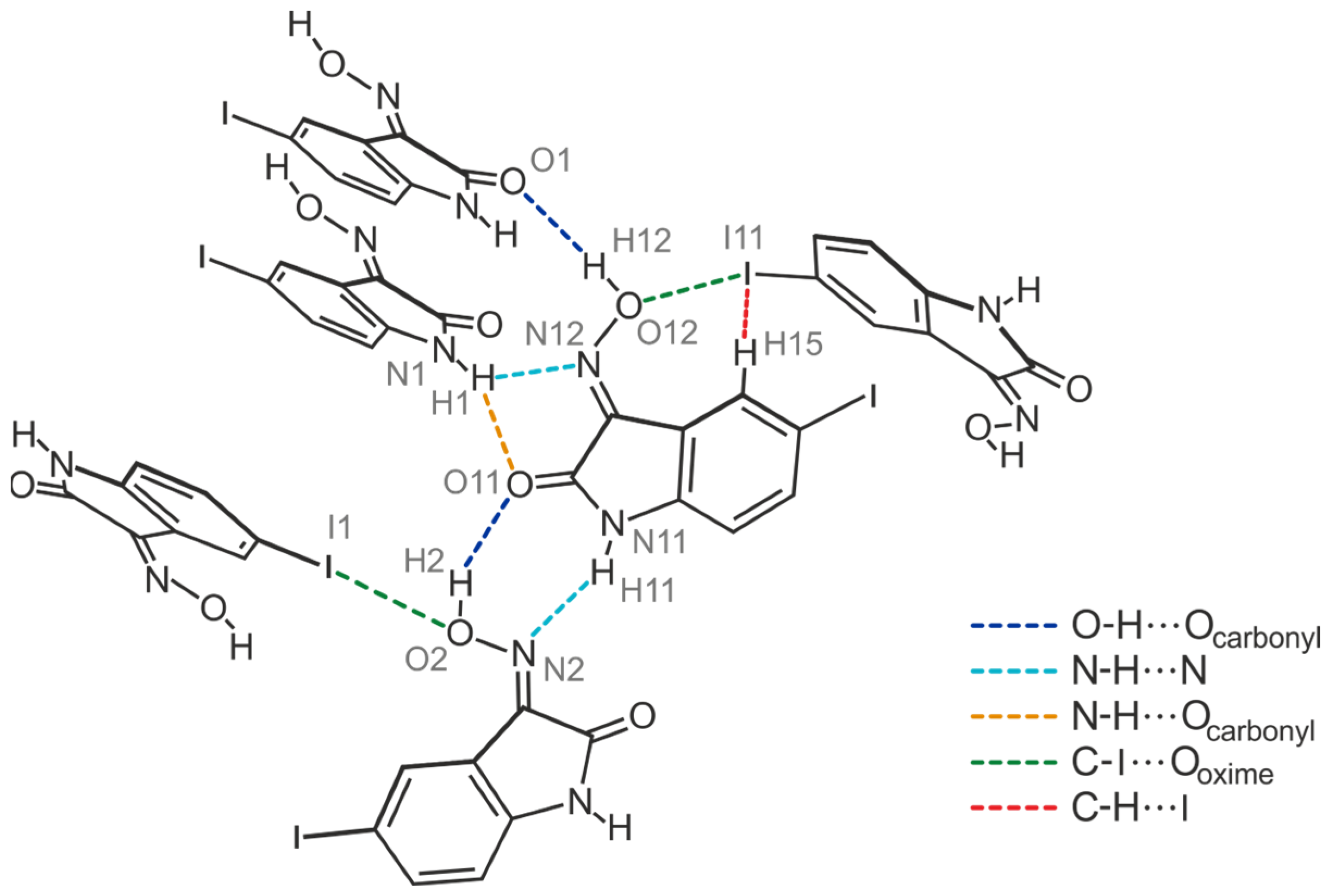
2.4.1. Polymorph 4-(I)
2.4.2. Polymorph 4-(II)
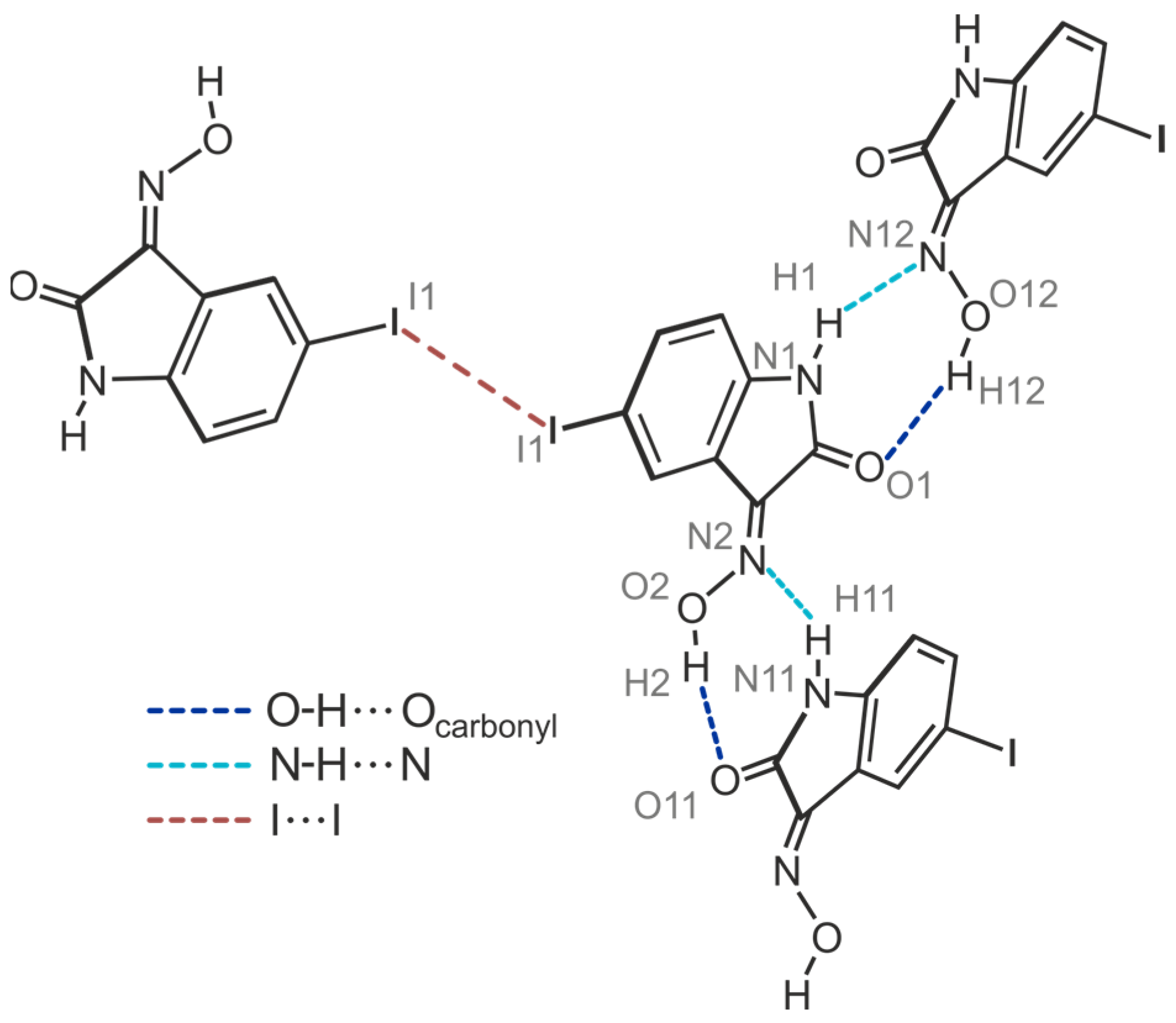
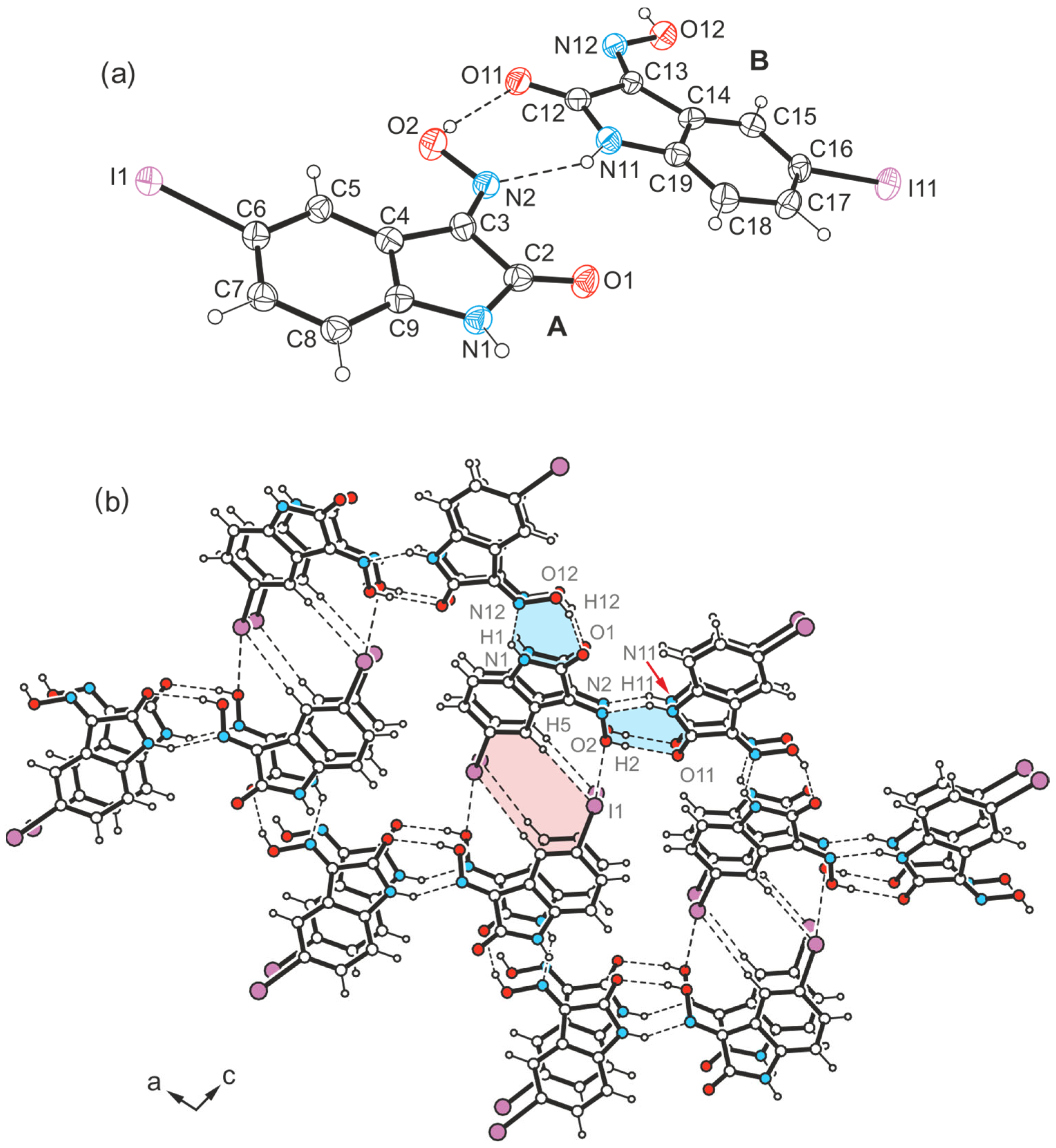
2.4.3. Polymorph 4-(III)
2.4.4. Polymorph 4-(IV)
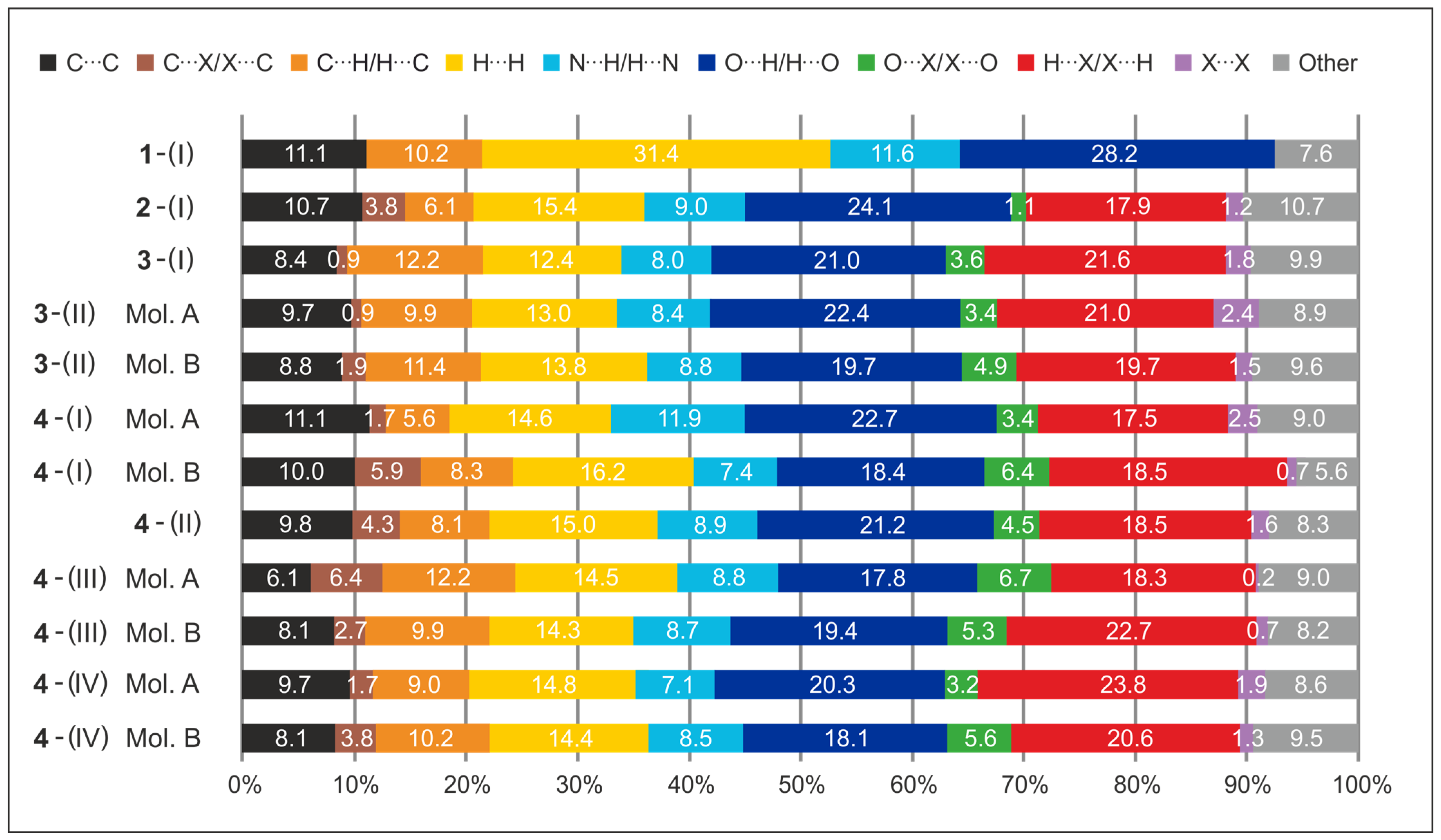
2.5. Hirshfeld Surface Analysis
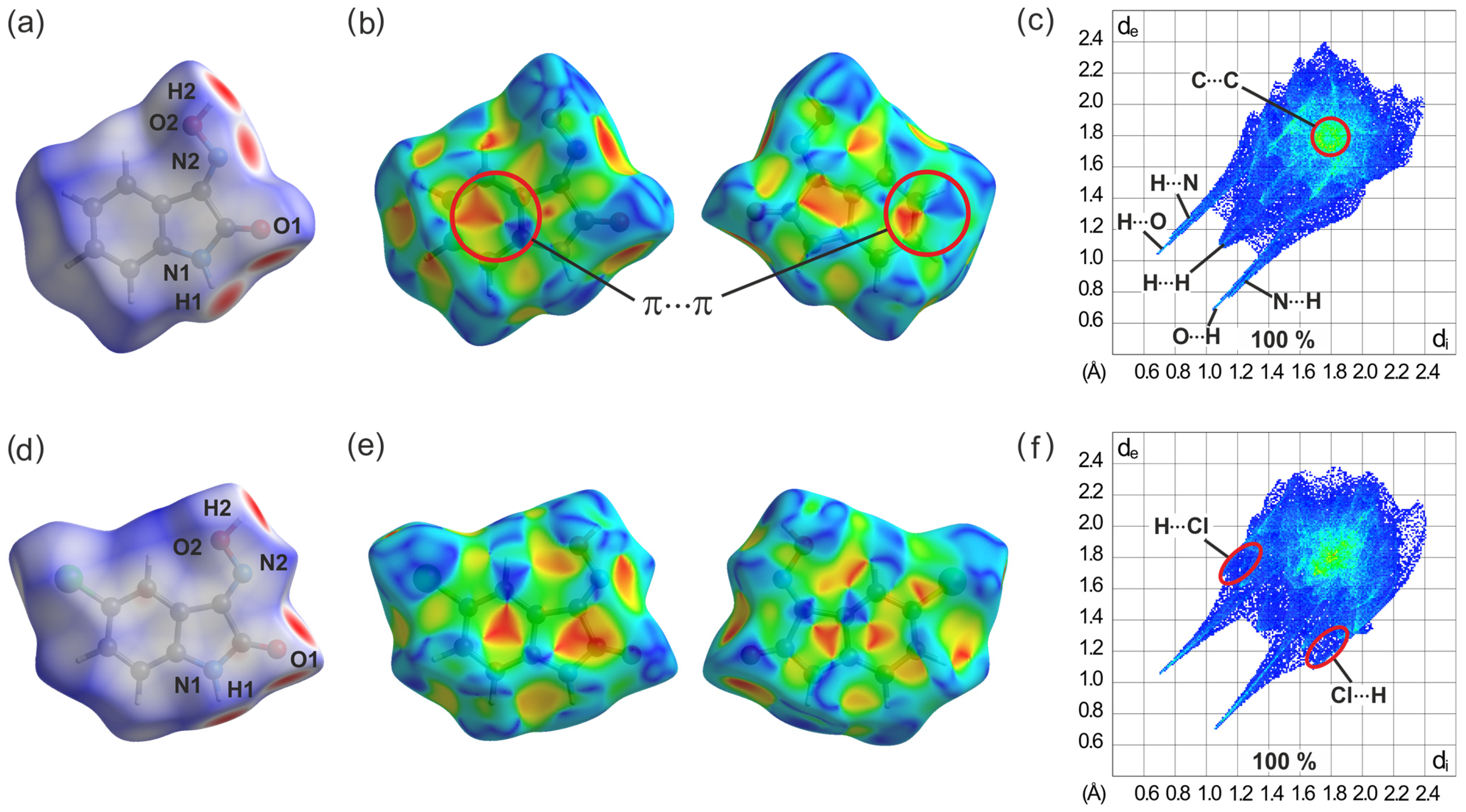
2.5.1. Hirshfeld Surface Analysis of the Crystal Structures of 1 and 2
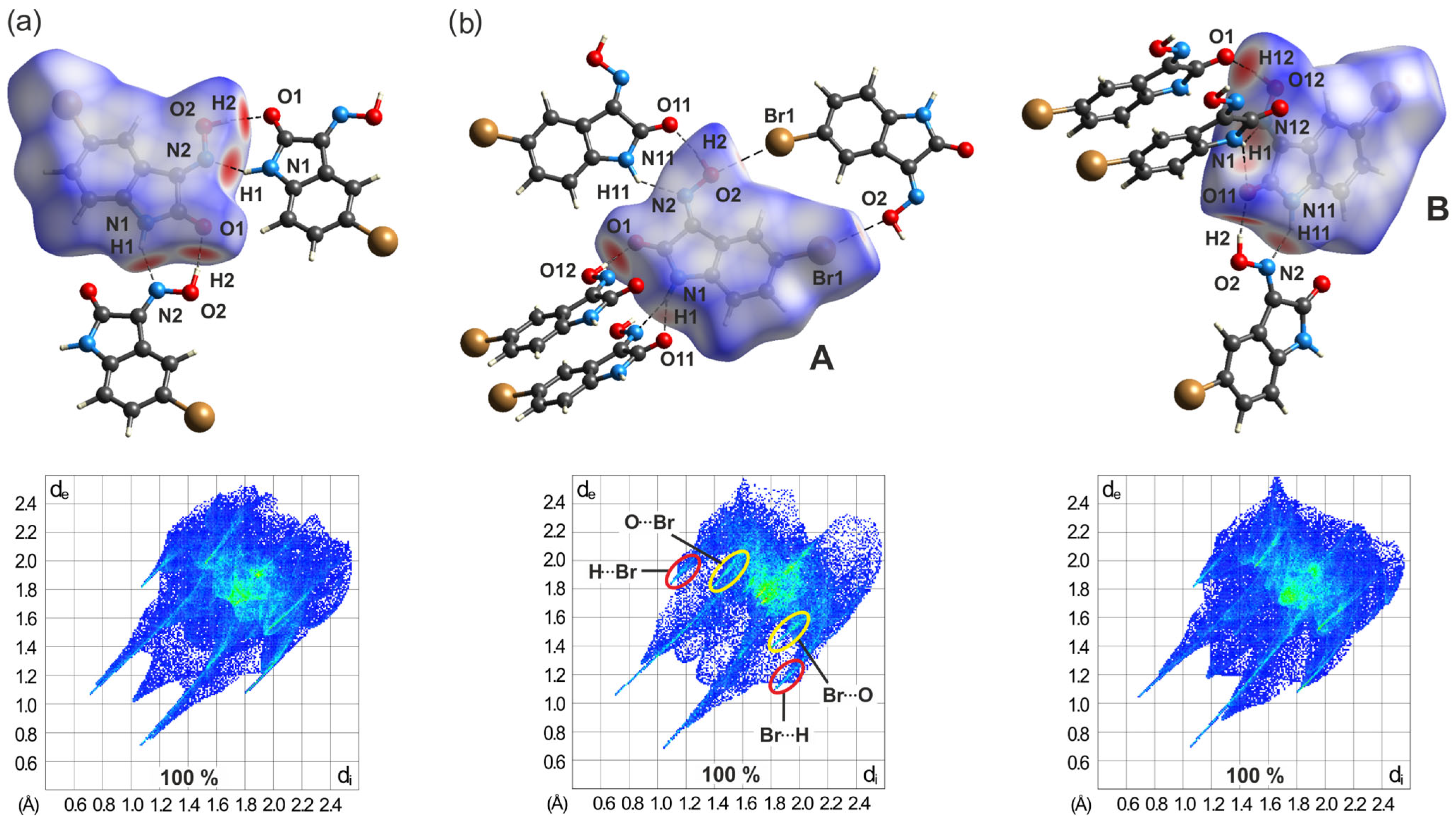
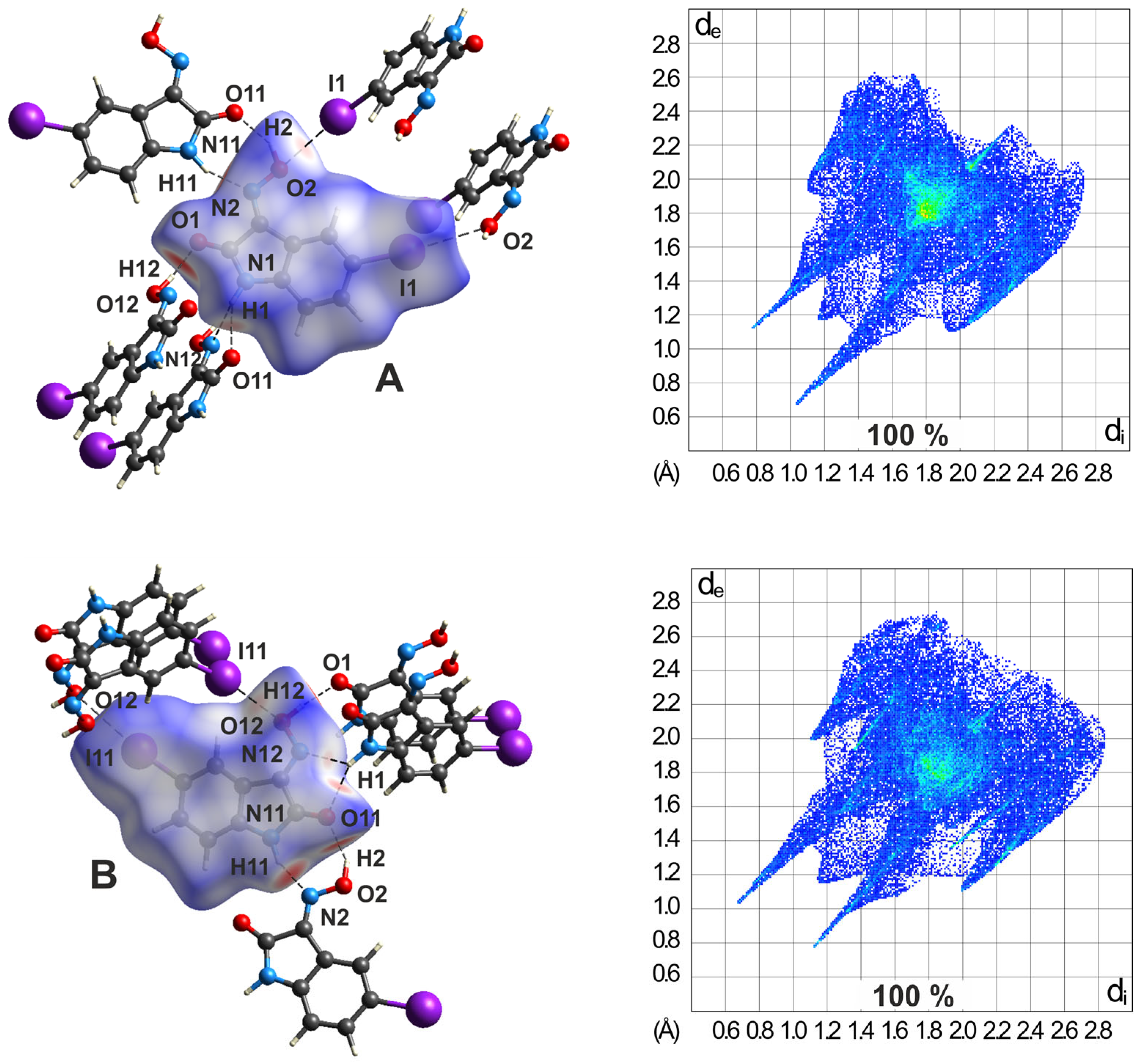
2.5.2. Hirshfeld Surface Analysis of the Polymorphs of 3
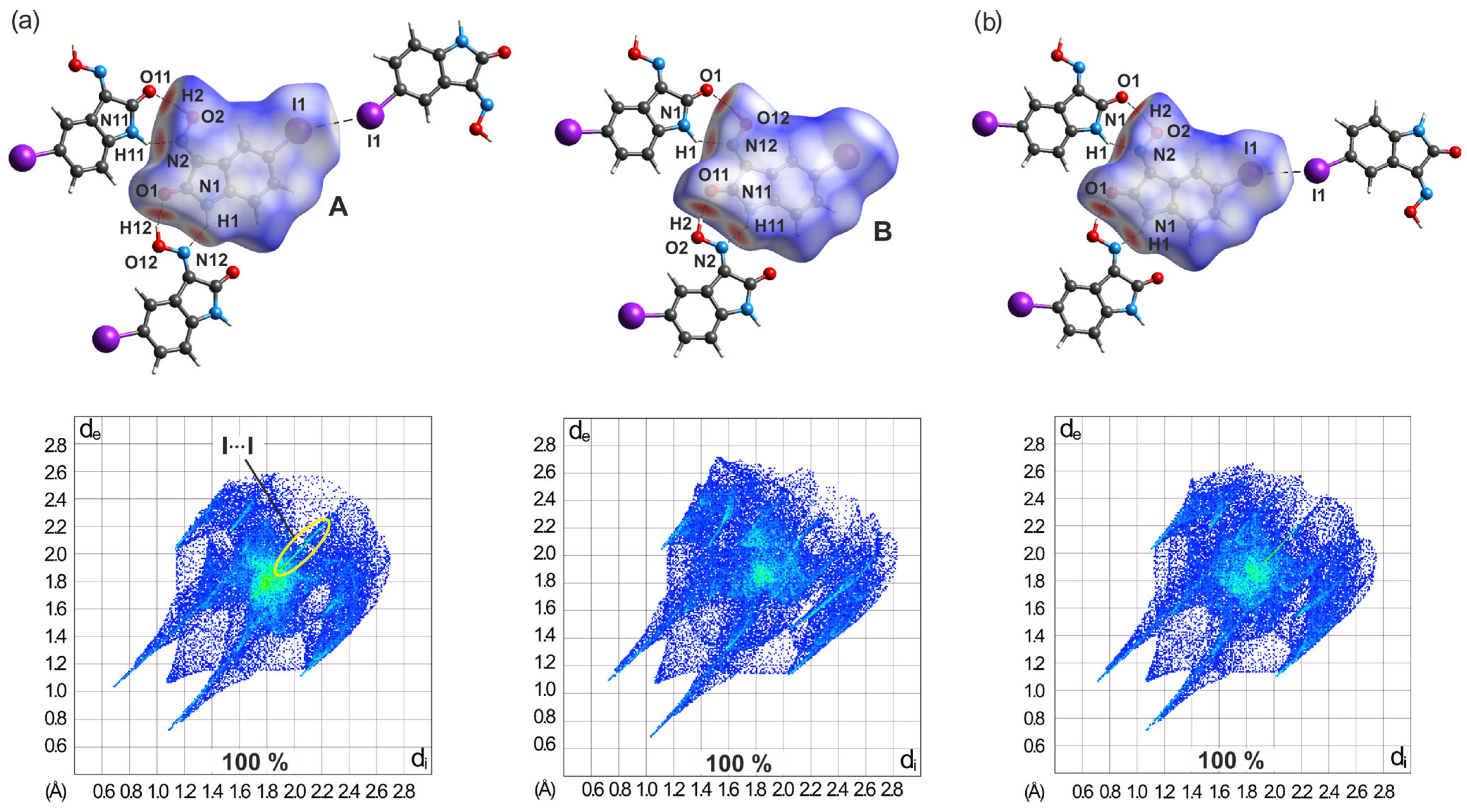
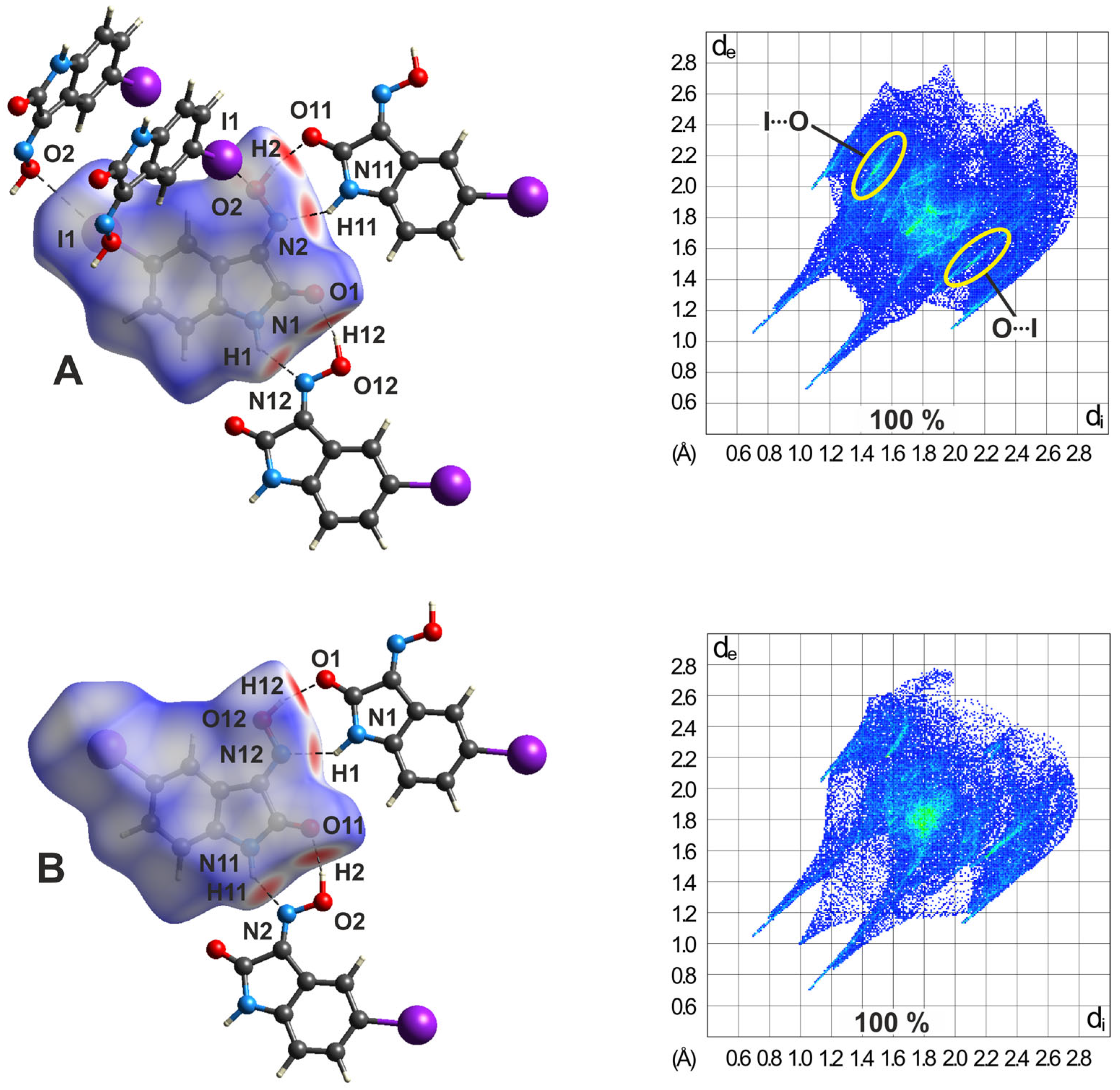
2.5.3. Hirshfeld Surfaces of the Polymorphs of 4
3. Conclusions
4. Materials and Methods
4.1. Crystal Structure Analysis
4.2. Synthesis of Compounds 1–4
4.2.1. Synthesis of Compounds 1–3
4.2.2. Synthesis of 5-Iodo-1H-isatin-3-oxime (4)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cavallo, G.; Metrangolo, P.; Milani, R.; Pilati, T.; Priimagi, A.; Resnati, G.; Terraneo, G. The Halogen Bond. Chem. Rev. 2016, 116, 2478–2601. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Beer, P.D. Halogen Bonding Anion Recognition. Chem. Commun. 2016, 52, 8645–8658. [Google Scholar] [CrossRef] [PubMed]
- Gilday, L.C.; Robinson, S.W.; Barendt, T.A.; Langton, M.J.; Mullaney, B.R.; Beer, P.D. Halogen Bonding in Supramolecular Chemistry. Chem. Rev. 2015, 115, 7118–7195. [Google Scholar] [CrossRef] [PubMed]
- Politzer, P.; Murray, J.S. Halogen Bonding: An Interim Discussion. ChemPhysChem 2013, 14, 278–294. [Google Scholar] [CrossRef] [PubMed]
- Wolters, L.P.; Schyman, P.; Pavan, M.J.; Jorgensen, W.L.; Bickelhaupt, F.M.; Kozuch, S. The Many Faces of Halogen Bonding: A Review of Theoretical Models and Methods. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2014, 4, 523–540. [Google Scholar] [CrossRef]
- Mukherjee, A.; Tothadi, S.; Desiraju, G.R. Halogen Bonds in Crystal Engineering: Like Hydrogen Bonds yet Different. Acc. Chem. Res. 2014, 47, 2514–2524. [Google Scholar] [CrossRef] [PubMed]
- Priimagi, A.; Cavallo, G.; Metrangolo, P.; Resnati, G. The Halogen Bond in the Design of Functional Supramolecular Materials: Recent Advances. Acc. Chem. Res. 2013, 46, 2686–2695. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Dubois, P. Halogen Bonding at Work: Recent Applications in Synthetic Chemistry and Materials Science. CrystEngComm 2013, 15, 3058–3071. [Google Scholar] [CrossRef]
