Synthesis and In Vitro Antitumor Activity Evaluation of Gefitinib-1,2,3-Triazole Derivatives
Abstract
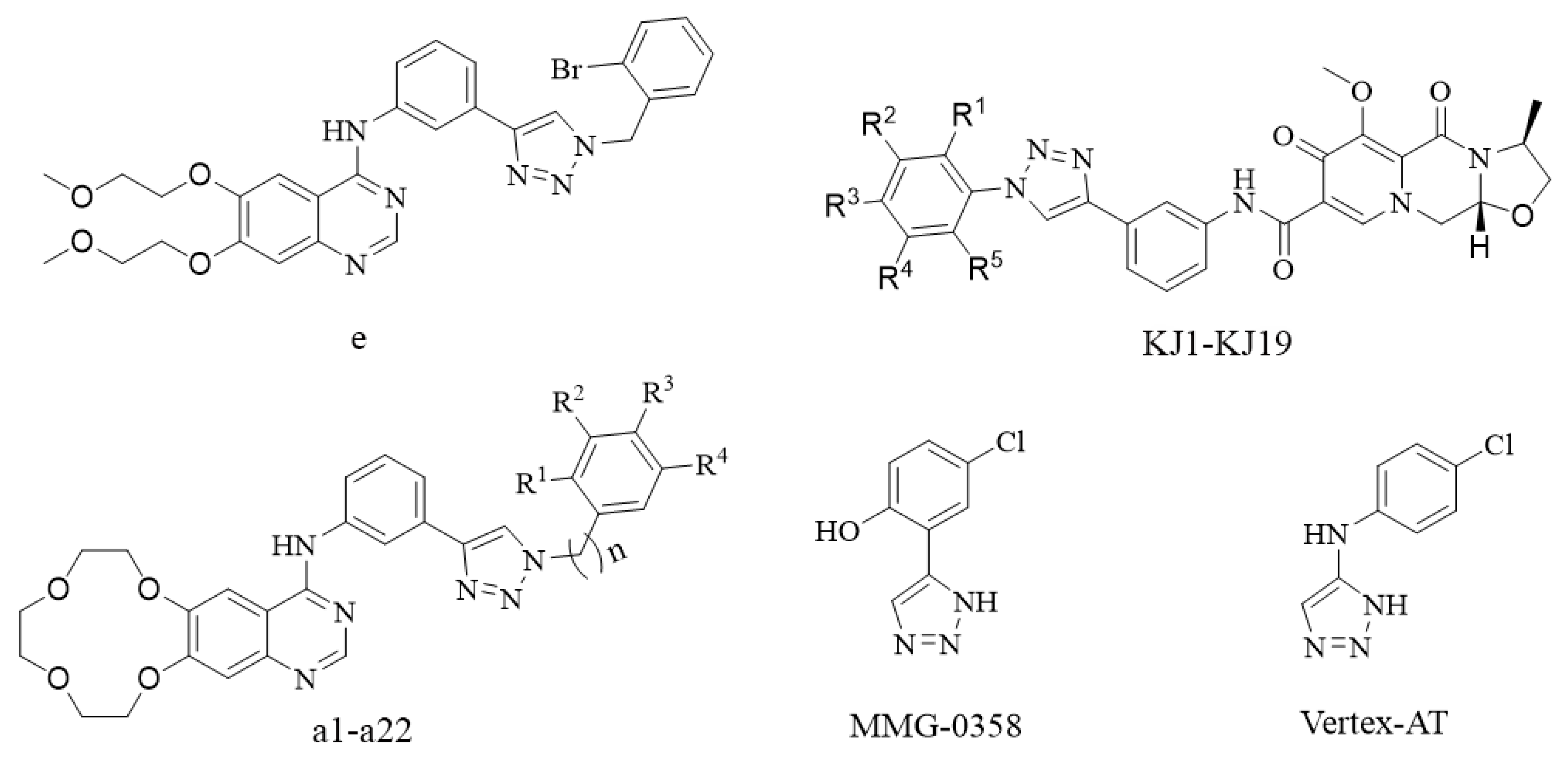
1. Introduction
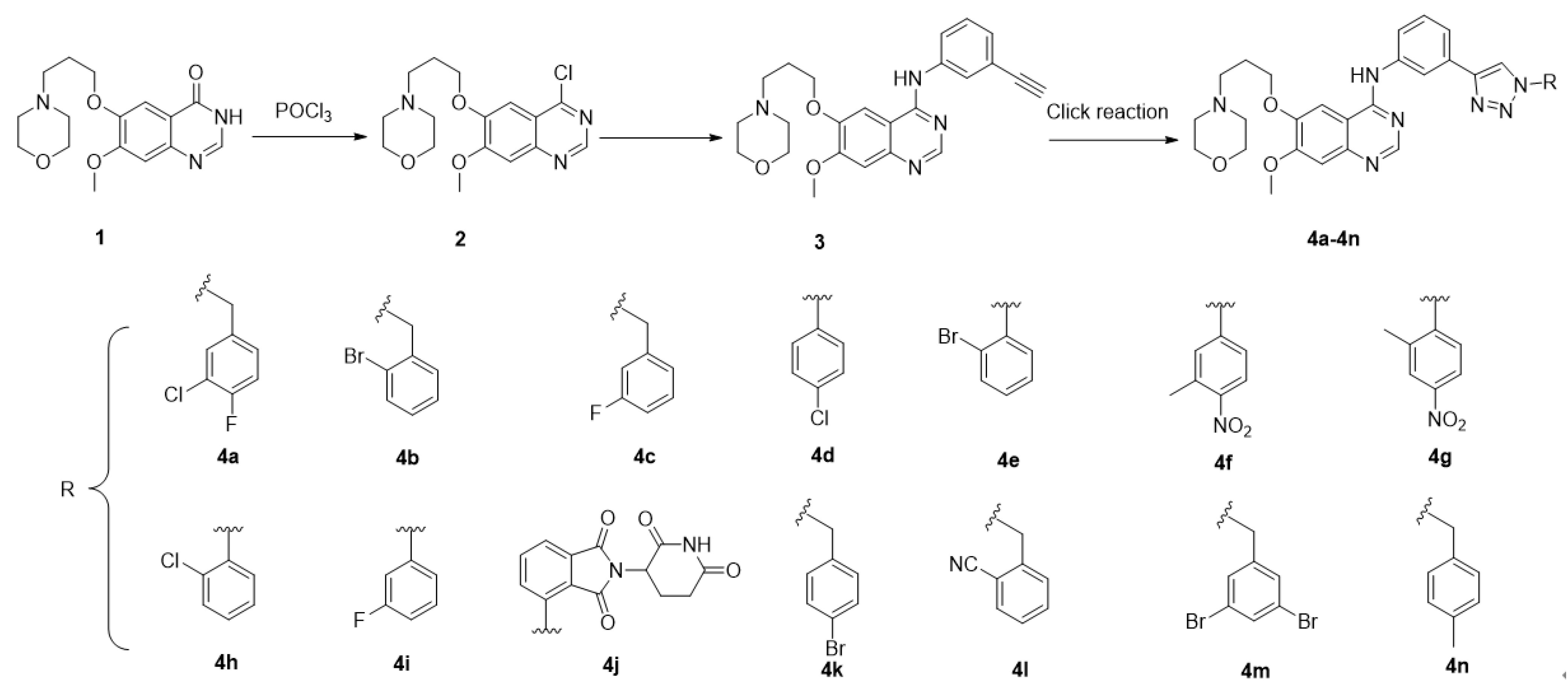
2. Chemistry
3. Results and Discussion
3.1. Compounds 4a–4n Suppress Cancer Cell Viability
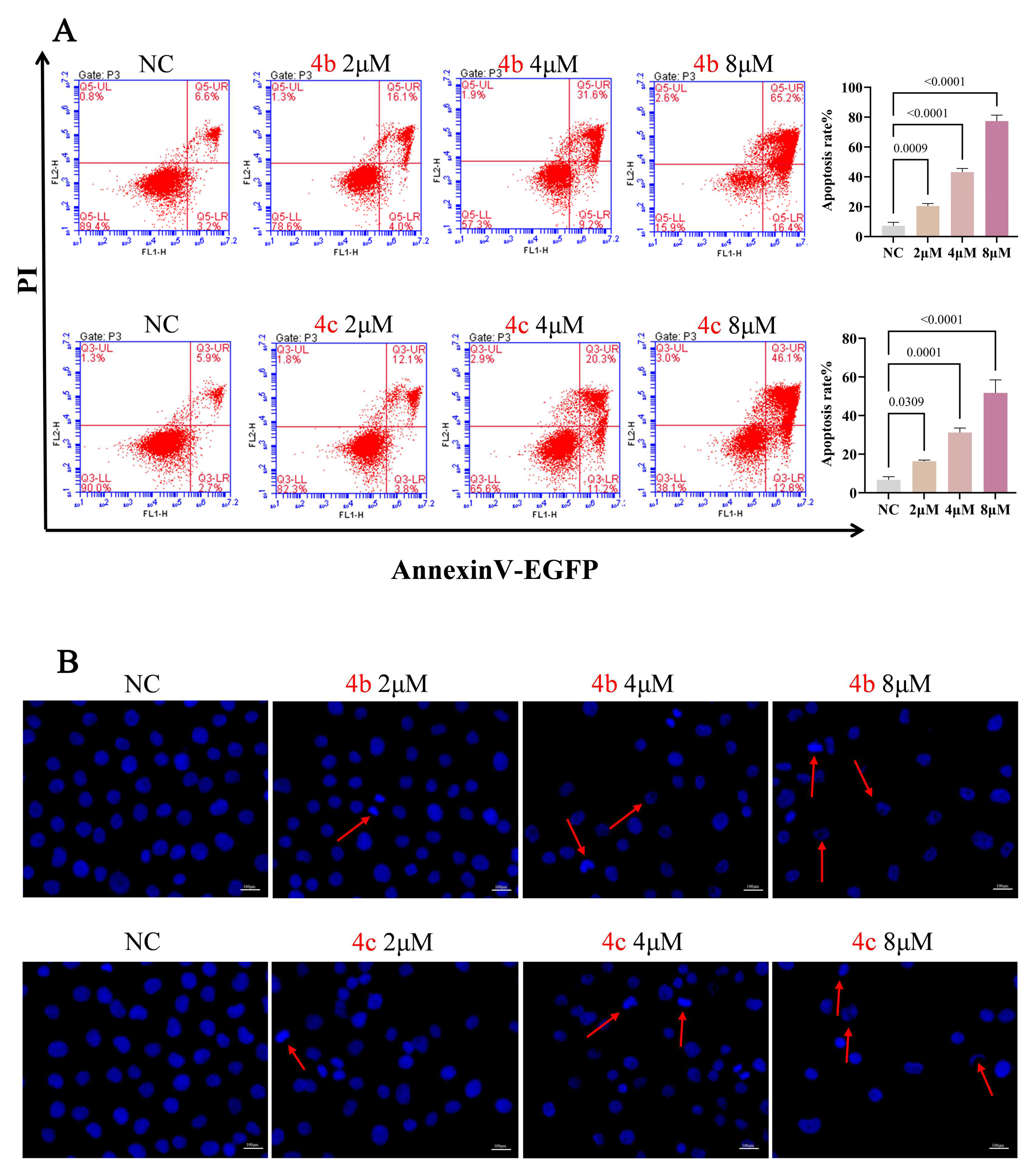
3.2. 4b and 4c Induce Apoptosis in H1299 Cells
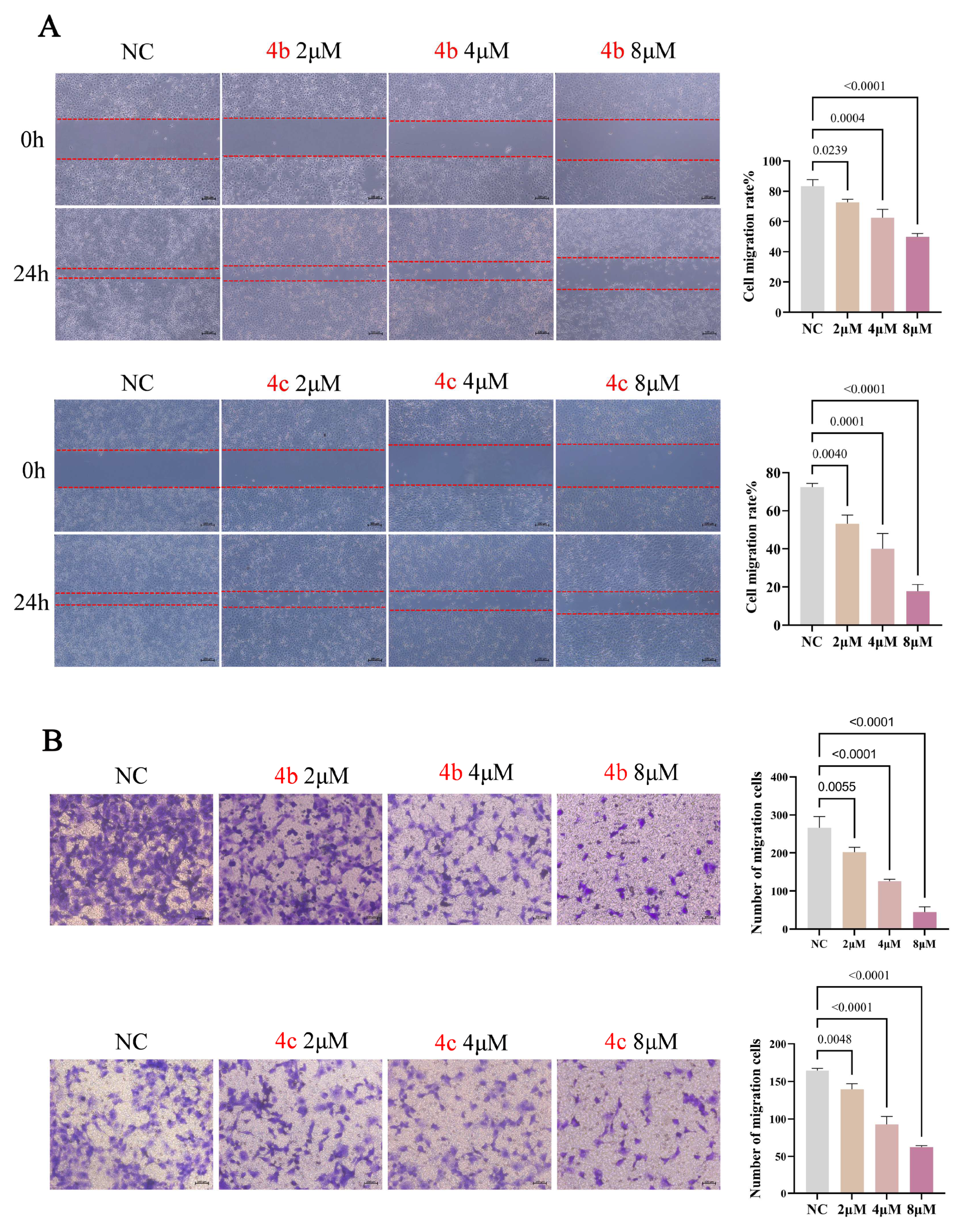
3.3. 4b and 4c Suppress Metastasis of H1299 Cells
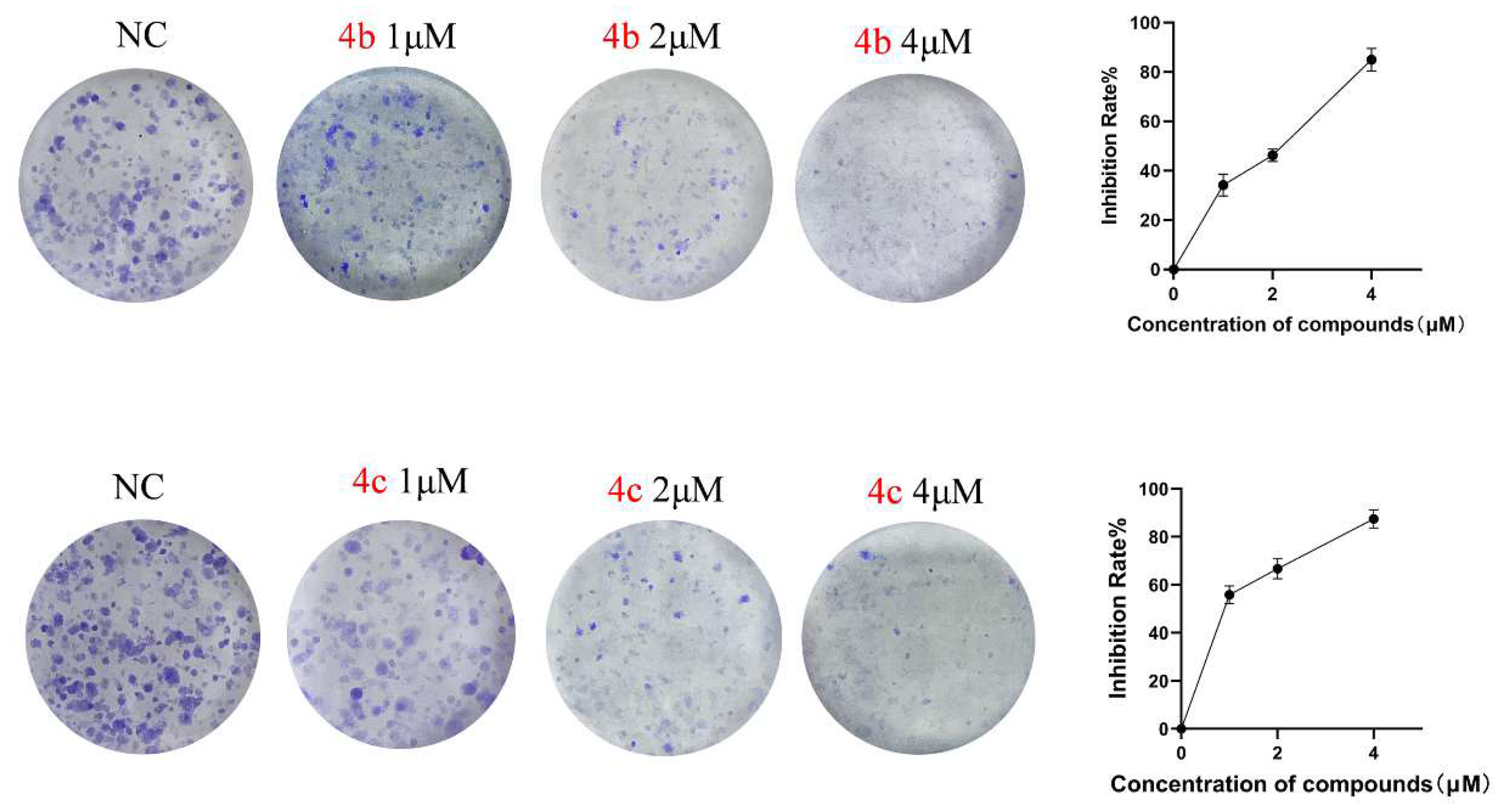
3.4. 4b and 4c Inhibit the Clonogenic Ability of H1299 Cells
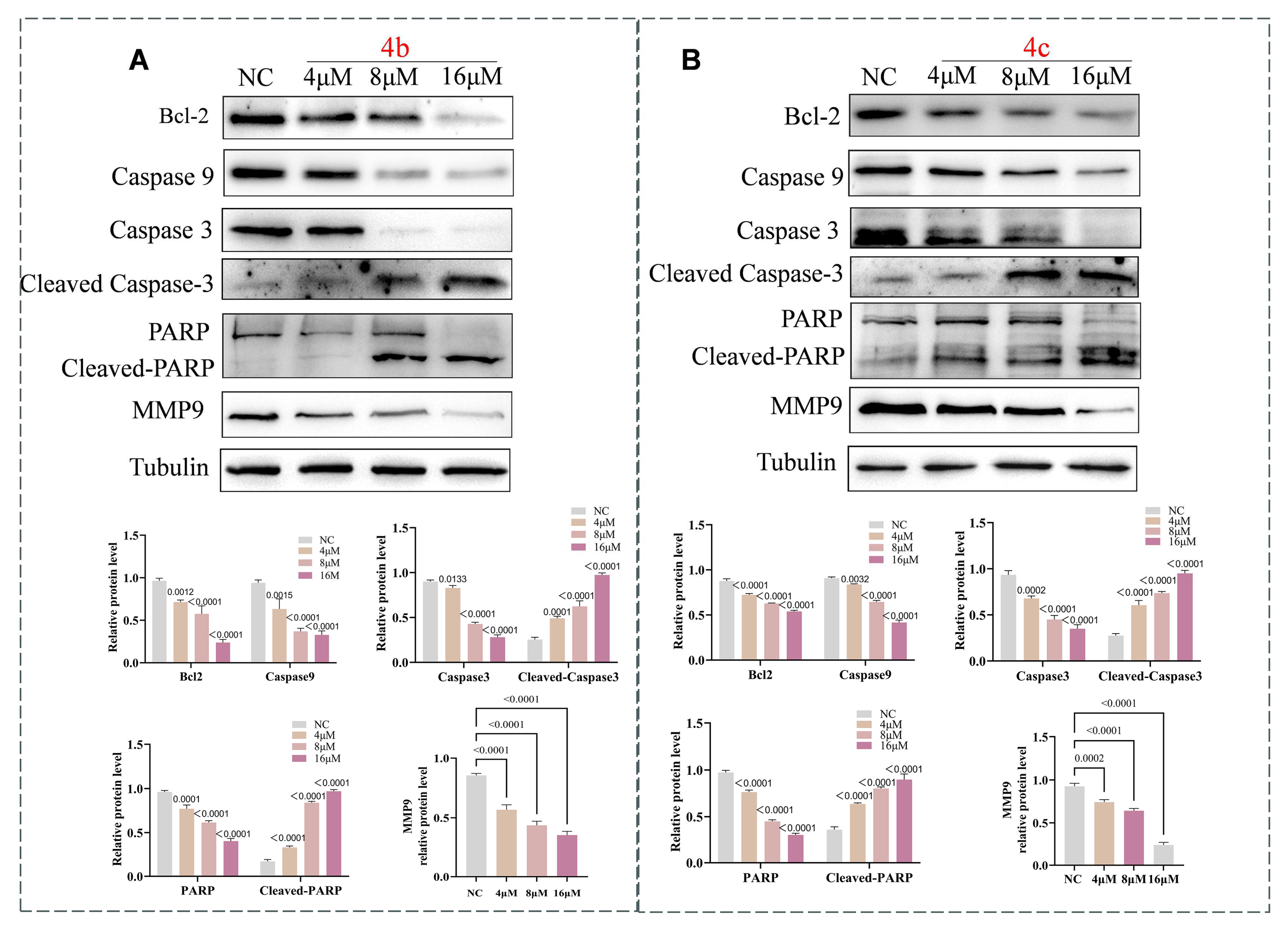
3.5. 4b and 4c Trigger Apoptosis via the Apoptosis Signaling Pathway
4. Conclusions
5. Experimental
5.1. Materials and Chemistry
5.1.1. Preparation of 4-(3-((4-chloro-7-methoxyquinazolin-6yl)oxy)propyl)morpholine
5.1.2. Preparation of N-(3-ethynylphenyl)-7-methoxy-6-(3-morpholinopropoxy)quinazolin-4-amine
5.1.3. General Procedure for the Preparation of Compound 4
5.2. Synthesis of Snalogues 4a–4n
5.3. Biological Study
5.3.1. Cell Culture
5.3.2. CCK-8 Assay to Assess Cell Proliferation and Cytotoxicity
5.3.3. Flow Cytometry Detection for Cell Apoptosis
5.3.4. DAPI Staining for Cell Apoptosis
5.3.5. Wound Healing Assay
5.3.6. Transwell Migration Assay
5.3.7. Colony Formation Assay
5.3.8. Western Blot Analysis
