Sesquiterpene Lactones and Flavonoid from the Leaves of Basin Big Sagebrush (Artemisia tridentata subsp. tridentata): Isolation, Characterization and Biological Activities
Abstract
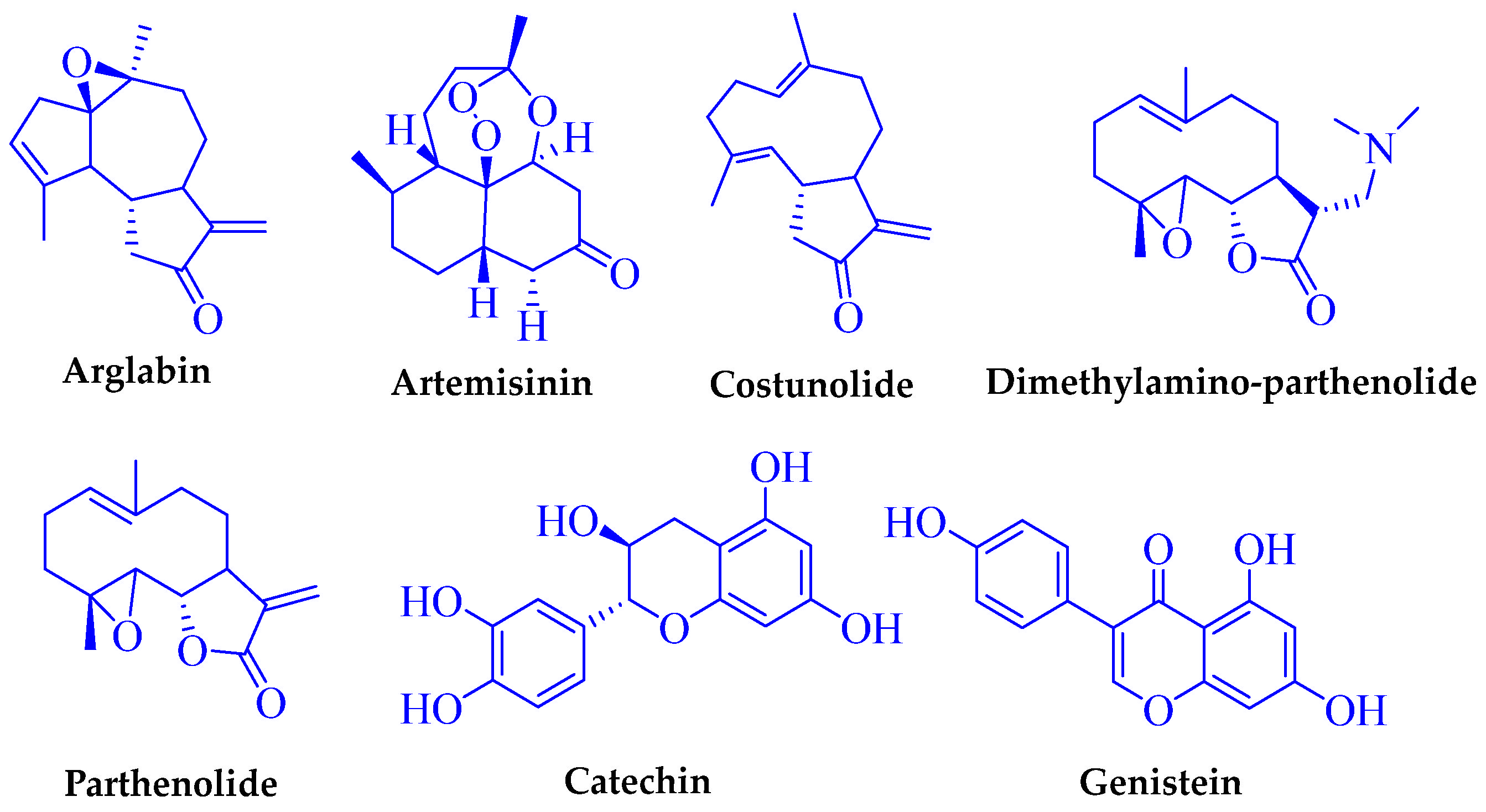
1. Introduction
2. Results
2.1. Thin-Layer Chromatography (TLC) Analysis
2.2. Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC) Analysis
2.3. Liquid Chromatography Mass Spectrometry (LC-MS) Analysis
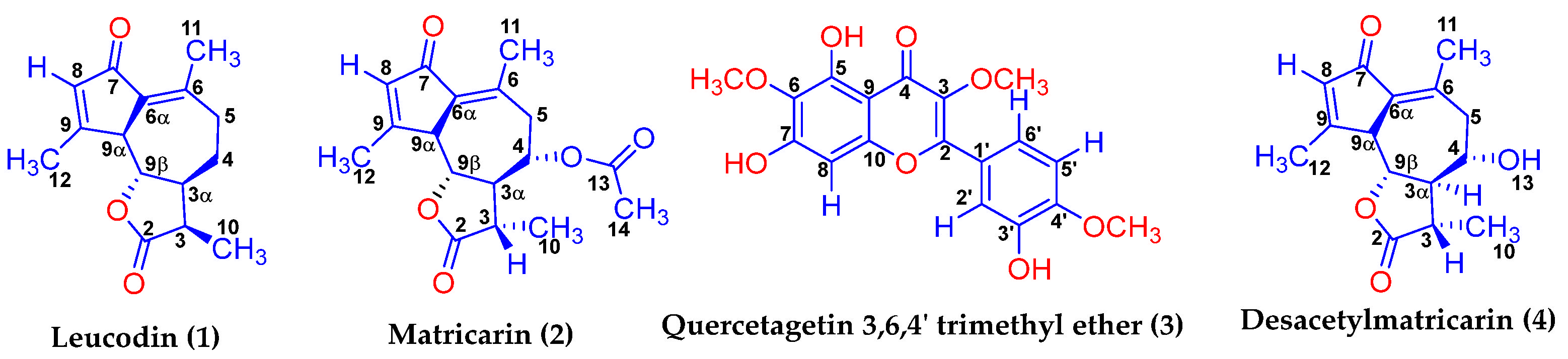
2.4. Nuclear Magnetic Resonance (NMR) and Fourier Transform Infrared Spectroscopy (FTIR) Studies
2.4.1. Matricarin (2)