- Metrangolo, P.; Meyer, F.; Pilati, T.; Resnati, G. Halogen Bonding in Supramolecular Chemistry. Angew. Chem. Int. Ed. 2008, 47, 6114–6127. [Google Scholar] [CrossRef]
- Metrangolo, P.; Neukirch, H.; Pilati, T.; Resnati, G. Halogen Bonding Based Recognition Processes: A World Parallel to Hydrogen Bonding. Acc. Chem. Res. 2005, 38, 386–395. [Google Scholar] [CrossRef]
- Ivanov, D.M.; Bokach, N.A.; Kukushkin, V.Y.; Frontera, A. Metal Centers as Nucleophiles: Oxymoron of Halogen Bond-Involving Crystal Engineering. Chem. Eur. J. 2022, 28, e202103173. [Google Scholar] [CrossRef]
- Teyssandier, J.; Mali, K.S.; De Feyter, S. Halogen Bonding in Two-Dimensional Crystal Engineering. ChemistryOpen 2020, 9, 225–241. [Google Scholar] [CrossRef]
- Sharber, S.A.; Mullin, W.J.; Thomas, S.W., III. Bridging the Void: Halogen Bonding and Aromatic Interactions to Program Luminescence and Electronic Properties of π-Conjugated Materials in the Solid State. Chem. Mater. 2021, 33, 6640–6661. [Google Scholar] [CrossRef]
- Sutar, R.L.; Huber, S.M. Catalysis of Organic Reactions through Halogen Bonding. ACS Catal. 2019, 9, 9622–9639. [Google Scholar] [CrossRef]
- Troff, R.W.; Mäkelä, T.; Topić, F.; Valkonen, A.; Raatikainen, K.; Rissanen, K. Alternative Motifs for Halogen Bonding. Eur. J. Org. Chem. 2013, 2013, 1617–1637. [Google Scholar] [CrossRef]
- Takemura, A.; McAllister, L.J.; Karadakov, P.B.; Pridmore, N.E.; Whitwood, A.C.; Bruce, D.W. Competition and Cooperation: Hydrogen and Halogen Bonding in Co-Crystals Involving 4-Iodotetrafluorobenzoic Acid, 4-Iodotetrafluorophenol and 4-Bromotetrafluorophenol. CrystEngComm 2014, 16, 4254–4264. [Google Scholar] [CrossRef]
- Robertson, C.C.; Perutz, R.N.; Brammer, L.; Hunter, C.A. A Solvent-Resistant Halogen Bond. Chem. Sci. 2014, 5, 4179–4183. [Google Scholar] [CrossRef]
- Pinfold, H.; Sacchi, M.; Pattison, G.; Costantini, G. Determining the Relative Structural Relevance of Halogen and Hydrogen Bonds in Self-Assembled Monolayers. J. Phys. Chem. C 2021, 125, 27784–27792. [Google Scholar] [CrossRef]
- Rowe, R.K.; Ho, P.S. Relationships between Hydrogen Bonds and Halogen Bonds in Biological Systems. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2017, 73, 255–264. [Google Scholar] [CrossRef]