5.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sabbah, D.A.; Hajjo, R.; Sweidan, K. Review on Epidermal Growth Factor Receptor (Egfr) Structure, Signaling Pathways, Interactions, and Recent Updates of Egfr Inhibitors. Curr. Top. Med. Chem. 2020, 20, 815–834. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Hackshaw, A.; Feng, Q.; Fu, X.; Zhang, Y.; Mao, C.; Tang, J. Comparison of Gefitinib, Erlotinib and Afatinib in Non-Small Cell Lung Cancer: A Meta-Analysis. Int. J. Cancer 2017, 140, 2805–2819. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Guo, S.; Liu, Y.; Jiang, H.; Liao, Y.-X.; Shen, J.; Song, W.; Hou, J.-T. A Stable Nir Fluorescent Probe for Imaging Lipid Droplets in Triple-Negative Breast Cancer. Sens. Actuators B Chem. 2024, 398, 134740. [Google Scholar] [CrossRef]
- Chen, H.; Jiang, H.; Xue, B.L.; Li, H.; Chen, J. One-Step Preparation of Thiol-Responsive Methylene Blue-Encapsulated Trimanganese Tetroxide for Pesticide Highly Sensitive Homogeneous Electrochemical Sensing. Microchem. J. 2024, 197, 109781. [Google Scholar] [CrossRef]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. Egfr Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef]
- Pao, W.; Miller, V.; Zakowski, M.; Doherty, J.; Politi, K.; Sarkaria, I.; Singh, B.; Heelan, R.; Rusch, V.; Fulton, L.; et al. Egf Receptor Gene Mutations Are Common in Lung Cancers from “Never Smokers” and Are Associated with Sensitivity of Tumors to Gefitinib and Erlotinib. Proc. Natl. Acad. Sci. USA 2004, 101, 13306–13311. [Google Scholar] [CrossRef] [PubMed]
