Ganoderma lucidum-Derived Meroterpenoids Show Anti-Inflammatory Activity In Vitro
Abstract
1. Introduction
2. Results and Discussion
2.1. Structure Elucidation of the Compounds
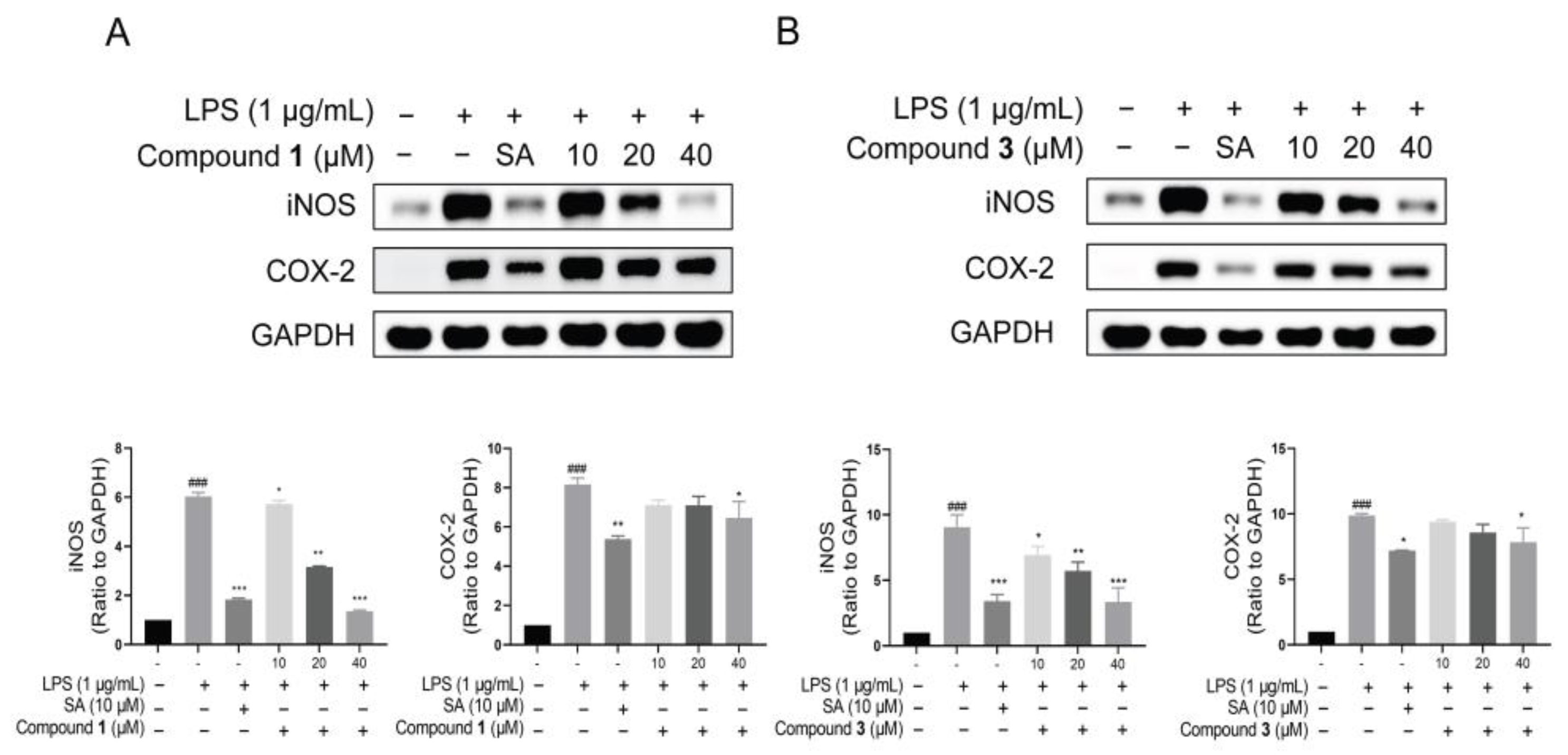
2.2. Biological Activity toward Inflammation
3. Experimental Section
3.1. General Procedures
3.2. Fungal Material
3.3. Extraction and Isolation
3.4. Compound Characterization Data
- Chizhiene A (1): yellow oils. UV (MeOH) λmax (logε) 365 (3.41), 257 (3.71), 212 (4.10) nm; HRESIMS: m/z 309.1097 [M + Na]+ (calcd for C17H18O4Na+, 309.1097); 1H and 13C NMR data; see Table 1.
- Chizhiene B (2): yellow oils. UV (MeOH) λmax (logε) 385 (3.50), 317 (4.02), 228 (3.78) nm; HRESIMS: m/z 289.1432 [M + H]+ (calcd for C17H21O4+, 289.1434); 1H and 13C NMR data; see Table 1.