- Voth, A.R.; Khuu, P.; Oishi, K.; Ho, P.S. Halogen Bonds as Orthogonal Molecular Interactions to Hydrogen Bonds. Nat. Chem. 2009, 1, 74–79. [Google Scholar] [CrossRef]
- Vasylyeva, V.; Nayak, S.K.; Terraneo, G.; Cavallo, G.; Metrangolo, P.; Resnati, G. Orthogonal Halogen and Hydrogen Bonds Involving a Peptide Bond Model. CrystEngComm 2014, 16, 8102–8105. [Google Scholar] [CrossRef]
- Riel, A.M.S.; Rowe, R.K.; Ho, E.N.; Carlsson, A.C.C.; Rappé, A.K.; Berryman, O.B.; Ho, P.S. Hydrogen Bond Enhanced Halogen Bonds: A Synergistic Interaction in Chemistry and Biochemistry. Acc. Chem. Res. 2019, 52, 2870–2880. [Google Scholar] [CrossRef]
- Decato, D.A.; Riel, A.M.S.; May, J.H.; Bryantsev, V.S.; Berryman, O.B. Theoretical, Solid-State, and Solution Quantification of the Hydrogen Bond-Enhanced Halogen Bond. Angew. Chem. Int. Ed. 2021, 60, 3685–3692. [Google Scholar] [CrossRef]
- Aakeröy, C.B.; Panikkattu, S.; Chopade, P.D.; Desper, J. Competing Hydrogen-Bond and Halogen-Bond Donors in Crystal Engineering. CrystEngComm 2013, 15, 3125–3136. [Google Scholar] [CrossRef]
- Aakeröy, C.B.; Sinha, A.S.; Epa, K.N.; Chopade, P.D.; Smith, M.M.; Desper, J. Structural Chemistry of Oximes. Cryst. Growth Des. 2013, 13, 2687–2695. [Google Scholar] [CrossRef]
- Rubin-Preminger, J.M.; Englert, U. Halogen Bonding in Substituted Cobaloximes. Inorg. Chim. Acta 2009, 362, 1135–1142. [Google Scholar] [CrossRef]
- Bedeković, N.; Martinez, V.; Topić, E.; Stilinović, V.; Cinčić, D. Cobaloximes as Building Blocks in Halogen-Bonded Cocrystals. Materials 2020, 13, 2370. [Google Scholar] [CrossRef]
- Mazik, M.; Buthe, A. Oxime-Based Receptors for Mono- and Disaccharides. J. Org. Chem. 2007, 72, 8319–8326. [Google Scholar] [CrossRef] [PubMed]
- Mazik, M.; Buthe, A. Highly effective receptors showing Di- vs. Monosaccharide Preference. Org. Biomol. Chem. 2008, 6, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Bruton, E.A.; Brammer, L.; Pigge, F.C.; Aakeröy, C.B.; Leinen, D.S. Hydrogen Bond Patterns in Aromatic and Aliphatic Dioximes. N. J. Chem. 2003, 27, 1084–1094. [Google Scholar] [CrossRef]