- Asahina, H.; Yamazaki, K.; Kinoshita, I.; Sukoh, N.; Harada, M.; Yokouchi, H.; Ishida, T.; Ogura, S.; Kojima, T.; Okamoto, Y.; et al. A Phase Ii Trial of Gefitinib as First-Line Therapy for Advanced Non-Small Cell Lung Cancer with Epidermal Growth Factor Receptor Mutations. Br. J. Cancer 2006, 95, 998–1004. [Google Scholar] [CrossRef]
- Akkurt, M.; Jarrahpour, A.; Chermahini, M.M.; Shiri, P.; Tahir, M.N. 4-(1-Methylethyl)-N-((E)-4-{[1-(Prop-2-En-1-Yl)-1h-1,2,3-Triazol-4-Yl]Methoxy}Benzylidene)Aniline. Acta Crystallogr. Sect. E Struct. Rep. Online 2013, 69 Pt 2, o247. [Google Scholar] [CrossRef]
- Mohammed, H.H.H.; El-Hafeez, A.A.A.; Ebeid, K.; Mekkawy, A.I.; Abourehab, M.A.S.; Wafa, E.I.; Alhaj-Suliman, S.O.; Salem, A.K.; Ghosh, P.; Abuo-Rahma, G.E.A.; et al. New 1,2,3-Triazole Linked Ciprofloxacin-Chalcones Induce DNA Damage by Inhibiting Human Topoisomerase I& Ii and Tubulin Polymerization. J. Enzym. Inhib. Med. Chem. 2022, 37, 1346–1363. [Google Scholar]
- Liang, T.; Sun, X.; Li, W.; Hou, G.; Gao, F. 1,2,3-Triazole-Containing Compounds as Anti-Lung Cancer Agents: Current Developments, Mechanisms of Action, and Structure-Activity Relationship. Front. Pharmacol. 2021, 12, 661173. [Google Scholar] [CrossRef]
- Zhang, B. Comprehensive Review on the Anti-Bacterial Activity of 1,2,3-Triazole Hybrids. Eur. J. Med. Chem. 2019, 168, 357–372. [Google Scholar] [CrossRef]