- Chizhiene E (3): yellow solids. UV (MeOH) λmax (logε) 365 (3.36), 257 (3.61), 213 (3.96) nm; HRESIMS: m/z 311.1254 [M + Na]+ (calcd for C17H20O4Na+, 311.1254); 1H and 13C NMR data; see Table 1.
- Chizhiene F (4): yellow solids. [α]D20 +3.6 (c 0.28, MeOH), (+)-4; [α]D20 –14.3 (c 0.21, MeOH), (–)-4; UV (MeOH) λmax (logε) 319 (4.10), 227 (3.95) nm; HRESIMS: m/z 275.1278 [M + H]+ (calcd. for C16H19O4+, 275.1278); 1H and 13C NMR data; see Table 2.
- Chizhiene C (5): yellow solids. [α]D25 +75.0 (c 0.32, MeOH), (+)-5; [α]D25 –57.7 (c 0.26, MeOH), (–)-5; UV (MeOH) λmax (logε) 364 (4.01), 256 (4.24), 227 (4.54) nm; HRESIMS: m/z 321.1331 [M + H]+ (calcd. for C17H21O6+, 321.1333); 1H and 13C NMR data; see Table 2.
- Chizhiene D (6): yellow solids. UV (MeOH) λmax (logε) 368 (3.63), 231 (4.37), 205 (4.50) nm; HRESIMS: m/z 323.0893 [M + Na]+ (calcd. for C17H16O5Na+, 323.0890); 1H and 13C NMR data; see Table 2.
3.5. ECD Calculations for Compounds 4 and 5
3.6. Cell Culture
3.7. Cell Viability Assay
3.8. Measurement of NO Production
3.9. Western Blotting Analysis
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, S.J.; Zhang, S.Y.; Peng, B.; Tan, D.C.; Wu, M.Y.; Wei, J.C.; Wang, Y.T.; Luo, H. Ganoderma lucidum: A comprehensive review of phytochemistry, efficacy, safety and clinical study. Food Sci. Hum. Well. 2024, 13, 568–596. [Google Scholar] [CrossRef]
- Guo, J.C.; Yang, L.; Ma, Q.Y.; Ge, Y.Z.; Kong, F.D.; Zhou, L.M.; Zhang, F.; Xie, Q.Y.; Yu, Z.F.; Dai, H.F.; et al. Triterpenoids and meroterpenoids with α-glucosidase inhibitory activities from the fruiting bodies of Ganoderma australe. Bioorg. Chem. 2021, 117, 105448–105457. [Google Scholar] [CrossRef]
- Elhassaneen, Y.A.; Ragab, S.S.; Salman, M.S. The potential effects of reishi mushroom (Ganoderma lucidum) consumption on bone health indices and serum minerals profile disorders induced by CCl4 on rats. Pyrx J. Med. Plant Res. 2016, 2, 001–007. [Google Scholar]
- Venkatesh, T.; Mainkar, P.S.; Chandrasekhar, S. Diastereoselective formal synthesis of polycyclic meroterpenoid (±)-cochlearol A. J. Org. Chem. 2021, 86, 5412–5416. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Gao, X.X.; Long, G.Q.; Yang, Y.C.; Chen, G.; Hou, G.L.; Huo, X.T.; Jia, J.M.; Wang, A.H.; Hu, G.S. Lanostane-type triterpenoids from the mycelial mat of Ganoderma lucidum and their hepatoprotective activities. Phytochemistry 2022, 198, 113131–113141. [Google Scholar] [CrossRef]