- Etter, M.C.; MacDonald, J.C.; Bernstein, J. Graph-set Analysis of Hydrogen-bond Patterns in Organic Crystals. Acta Crystallogr. Sect. B 1990, 46, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.L. Patterns in Hydrogen Bonding: Functionality and Graph Set Analysis in Crystals. Angew. Chemie Int. Ed. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Maurin, J.K. Resonance-Assisted Hydrogen Bonds Between Oxime and Carboxyl Groups. II. The Tetrameric Structure of Pyruvic Acid Oxime. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1995, 51, 2111–2113. [Google Scholar] [CrossRef]
- Maurin, J.K. Oxime-Carboxyl Hydrogen Bonds: The Preferred Interaction Determining Crystal Packing of “Carboxyoximes”. Acta Crystallogr. Sect. B Struct. Sci. 1998, 54, 866–871. [Google Scholar] [CrossRef]
- Kubicki, M.; Borowiak, T.; Antkowiak, W.Z. Hydrogen Bonds in “Carboxyoximes”: The Case of Bornane Derivatives. Z. Naturforsch. B 2000, 55, 677–684. [Google Scholar] [CrossRef]
- Maurin, J.K. Structure of (E)-4-Benzoylbutyramide Oxime. Acta Cryst. 1992, C48, 1819–1820. [Google Scholar] [CrossRef]
- Mazik, M.; Bläser, D.; Boese, R. Programmed Construction of Discrete Self-Assembled Cyclic Aggregates. Chem. Eur. J. 2000, 6, 2865–2873. [Google Scholar] [CrossRef]
- Mazik, M.; Bläser, D.; Boese, R. α,ß -Unsaturated Ketoximes Carrying a Terminal Pyridine or Quinoline Subunit as Building Blocks for Supramolecular Syntheses. J. Org. Chem. 2005, 70, 9115–9122. [Google Scholar] [CrossRef]
- Mazik, M.; Bläser, D.; Boese, R. New Helical Hydrogen-Bonded Assemblies Forming Channel-Inclusion Complexes. Tetrahedron Lett. 1999, 40, 4783–4786. [Google Scholar] [CrossRef]
- Aakeröy, C.B.; Beatty, A.M.; Leinen, D.S. Syntheses and Crystal Structures of New “Extended” Building Blocks for Crystal Engineering: (Pyridylmethylene)Aminoacetophenone Oxime Ligands. Cryst. Growth Des. 2001, 1, 47–52. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sect. B 2016, 72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Bruno, I.J.; Cole, J.C.; Edgington, P.R.; Kessler, M.; Macrae, C.F.; McCabe, P.; Pearson, J.; Taylor, R. New Software for Searching the Cambridge Structural Database and Visualizing Crystal Structures. Acta Crystallogr. Sect. B 2002, 58, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Haleblian, J.; McCrone, W. Pharmaceutical Applications of Polymorphism. J. Pharm. Sci. 1969, 58, 911–929. [Google Scholar] [CrossRef] [PubMed]
- Dunitz, J.D.; Bernstein, J. Disappearing Polymorphs. Acc. Chem. Res. 1995, 28, 193–200. [Google Scholar] [CrossRef]