- Chu, X.M.; Wang, C.; Wang, W.L.; Liang, L.L.; Liu, W.; Gong, K.K.; Sun, K.L. Triazole Derivatives and Their Antiplasmodial and Antimalarial Activities. Eur. J. Med. Chem. 2019, 166, 206–223. [Google Scholar] [CrossRef] [PubMed]
- Emami, S.; Ghobadi, E.; Saednia, S.; Hashemi, S.M. Current Advances of Triazole Alcohols Derived from Fluconazole: Design, In vitro and in Silico Studies. Eur. J. Med. Chem. 2019, 170, 173–194. [Google Scholar] [CrossRef]
- Biliavska, L.; Pankivska, Y.; Povnitsa, O.; Zagorodnya, S.; Gudz, G.; Shermolovich, Y. Anti-Adenoviral Activity of 2-(3-Chlorotetrahydrofuran-2-Yl)-4-Tosyl-5-(Perfluoropropyl)-1,2,3-Triazole. Medicina 2018, 54, 81. [Google Scholar] [CrossRef]
- Bakale, R.D.; Sulakhe, S.M.; Kasare, S.L.; Sathe, B.P.; Rathod, S.S.; Choudhari, P.B.; Rekha, E.M.; Sriram, D.; Haval, K.P. Design, Synthesis and Antitubercular Assessment of 1, 2, 3-Triazole Incorporated Thiazolylcarboxylate Derivatives. Bioorg. Med. Chem. Lett. 2024, 97, 129551. [Google Scholar] [CrossRef]
- Alam, M.M. 1,2,3-Triazole Hybrids as Anticancer Agents: A Review. Arch. Pharm. 2022, 355, e2100158. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhao, S.J.; Liu, Y. 1,2,3-Triazole-Containing Hybrids as Potential Anticancer Agents: Current Developments, Action Mechanisms and Structure-Activity Relationships. Eur. J. Med. Chem. 2019, 183, 111700. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.Q.; Gong, X.Q.; Zhu, Y.Y.; Yao, X.J.; Peng, L.Z.; Sun, G.; Yang, J.X.; Mao, L.F. Novel 1,2,3-Triazole Erlotinib Derivatives as Potent Ido1 Inhibitors: Design, Drug-Target Interactions Prediction, Synthesis, Biological Evaluation, Molecular Docking and Adme Properties Studies. Front. Pharmacol. 2022, 13, 854965. [Google Scholar] [CrossRef]
- Xie, H.; Mao, L.; Fan, G.; Wu, Z.; Wang, Y.; Hou, X.; Wang, J.; Wang, H.; Liu, L.; Li, S. Design and Synthesis of Cabotegravir Derivatives Bearing 1,2,3-Triazole and Evaluation of Anti-Liver Cancer Activity. Front. Pharmacol. 2023, 14, 1265289. [Google Scholar] [CrossRef]
- Mao, L.; Sun, G.; Zhao, J.; Xu, G.; Yuan, M.; Li, Y.M. Design, Synthesis and Antitumor Activity of Icotinib Derivatives. Bioorg. Chem. 2020, 105, 104421. [Google Scholar] [CrossRef]
- Chen, X.; Mao, L.F.; Tian, S.; Tian, X.; Meng, X.; Wang, M.K.; Xu, W.; Li, Y.M.; Liu, K.; Dong, Z. Icotinib Derivatives as Tyrosine Kinase Inhibitors with Anti-Esophageal Squamous Carcinoma Activity. Front. Pharmacol. 2022, 13, 1028692. [Google Scholar] [CrossRef] [PubMed]
- Röhrig, U.F.; Reynaud, A.; Majjigapu, S.R.; Vogel, P.; Pojer, F.; Zoete, V. Inhibition Mechanisms of Indoleamine 2,3-Dioxygenase 1 (Ido1). J. Med. Chem. 2019, 62, 8784–8795. [Google Scholar] [CrossRef] [PubMed]
- Lipunova, G.N.; Nosova, E.V.; Charushin, V.N.; Chupakhin, O.N. Synthesis and Antitumour Activity of 4-Aminoquinazoline Derivatives. Russ. Chem. Rev. 2016, 85, 759–793. [Google Scholar] [CrossRef]