- Yang, L.; Kong, D.X.; Xiao, N.; Ma, Q.Y.; Xie, Q.Y.; Guo, J.C.; Deng, C.Y.; Ma, H.X.; Hua, Y.; Dai, H.F.; et al. Antidiabetic lanostane triterpenoids from the fruiting bodies of Ganoderma weberianum. Bioorg. Chem. 2022, 127, 106025–106034. [Google Scholar] [CrossRef]
- Baby, S.; Johnson, A.J.; Govindan, B. Secondary metabolites from Ganoderma. Phytochemistry 2015, 114, 66–101. [Google Scholar] [CrossRef]
- Zhang, J.J.; Qin, F.Y.; Cheng, Y.X. Insights into Ganoderma fungi meroterpenoids opening a new era of racemic natural products in mushrooms. Med. Res. Rev. 2024; early view. [Google Scholar] [CrossRef]
- Mothana, R.A.; Jansen, R.; Julich, W.D.; Lindequist, U. Ganomycins A and B, new antimicrobial farnesyl hydroquinones from the basidiomycete Ganoderma pfeifferi. J. Nat. Prod. 2000, 63, 416–418. [Google Scholar] [CrossRef]
- Zhang, D.W.; Fan, H.L.; Zhang, W.; Li, C.J.; Luo, S.; Qin, H.B. Collective enantioselective total synthesis of (+)-sinensilactam A, (+)-lingzhilactone B and (–)-lingzhiol: Divergent reactivity of styrene. Chem. Commun. 2020, 56, 10066–10069. [Google Scholar] [CrossRef]
- Riehl, P.S.; Richardson, A.D.; Sakamoto, T.; Schindler, C.S. Eight-step enantiodivergent synthesis of (+)- and (–)-Lingzhiol. Org. Lett. 2020, 22, 290–294. [Google Scholar] [CrossRef]
- Li, X.Y.; Liu, X.Y.; Jiao, X.Z.; Yang, H.G.; Yao, Y.Y.; Xie, P. An approach to (±)-Lingzhiol. Org. Lett. 2016, 18, 1944–1946. [Google Scholar] [CrossRef]
- Gautam, S.K.; Birman, V.B. Biogenetically inspired synthesis of lingzhiol. Org. Lett. 2016, 18, 1499–1501. [Google Scholar] [CrossRef] [PubMed]
- Long, R.; Huang, J.; Shao, W.B.; Liu, S.; Lan, Y.; Gong, J.X.; Yang, Z. Asymmetric total synthesis of (–)-lingzhiol via a Rh-catalysed [3+2] cycloaddition. Nat. Commun. 2014, 5, 5707–5716. [Google Scholar] [CrossRef]
- Mehl, L.M.; Maier, M.E. A radical-based synthesis of lingzhiol. J. Org. Chem. 2017, 82, 9844–9850. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.L.; Ma, Q.Y.; Huang, S.Z.; Guo, Z.K.; Guo, J.C.; Dai, H.F.; Zhao, Y.X. Two new phenolic compounds from the fruiting bodies of Ganoderma tropicum. B. Korean Chem. Soc. 2013, 34, 884–886. [Google Scholar] [CrossRef]