- Bernstein, J.; Davey, R.J.; Henck, J.-O. Concomitant Polymorphs. Angew. Chem. Int. Ed. 1999, 38, 3440–3461. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Spackman, M.A.; Mitchell, A.S. Novel Tools for Visualizing and Exploring Intermolecular Interactions in Molecular Crystals. Acta Crystallogr. Sect. B Struct. Sci. 2004, 60, 627–668. [Google Scholar] [CrossRef] [PubMed]
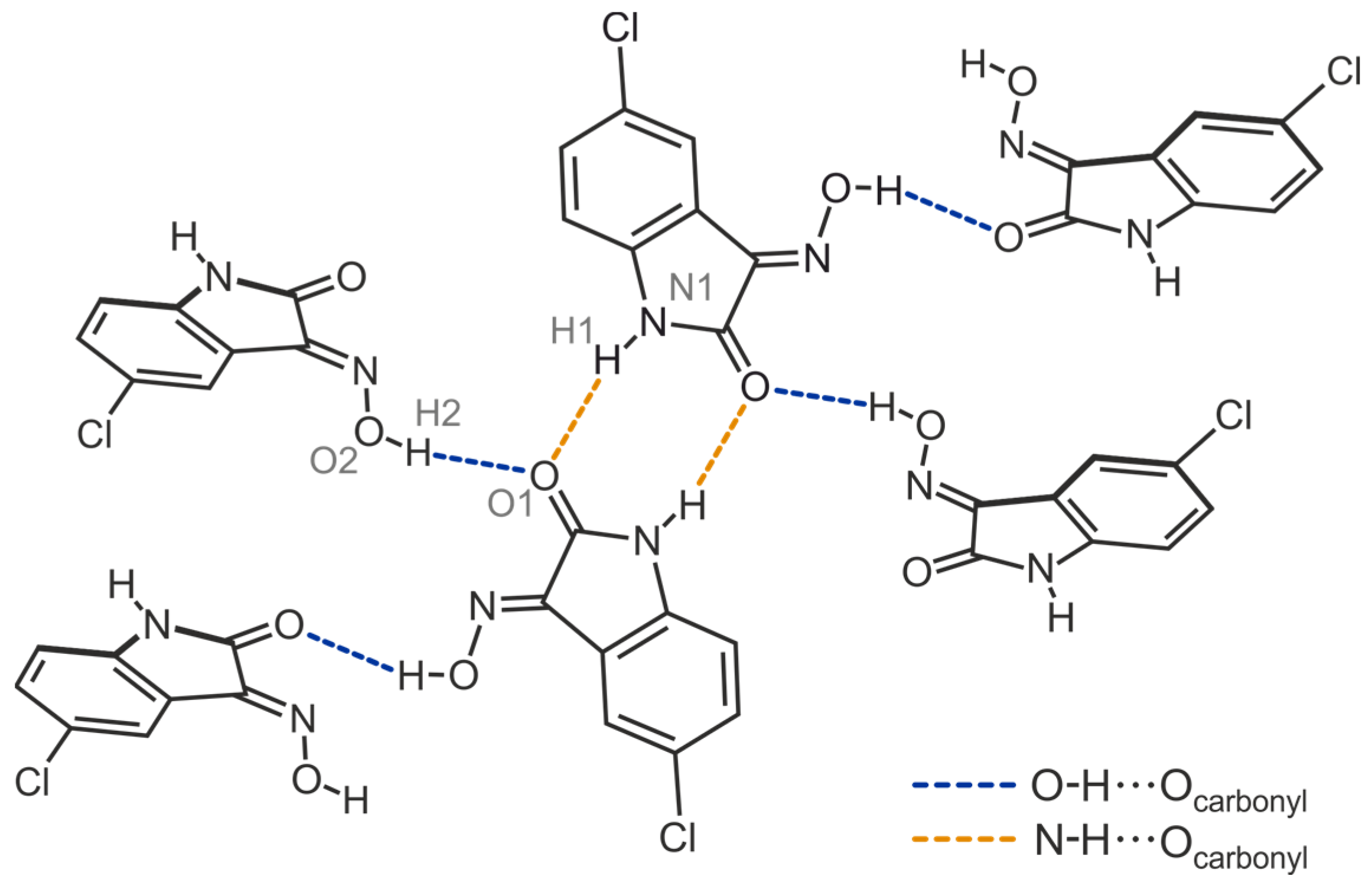
- Mo, X.L.; Chen, C.H.; Liang, C.; Mo, D.L. Copper-Catalyzed Carbonyl Group Controlled Coupling of Isatin Oximes with Arylboronic Acids To Prepare N-Aryloxindole Nitrones. Eur. J. Org. Chem. 2018, 2018, 150–159. [Google Scholar] [CrossRef]
- Dance, I. π-π Interactions: Theory and Scope. In Encyclopedia of Supramolecular Chemistry; Atwood, J.L., Steed, J.M., Eds.; Dekker: New York, NY, USA, 2004; pp. 1076–1092. [Google Scholar]
- James, S.L. π-π Stacking as a Crystal Engineering Tool. In Encyclopedia of Supramolecular Chemistry; Atwood, J.L., Steed, J.W., Eds.; Dekker: New York, NY, USA, 2004; pp. 1093–1099. [Google Scholar]
- Salonen, L.M.; Ellermann, M.; Diederich, F. Aromatic Rings in Chemical and Biological Recognition: Energetics and Structures. Angew. Chem. Int. Ed. 2011, 50, 4808–4842. [Google Scholar] [CrossRef]
- Martins, B.B.; Gervini, V.C.; Pires, F.C.; Bortoluzzi, A.J.; de Oliveira, A.B. (3Z)-5-Chloro-3-(Hydroxyimino)Indolin-2-One. IUCrData 2016, 1, 161505–161506. [Google Scholar] [CrossRef]
- Steiner, T.; Desiraju, G.R. The Weak Hydrogen Bond in Chemistry and Structural Biology; Oxford University Press: New York, NY, USA, 1999. [Google Scholar]
- Bondi, A. Van Der Waals Volumes and Radii. J. Phys. Chem. 1964, 68, 441–451. [Google Scholar] [CrossRef]
- Saha, B.K.; Rather, S.A.; Saha, A. Interhalogen Interactions in the Light of Geometrical Correction. Cryst. Growth Des. 2016, 16, 3059–3062. [Google Scholar] [CrossRef]
- Pedireddi, V.R.; Reddy, D.S.; Goud, B.S.; Craig, D.C.; Rae, A.D.; Desiraju, G.R. The Nature of Halogen...Halogen Interactions and the Crystal Structure of 1,3,5,7-Tetraiodoadamantane. J. Chem. Soc. Perkin Trans. 2 1994, 2, 2353–2360. [Google Scholar] [CrossRef]
- Awwadi, F.F.; Willett, R.D.; Peterson, K.A.; Twamley, B. The Nature of Halogen...Halogen Synthons: Crystallographic and Theoretical Studies. Chemistry 2006, 12, 8952–8960. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.A.; Saeed, R.R.A.; Shehata, M.N.I.; Ahmed, M.N.; Shawky, A.M.; Khowdiary, M.M.; Elkaeed, E.B.; Soliman, M.E.S.; Moussa, N.A.M. Type I–IV Halogen⋯Halogen Interactions: A Comparative Theoretical Study in Halobenzene⋯Halobenzene Homodimers. Int. J. Mol. Sci. 2022, 23, 3114. [Google Scholar] [CrossRef] [PubMed]