- Hong-Xia, L.I.; Qian, Y.-M.; Li-Sheng, X.U. Design, Synthesis and Anticancer Activity Evaluation of Novel Quinazoline Derivatives as Efgr Inhibitors. Chin. J. Struct. Chem. 2021, 40, 10. [Google Scholar]
- Abdel-Aziz, A.A.; El-Azab, A.S.; AlSaif, N.A.; Alanazi, M.M.; El-Gendy, M.A.; Obaidullah, A.J.; Alkahtani, H.M.; Almehizia, A.A.; Al-Suwaidan, I.A. Synthesis, Anti-Inflammatory, Cytotoxic, and Cox-1/2 Inhibitory Activities of Cyclic Imides Bearing 3-Benzenesulfonamide, Oxime, and Β-Phenylalanine Scaffolds: A Molecular Docking Study. J. Enzym. Inhib. Med. Chem. 2020, 35, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Wu, Z.; Qi, Y.; Wu, B.; Zhu, X. The Metastasizing Mechanisms of Lung Cancer: Recent Advances and Therapeutic Challenges. Biomed. Pharmacother. 2021, 138, 111450. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, C.; Manguinhas, R.; Costa, J.G.; Gil, N.; Codony-Servat, J.; Castro, M.; Miranda, J.P.; Fernandes, A.S.; Rosell, R.; Oliveira, N.G. A Narrative Review of the Migration and Invasion Features of Non-Small Cell Lung Cancer Cells Upon Xenobiotic Exposure: Insights from In Vitro Studies. Transl. Lung Cancer Res. 2021, 10, 2698–2714. [Google Scholar] [CrossRef] [PubMed]
- Oike, T.; Komatsu, S.; Komatsu, Y.; Nachankar, A.; Darwis, N.D.M.; Shibata, A.; Ohno, T. Reporting of Methodologies Used for Clonogenic Assays to Determine Radiosensitivity. J. Radiat. Res. 2020, 61, 828–831. [Google Scholar] [CrossRef]