- Huang, S.Z.; Cheng, B.H.; Ma, Q.Y.; Wang, Q.; Kong, F.D.; Dai, H.F.; Qiu, S.Q.; Zheng, P.Y.; Liu, Z.Q.; Zhao, Y.X. Anti-allergic prenylated hydroquinones and alkaloids from the fruiting body of Ganoderma calidophilum. RSC Adv. 2016, 6, 21139–21147. [Google Scholar] [CrossRef]
- Richardson, A.D.; Vogel, T.R.; Traficante, E.F.; Glover, K.J.; Schindler, C.S. Total synthesis of (+)-cochlearol B by an approach based on a catellani reaction and visible-light-enabled [2+2] cycloaddition. Angew. Chem. Int. Ed. 2022, 61, e202201213. [Google Scholar] [CrossRef]
- Zhang, L.J.; Xie, Y.; Wang, Y.Q.; Xu, Y.Y.; Mei, R.Q. Triterpene-farnesyl hydroquinone conjugates from Ganoderma calidophilum. Nat. Prod. Res. 2021, 35, 2199–2204. [Google Scholar] [CrossRef]
- Wang, C.F.; Liu, X.M.; Lian, C.L.; Ke, J.Y.; Liu, J.Q. Triterpenes and aromatic meroterpenoids with antioxidant activity and neuroprotective effects from Ganoderma lucidum. Molecules 2019, 24, 4353. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, P.; Zhang, C.Y.; Bao, F.Y.; Li, H.; Chen, L.X. Meroterpenoids from Ganoderma sinense protect hepatocytes and cardiomyocytes from oxidative stress induced injuries. Fitoterapia 2018, 131, 73–79. [Google Scholar] [CrossRef]
- Šimek, M.; Bártová, K.; Issad, S.; Hájek, M.; Císařová, I.; Jahn, U. Unified total synthesis of diverse meroterpenoids from Ganoderma Applanatum. Org. Lett. 2022, 24, 4552–4556. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.C.; Kong, F.D.; Ma, Q.Y.; Xie, Q.Y.; Zhang, R.S.; Dai, H.F.; Wu, Y.G.; Zhao, Y.X. Meroterpenoids with protein tyrosine phosphatase 1B inhibitory activities from the fruiting bodies of Ganoderma ahmadii. Front. Chem. 2020, 8, 279–286. [Google Scholar] [CrossRef]
- Wang, K.; Bao, L.; Ma, K.; Zhang, J.J.; Chen, B.S.; Han, J.J.; Ren, J.W.; Luo, H.J.; Liu, H.W. A novel class of α-glucosidase and HMG-CoA reductase inhibitors from Ganoderma leucocontextum and the anti-diabetic properties of ganomycin I in KK-Ay mice. Eur. J. Med. Chem. 2017, 127, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Bao, L.; Zhou, N.; Zhang, J.J.; Liao, M.F.; Zheng, Z.Y.; Wang, Y.J.; Liu, C.; Wang, J.; Wang, L.F.; et al. Structural modification of