- Spackman, M.A.; Byrom, P.G. A Novel Definition of a Molecule in a Crystal. Chem. Phys. Lett. 1997, 4, 215–220. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Mitchell, A.S.; Spackman, M.A. Hirshfeld Surfaces: A New Tool for Visualising and Exploring Molecular Crystals. Chem. Eur. J. 1998, 4, 2136–2141. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards Quantitative Analysis of Intermolecular Interactions with Hirshfeld Surfaces. Chem. Commun. 2007, 3814–3816. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting Intermolecular Interactions in Molecular Crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld Surface Analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A Program for Hirshfeld Surface Analysis, Visualization and Quantitative Analysis of Molecular Crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Fabbiani, F.P.A.; Spackman, M.A. Comparison of Polymorphic Molecular Crystal Structures through Hirshfeld Surface Analysis. Cryst. Growth Des. 2007, 7, 755–769. [Google Scholar] [CrossRef]
- Baeyer, A.; Comstock, W. Über Oxindol Und Isatoxim. Ber. Dtsch. Chem. Ges. 1883, 16, 1704–1711. [Google Scholar] [CrossRef]
- Borsche, W.; Sander, W. Untersuchungen Über Isatin Und Seine Derivate II: Isatoxim -> o-Cyan-Phenyl-Isocyanat Und Verwandte Reaktionen. Chem. Ber. 1914, 47, 2815–2826. [Google Scholar] [CrossRef]
- Jeankumar, V.U.; Alokam, R.; Sridevi, J.P.; Suryadevara, P.; Matikonda, S.S.; Peddi, S.; Sahithi, S.; Alvala, M.; Yogeeswari, P.; Sriram, D. Discovery and Structure Optimization of a Series of Isatin Derivatives as Mycobacterium Tuberculosis Chorismate Mutase Inhibitors. Chem. Biol. Drug Des. 2014, 83, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Schunck, E.; Marchlewski, L. Zur Kenntniss Der Rothen Isomeren Des Indigotins Und Über Einige Derivate Des Isatins. Ber. Dtsch. Chem. Ges. 1895, 28, 539–547. [Google Scholar] [CrossRef]
- Kearney, T.; Harris, P.A.; Jackson, A.; Joule, J.A. Synthesis of Isatin-3-Oximes from 2-Nitroacetanilides. Synthesis 1992, 8, 769–772. [Google Scholar] [CrossRef]