- Wan Mohd Tajuddin, W.N.B.; Lajis, N.H.; Abas, F.; Othman, I.; Naidu, R. Mechanistic Understanding of Curcumin’s Therapeutic Effects in Lung Cancer. Nutrients 2019, 11, 2989. [Google Scholar] [CrossRef]
- Hu, Q.; Zhang, W.; Wu, Z.; Tian, X.; Xiang, J.; Li, L.; Li, Z.; Peng, X.; Wei, S.; Ma, X.; et al. Baicalin and the Liver-Gut System: Pharmacological Bases Explaining Its Therapeutic Effects. Pharmacol. Res. 2021, 165, 105444. [Google Scholar] [CrossRef]
- Augoff, K.; Hryniewicz-Jankowska, A.; Tabola, R.; Stach, K. Mmp9: A Tough Target for Targeted Therapy for Cancer. Cancers 2022, 14, 1847. [Google Scholar] [CrossRef]
- Gong, L.; Wu, D.; Zou, J.; Chen, J.; Chen, L.; Chen, Y.; Ni, C.; Yuan, H. Prognostic Impact of Serum and Tissue Mmp-9 in Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. Oncotarget 2016, 7, 18458–18468. [Google Scholar] [CrossRef]


| Compd No. | IC50 (μM) | ||
|---|---|---|---|
| H1299 | A549 | NCI-H1437 | |
| 4a | 7.59 ± 0.19 | 6.81 ± 0.33 | 2.05 ± 0.03 |
| 4b | 4.42 ± 0.24 | 3.94 ± 0.01 | 1.56 ± 0.06 |
| 4c | 4.60 ± 0.18 | 4.00 ± 0.08 | 3.51 ± 0.05 |
| 4d | 11.44 ± 0.97 | 9.04 ± 0.41 | 6.41 ± 0.54 |
| 4e | 11.88 ± 0.32 | 8.62 ± 0.76 | 11.16 ± 0.95 |
| 4f | 13.26 ± 0.52 | 13.65 ± 0.71 | 9.27 ± 0.40 |
| 4g | 6.02 ± 0.12 | 4.65 ± 0.08 | 2.15 ± 0.10 |
| 4h | 7.69 ± 0.11 | 9.81 ± 0.34 | 3.09 ± 0.13 |
| 4i | 9.35 ± 0.31 | 12.96 ± 0.29 | 4.77 ± 0.32 |
| 4j | 12.02 ± 0.33 | 11.41 ± 0.89 | 6.60 ± 0.07 |
| 4k | 7.15 ± 0.12 | 8.95 ± 0.99 | 5.95 ± 0.72 |
| 4l | 5.43 ± 0.24 | 7.53 ± 0.66 | 2.23 ± 0.26 |