natural product ganomycin I leading to discovery of a α-Glucosidase and HMG-CoA reductase dual inhibitor improving obesity and metabolic dysfunction in vivo. J. Med. Chem. 2018, 61, 3609–3625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.X.; Xiao, D.H.; Wang, B. A concise total synthesis of cochlearoid B. Org. Biomol. Chem. 2018, 16, 3358–3361. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.S.; Tian, J.; Zhang, J.J.; Wang, K.; Liu, L.; Yang, B.; Bao, L.; Liu, H.W. Triterpenes and meroterpenes from Ganoderma lucidum with inhibitory activity against HMGs reductase, aldose reductase and α-glucosidase. Fitoterapia 2017, 120, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Ma, Q.Y.; Kong, F.D.; Xie, Q.Y.; Huang, S.Z.; Zhou, L.M.; Dai, H.F.; Yu, Z.F.; Zhao, Y.X. Two new compounds from the fruiting bodies of Ganoderma philippii. J. Asian Nat. Prod. Res. 2018, 20, 249–254. [Google Scholar] [CrossRef]
- Wang, M.; Wang, F.; Xu, F.; Ding, L.Q.; Zhang, Q.; Li, H.X.; Zhao, F.; Wang, L.Q.; Zhu, L.H.; Chen, L.X.; et al. Two pairs of farnesyl phenolic enantiomers asnatural nitric oxide inhibitors from Ganoderma sinense. Bioorg. Med. Chem. Lett. 2016, 26, 3342–3345. [Google Scholar] [CrossRef]
- Wang, D.; Wang, Y.L.; Zhang, P.; Li, X.Z.; Li, H.; Chen, L.X. New sesquiterpenoid derivatives from Ganoderma sinense with nitric oxide inhibitory activity. Phytochem. Lett. 2020, 35, 84–87. [Google Scholar] [CrossRef]
- Li, Q.Z.; Chen, X.; Mao, P.W.; Jin, M.Y.; Wu, Q.; Zhou, X.W. N-Glycosylated Ganoderma lucidum immunomodulatory protein improved anti-inflammatory activity via inhibition of the p38 MAPK pathway. Food Funct. 2021, 12, 3393–3404. [Google Scholar] [CrossRef]
- Zhu, L.P.; Wu, M.; Li, P.; Zhou, Y.F.; Zhong, J.Y.; Zhang, Z.Q.; Li, Y.; Yao, W.X.; Xu, J.H. High-pressure supercritical CO2 extracts of Ganoderma lucidum fruiting body and their anti-hepatoma effect associated with the Ras/Raf/MEK/ERK signaling pathway. Front. Pharmacol. 2020, 11, 602702. [Google Scholar] [CrossRef] [PubMed]
- Shaher, F.; Wang, S.Q.; Qiu, H.B.; Hu, Y.; Zhang, Y.; Wang, W.Q.; Al-Ward, H.; Abdulghani, M.A.M.; Baldi, S.; Zhou, S.B. Effect and mechanism of Ganoderma lucidum spores on alleviation of diabetic cardiomyopathy in a Pilot in vivo study. Diabetes Metab. Syndr. Obes. 2020, 13, 4809–4822. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.L.; Guo, P.X.; Luo, Q.; Yan, H.; Cheng, Y.X. Petchienes A–E, meroterpenoids from Ganoderma petchii. Nat. Prod. Commun. 2015, 10, 2019–2022. [Google Scholar] [PubMed]