- Wei, W.T.; Zhu, W.M.; Ying, W.W.; Wu, Y.; Huang, Y.L.; Liang, H. Metal-Free Synthesis of Isatin Oximes: Via Radical Coupling Reactions of Oxindoles with t-BuONO in Water. Org. Biomol. Chem. 2017, 15, 5254–5257. [Google Scholar] [CrossRef]
- Baeyer, A.; Knop, C.A. Untersuchungen Über Die Gruppe Des Indigblaus. Justus Liebigs Ann. Chem. 1866, 140, 1–38. [Google Scholar] [CrossRef]
- Jensen, B.S.; Jorgensen, T.D.; Ahring, P.K.; Christophersen, P.; Strobaek, D.; Teuber, L.; Olesen, S.P. Use of Isatin Derivatives as Ion Channel Activating Agents. WO Patent 00/33834, 15 June 2000. [Google Scholar]
- Zhu, G.-D.; Gandhi, V.B.; Gong, J.; Luo, Y.; Liu, X.; Shi, Y.; Guan, R.; Magnone, S.R.; Klinghofer, V.; Johnson, E.F.; et al. Discovery and SAR of Oxindole–Pyridine-Based Protein Kinase B/Akt Inhibitors for Treating Cancers. Bioorg. Med. Chem. Lett. 2006, 16, 3424–3429. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S. Beitrag Zur Kenntnis Aromatischer Nitrosokörper. Ber. Dtsch. Chem. Ges. 1883, 16, 517–523. [Google Scholar] [CrossRef]
- X-AREA, version 1.75. Software Package for Data Collection and Data Evaluation. Stoe & Cie: Darmstadt, Germany, 2015.
- X-RED, version 1.61.1. Program for Data Reduction and Absorption Correction. Stoe & Cie: Darmstadt, Germany, 2014.
- LANA, version 1.63.1. Laue Analyzer. Stoe & Cie: Darmstadt, Germany, 2015.
- Sheldrick, G.M. SHELXT—Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. Sect. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- XStep-32, version 1.07e; Stoe & Cie: Darmstadt, Germany, 2000.
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, L.J. WinGX and ORTEP for Windows: An Update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Spek, A.L. checkCIF validation ALERTS: What they mean and how to respond. Acta Crystallogr. E 2020, 76, 1–11. [Google Scholar] [CrossRef]
- Golushko, A.A.; Sandzhieva, M.A.; Ivanov, A.Y.; Boyarskaya, I.A.; Khoroshilova, O.V.; Barkov, A.Y.; Vasilyev, A.V. Reactions of 3,3,3-Trihalogeno-1-Nitropropenes with Arenes in the Superacid CF3SO3H: Synthesis of (Z)-3,3,3-Trihalogeno-1,2-diarylpropan-1-one Oximes and Study on the Reaction Mechanism. J. Org. Chem. 2018, 83, 10142–10157. [Google Scholar] [CrossRef]
- Campbell, A.; Tasker, P.; Parsons, S. CCDC 1413226: Experimental Crystal Structure Determination. 2015. [Google Scholar] [CrossRef]

| Structure | Interaction | |||