| 4m | 4.79 ± 0.13 | 2.39 ± 0.30 | 2.53 ± 0.11 |
| 4n | 5.91 ± 0.44 | 10.41 ± 0.82 | 5.79 ± 0.88 |
| Gefitinib a | 14.23 ± 0.08 | 15.11 ± 0.05 | 20.44 ± 1.43 |
| Compound No. | IC50 (μM) | Inhibition Rate %, 48 h | ||||
|---|---|---|---|---|---|---|
| 2 μM | 4 μM | 8 μM | 16 μM | 32 μM | ||
| 4b | 20.25 ± 1.26 | 9.09 ± 1.00 | 12.76 ± 1.37 | 29.76 ± 1.27 | 39.08 ± 2.18 | 64.85 ± 1.84 |
| 4c | 29.38 ± 5.53 | 7.18 ± 1.22 | 20.72 ± 3.30 | 27.55 ± 2.79 | 30.56 ± 1.93 | 56.13 ± 5.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Liu, J.; Gao, E.; Mao, L.; Hu, S.; Li, S. Synthesis and In Vitro Antitumor Activity Evaluation of Gefitinib-1,2,3-Triazole Derivatives. Molecules 2024, 29, 837. https://doi.org/10.3390/molecules29040837
Liu Z, Liu J, Gao E, Mao L, Hu S, Li S. Synthesis and In Vitro Antitumor Activity Evaluation of Gefitinib-1,2,3-Triazole Derivatives. Molecules. 2024; 29(4):837. https://doi.org/10.3390/molecules29040837
Chicago/Turabian StyleLiu, Zijun, Jiancheng Liu, En Gao, Longfei Mao, Shu Hu, and Sanqiang Li. 2024. "Synthesis and In Vitro Antitumor Activity Evaluation of Gefitinib-1,2,3-Triazole Derivatives" Molecules 29, no. 4: 837. https://doi.org/10.3390/molecules29040837
APA StyleLiu, Z., Liu, J., Gao, E., Mao, L., Hu, S., & Li, S. (2024). Synthesis and In Vitro Antitumor Activity Evaluation of Gefitinib-1,2,3-Triazole Derivatives. Molecules, 29(4), 837. https://doi.org/10.3390/molecules29040837