- Wang, Y.X.; Peng, Y.L.; Qiu, B.; Lv, Q.; Huang, L.S.X.; Cheng, Y.X. Meroterpenoids with a large conjugated system from Ganoderma lucidum and their inhibitory activities against renal fibrosis. Fitoterapia 2022, 161, 105257–105264. [Google Scholar] [CrossRef]
- Yan, Y.M.; Wang, X.L.; Luo, Q.; Jiang, L.P.; Yang, C.P.; Hou, B.; Zuo, Z.L.; Chen, Y.B.; Cheng, Y.X. Metabolites from the mushroom Ganoderma lingzhi as stimulators of neural stem cell proliferation. Phytochemistry 2015, 114, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Yang, P.F.; Yang, Y.N.; Feng, Z.M.; Jiang, J.S.; Zhang, P.C. Direct assignment of the threo and erythro configurations in polyacetylene glycosides by 1H NMR spectroscopy. Org. Lett. 2017, 19, 686–689. [Google Scholar] [CrossRef]
- Antushevich, H. Interplays between inflammasomes and viruses, bacteria (pathogenic and probiotic), yeasts and parasites. Immunol. Lett. 2020, 228, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ryu, D.H.; Cho, J.Y.; Sadiq, N.B.; Kim, J.C.; Lee, B.; Hamayun, M.; Lee, T.S.; Kim, H.S.; Park, S.H.; Nho, C.W.; et al. Optimization of antioxidant, anti-diabetic, and anti-inflammatory activities and ganoderic acid content of differentially dried Ganoderma lucidum using response surface methodology. Food Chem. 2021, 335, 127645–127652. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Su, T.; Yu, H.; Kwan, H.Y.; Ma, X.Q.; Cao, H.H.; Cheng, C.Y.; Leung, A.K.; Chan, C.L.; Li, W.D.; Cao, H.; et al. Comparisons of the chemical profiles, cytotoxicities and anti-inflammatory effects of raw and rice wine-processed Herba Siegesbeckiae. J. Ethnopharmacol. 2014, 156, 365–369. [Google Scholar] [CrossRef]
- Guan, X.Q.; Yang, J.; Wang, W.Y.; Zhao, B.; Hu, S.Y.; Yu, D.H.; Yuan, L.; Shi, Y.F.; Xu, J.L.; Dong, J.Y.; et al. Dual inhibition of MYC and SLC39A10 by a novel natural product STAT3 inhibitor derived from Chaetomium globosum suppresses tumor growth and metastasis in gastric cancer. Pharmacol. Res. 2023, 189, 106703–106714. [Google Scholar] [CrossRef]