|---|---|---|---|---|
| C-X∙∙∙O | d(X∙∙∙O) | d(C∙∙∙O) | ∠(C-X∙∙∙O) | |
| D-H∙∙∙A | d(H∙∙∙O) | d(D···A) | ∠(D-H∙∙∙A) | |
| (X = Br, I; D, A = N, O) | in Å | in Å | in ° | |
| 3-(II) | C(6)-Br(1)∙∙∙O(2) | 3.183(2) | 5.038(3) | 164.1(1) |
| N(1)-H(1)···O(11) | 2.48(5) | 3.134(3) | 141(4) | |
| N(1)-H(1)···N(12) | 2.34(3) | 3.030(4) | 146(5) | |
| N(11)-H(11)···N(2) | 2.18(4) | 2.873(4) | 142(3) | |
| O(2)-H(2)···O(11) | 1.95(5) | 2.724(3) | 170(5) | |
| O(12)-H(12)···O(1) | 1.87(5) | 2.701(3) | 168(5) | |
| 4-(III) | C(6)-I(1)···O(2) | 3.383(2) | 5.564(4) | 169.4(1) |
| N(1)-H(1)···N(12) | 2.14(5) | 2.901(4) | 142(5) | |
| O(2)-H(2)···O(11) | 2.01(6) | 2.704(4) | 163(5) | |
| N(11)-H(11)···N(2) | 2.09(3) | 2.833(4) | 146(5) | |
| O(12)-H(12)···O(1) | 1.85(6) | 2.694(4) | 163(5) | |
| 4-(IV) | C(6)-I(1)···O(2) | 3.287(9) | 5.390(14) | 173.4(4) |
| C(16)-I(11)···O(12) | 3.285(9) | 5.353(14) | 168.8(4) | |
| N(1)-H(1)···N(12) | 2.55(14) | 3.081(17) | 120(12) | |
| N(1)-H(1)···O(11) | 2.41(5) | 3.205(14) | 151(11) | |
| N(11)-H(11)···N(2) | 2.03(14) | 2.859(19) | 158(14) | |
| O(2)-H(2)···O(11) | 2.01(18) | 2.758(13) | 148(17) | |
| O(12)-H(12)···O(1) | 1.84(8) | 2.665(13) | 167(16) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meier, E.; Seichter, W.; Mazik, M. Combination of Hydrogen and Halogen Bonds in the Crystal Structures of 5-Halogeno-1H-isatin-3-oximes: Involvement of the Oxime Functionality in Halogen Bonding. Molecules 2024, 29, 1174. https://doi.org/10.3390/molecules29051174
Meier E, Seichter W, Mazik M. Combination of Hydrogen and Halogen Bonds in the Crystal Structures of 5-Halogeno-1H-isatin-3-oximes: Involvement of the Oxime Functionality in Halogen Bonding. Molecules. 2024; 29(5):1174. https://doi.org/10.3390/molecules29051174
Chicago/Turabian StyleMeier, Eric, Wilhelm Seichter, and Monika Mazik. 2024. "Combination of Hydrogen and Halogen Bonds in the Crystal Structures of 5-Halogeno-1H-isatin-3-oximes: Involvement of the Oxime Functionality in Halogen Bonding" Molecules 29, no. 5: 1174. https://doi.org/10.3390/molecules29051174
APA StyleMeier, E., Seichter, W., & Mazik, M. (2024). Combination of Hydrogen and Halogen Bonds in the Crystal Structures of 5-Halogeno-1H-isatin-3-oximes: Involvement of the Oxime Functionality in Halogen Bonding. Molecules, 29(5), 1174. https://doi.org/10.3390/molecules29051174