- Wu, Y.L.; Han, F.; Luan, S.S.; Ai, R.; Zhang, P.; Li, H.; Chen, L.X. Triterpenoids from Ganoderma lucidum and their potential anti-inflammatory effects. J. Agric. Food Chem. 2019, 67, 5147–5158. [Google Scholar] [CrossRef] [PubMed]
- Kou, R.W.; Gao, Y.Q.; Xia, B.; Wang, J.Y.; Liu, X.N.; Tang, J.J.; Yin, X.; Gao, J.M. Ganoderterpene A, a new triterpenoid from Ganoderma lucidum, attenuates LPS-Induced inflammation and apoptosis via suppressing MAPK and TLR-4/NF-κB pathways in BV-2 cells. J. Agric. Food Chem. 2021, 69, 12730–12740. [Google Scholar] [CrossRef] [PubMed]

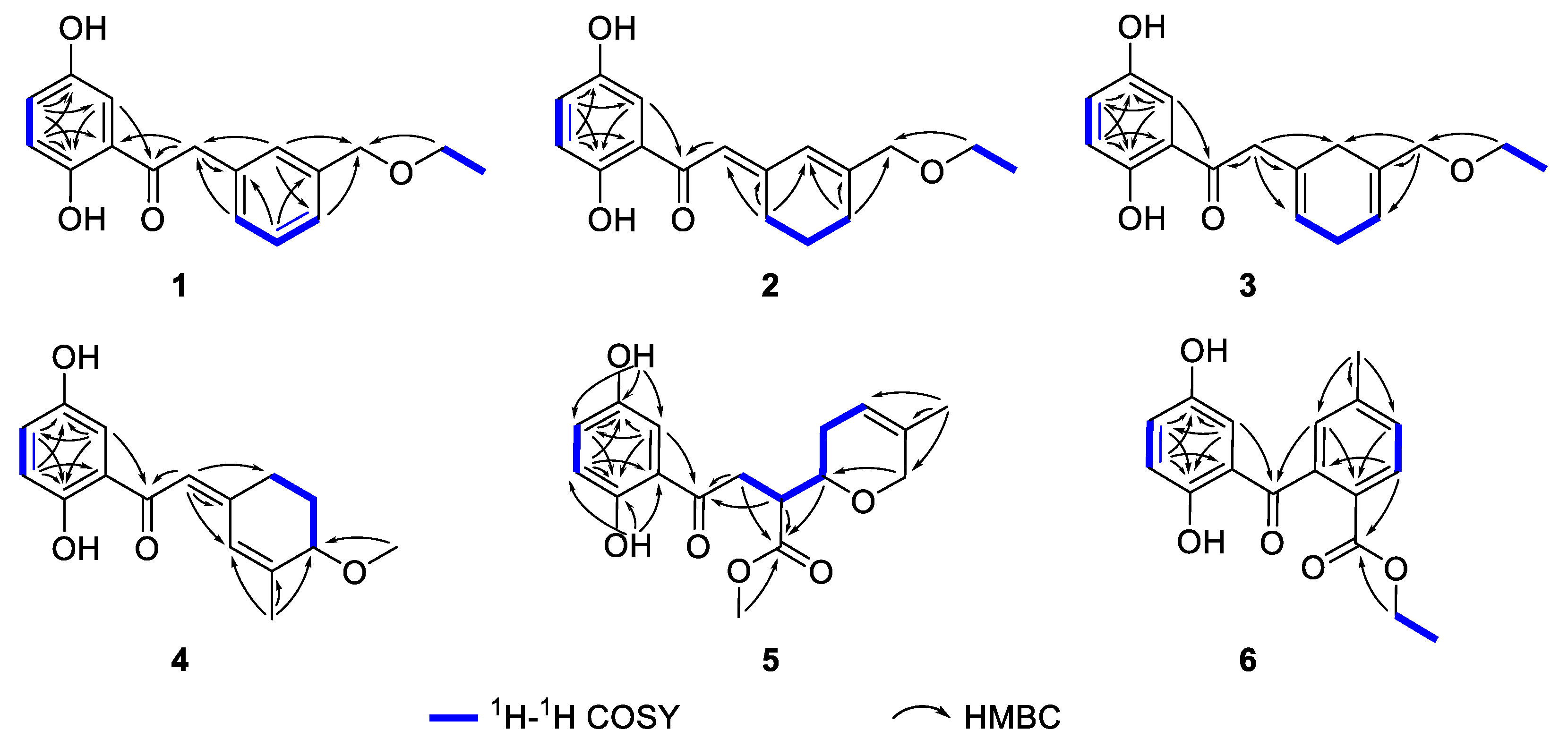
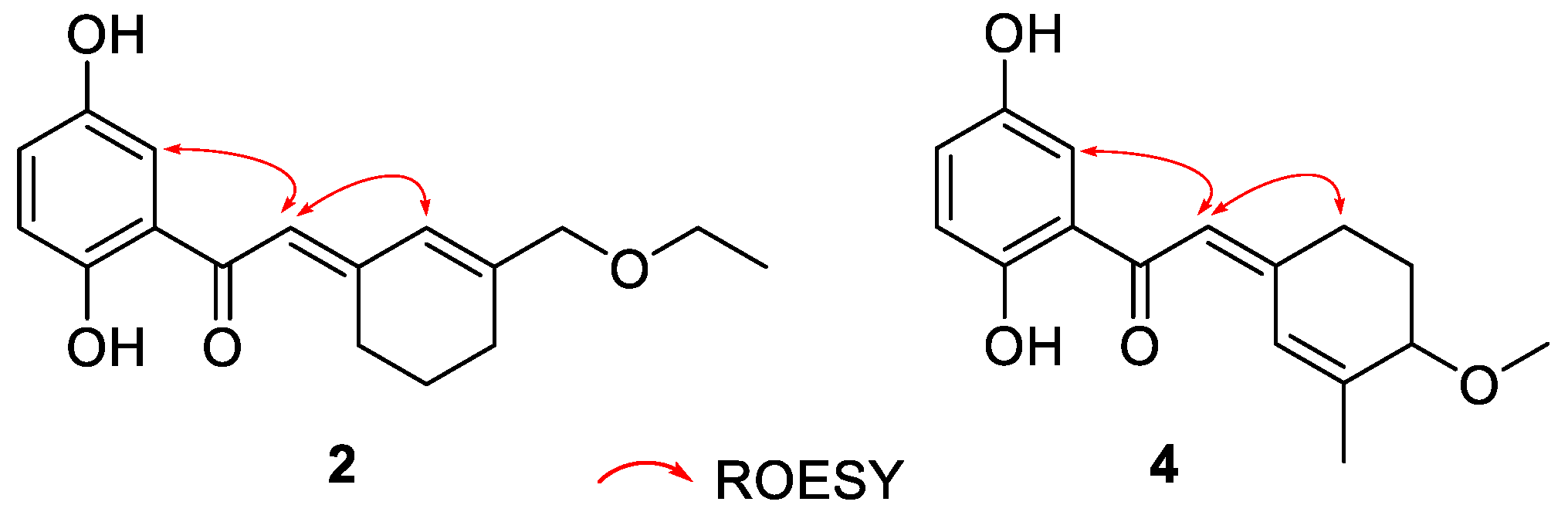
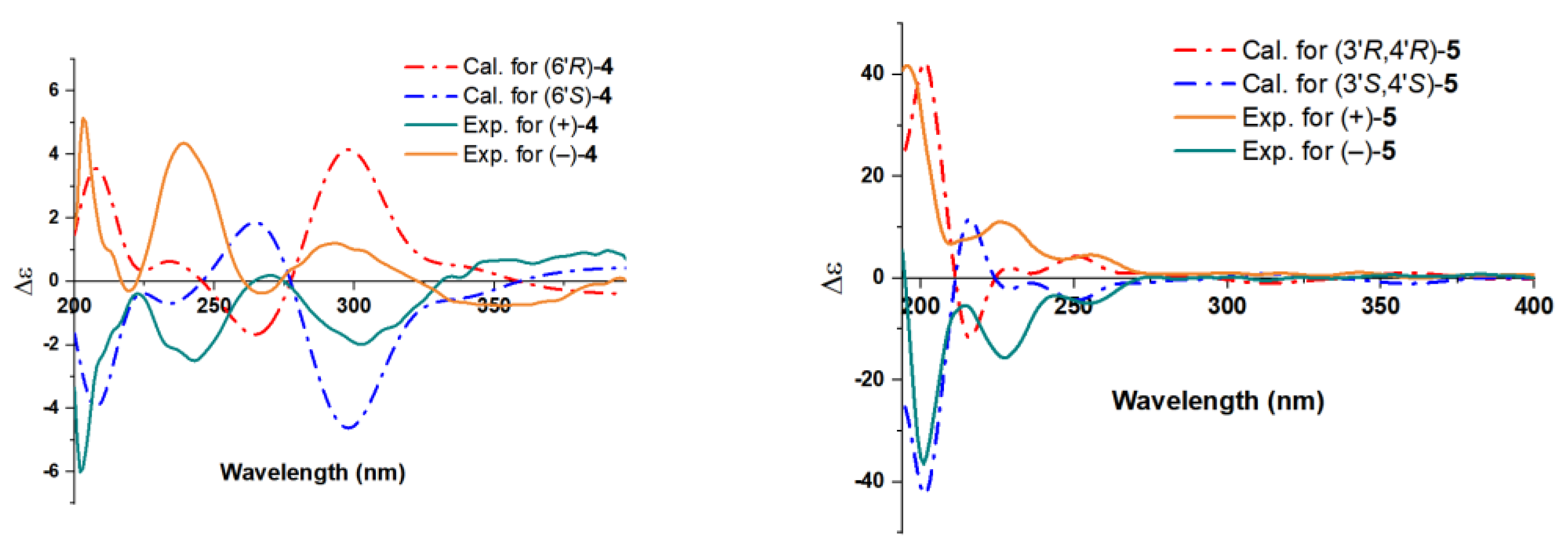


| 1 | 2 | 3 | ||||
|---|---|---|---|---|---|---|
| No. | δH | δC | δH | δC | δH | δC |
| 1 | 157.1, C | 157.3, C | 156.9, C | |||
| 2 | 120.2, C | 122.2, C | 120.4, C | |||
| 3 | 7.34 (d, 2.9) | 116.2, CH | 7.25 (d, 2.9) | 115.2, CH | 7.30 (d, 2.9) | 116.1, CH |
| 4 | 150.6, C | 150.4, C | 150.6, C | |||
| 5 | 7.00 (dd, 8.9, 2.9) | 126.0, CH | 6.97 (dd, 8.9, 2.9) | 125.1, CH | 7.01 (dd, 8.9, 2.9) | 126.0, CH |
| 6 | 6.80 (d, 8.9) | 119.8, CH | 6.77 (d, 8.9) | 119.5, CH | 6.79 (d, 8.9) | 119.7, CH |
| 1′ | 205.3, C | 197.1, C | 206.0, C | |||
| 2′ | 4.31 (s) | 46.2, CH2 | 6.76 (br s) | 119.6, CH | 3.68 (br s) | 47.9, CH2 |
| 3′ | 136.2, C | 157.3, C | 130.7, C | |||
| 4′ | 7.22 (br d, 7.6) | 130.0, CH | 3.00 (t-like, 6.3) | 28.7, CH2 | 5.62 (m) | 124.1, CH |
| 5′ | 7.31 (t, 7.6) | 129.7, CH | 1.78 (p, 6.3) | 23.2, CH2 | 2.77 (m) | 28.4, CH2 |
| 6′ | 7.23 (br d, 7.6) | 127.5, CH | 2.21 (t, 6.3) | 27.3, CH2 | 5.70 (m) | 122.7, CH |
| 7′ | 140.2, C | 151.3, C | 133.4, C | |||
| 8′ | 7.28 (br s) | 129.9, CH | 6.36 (br s) | 127.5, CH | Ha: 2.66 (d, 8.0) | 31.2, CH2 |
| Hb: 2.65 (d, 8.0) | ||||||
| 9′ | 4.49 (s) | 73.5, CH2 | 4.04 (br s) | 74.7, CH2 | 3.88 (br s) | 75.6, CH2 |
| 10′ | 3.54 (q, 7.0) | 66.8, CH2 | 3.53 (q, 7.0) | 67.1, CH2 | 3.45 (q, 7.0) | 66.2, CH2 |
| 11′ | 1.20 (t, 7.0) | 15.4, CH3 | 1.23 (t, 7.0) | 15.5, CH3 | 1.17 (t, 7.0) | 15.4, CH3 |
| 4 | 5 | 6 | ||||
|---|---|---|---|---|---|---|
| No. | δH | δC | δH | δC | δH | δC |
| 1 | 157.4, C | 153.2, C | 156.9, C | |||
| 2 | 122.1, C | 120.4, C | 121.4, C | |||
| 3 | 7.24 (d, 2.9) | 115.4, CH | 7.20 (d, 3.0) | 114.5, CH | 6.47 (d, 3.0) | 117.8, CH |
| 4 | 150.4, C | 149.5, C | 150.5, C | |||
| 5 | 6.97 (dd, 8.9, 2.9) | 125.2, CH | 6.98 (dd, 8.8, 3.0) | 124.2, CH | 7.00 (dd, 8.9, 3.0) | 125.9, CH |
| 6 | 6.78 (d, 8.9) | 119.7, CH | 6.81 (d, 8.8) | 118.4, CH | 6.87 (d, 8.9) | 119.6, CH |
| 1′ | 196.6, C | 203.0, C | 204.2, C | |||
| 2′ | 6.64 (br s) | 117.1, C | Ha: 3.35 (dd, 18.4, 3.6) | 37.6, CH2 | 141.6, C | |
| Hb: 3.44 (dd, 18.4, 10.3) | ||||||
| 3′ | 154.9, C | 3.03 (ddd, 10.3, 5.9, 3.6) | 44.9, CH | 127.3, C | ||
| 4′ | Ha: 2.66 (dddd, 15.4, 8.5, 4.0, 1.4) | 30.4, CH2 | 3.70 (ddd, 10.1, 5.9, 3.4) | 73.2, CH | 7.98 (d, 8.0) | 131.3, CH |
| Hb: 2.45 (dddd, 15.4, 8.9, 4.0, 1.4) | ||||||
| 5′ | Ha: 2.06 (ddt, 12.9, 8.5, 4.0) | 28.4, CH2 | Ha: 2.09 (m) | 27.9, CH2 | 7.48 (br d, 8.0) | 131.6, CH |
| Hb: 1.90 (dddd, 12.9, 8.9, 6.4, 4.0) | Hb: 1.91 (br d, 17.2) | |||||
| 6′ | 3.86 (t-like, 5.3) | 78.9, CH | 5.47 (m) | 117.7, CH | 145.2, C | |
| 7′ | 150.7, C | 132.6, C | 7.25 (br s) | 129.0, CH | ||
| 8′ | 7.41 (br s) | 125.4, CH | 3.96 (br s) | 68.7, CH2 | 2.47 (br s) | 21.5, CH3 |
| 9′ | 1.97 (br s) | 21.6, CH3 | 1.56 (br s) | 18.2, CH3 | 167.0, C | |
| 10′ | 3.44 (s) | 57.5, CH3 | 173.0, C | 4.11 (q, 7.1) | 62.5, CH2 | |
| 11′ | 3.60 (s) | 51.7, CH3 | 1.10 (t, 7.1) | 13.9, CH3 | ||
| 1-OH | 10.91 (s) | |||||
| 4-OH | 9.17 (s) | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.-Y.; Cai, D.; Tang, X.-P.; Cheng, Y.-X. Ganoderma lucidum-Derived Meroterpenoids Show Anti-Inflammatory Activity In Vitro. Molecules 2024, 29, 1149. https://doi.org/10.3390/molecules29051149
Liu Y-Y, Cai D, Tang X-P, Cheng Y-X. Ganoderma lucidum-Derived Meroterpenoids Show Anti-Inflammatory Activity In Vitro. Molecules. 2024; 29(5):1149. https://doi.org/10.3390/molecules29051149
Chicago/Turabian StyleLiu, Yun-Yun, Dan Cai, Xin-Ping Tang, and Yong-Xian Cheng. 2024. "Ganoderma lucidum-Derived Meroterpenoids Show Anti-Inflammatory Activity In Vitro" Molecules 29, no. 5: 1149. https://doi.org/10.3390/molecules29051149
APA StyleLiu, Y.-Y., Cai, D., Tang, X.-P., & Cheng, Y.-X. (2024). Ganoderma lucidum-Derived Meroterpenoids Show Anti-Inflammatory Activity In Vitro. Molecules, 29(5), 1149. https://doi.org/10.3390/molecules29051149