2.4.2. Desacetylmatricarin (4)
2.4.3. Leucodin (1)
2.4.4. Quercetagetin 3,6,4′-Trimethyl Ether (3)
2.5. Antioxidant Assay
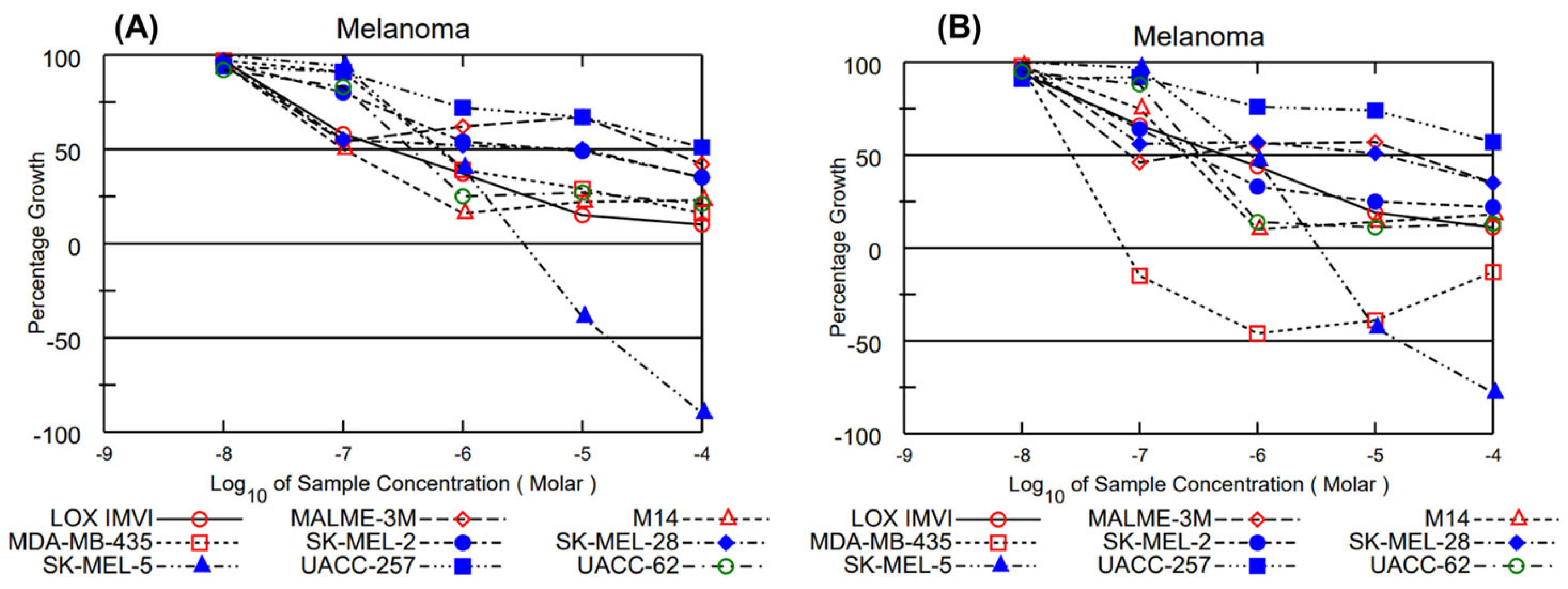
2.6. NCI—60 Cell Lines Evaluation
3. Discussion
4. Materials and Methods
4.1. Plant Collection
4.2. Materials
4.3. Methods
4.3.1. Extraction of Plant Material
4.3.2. TLC Profile
4.3.3. Fractionation and Isolation of Crude Chloroform Extract by Column Chromatography
4.3.4. Recrystallization of Isolated Components
4.3.5. Total Antioxidant Assay
FRAP Assay
DPPH Assay
4.3.6. National Cancer Institute (NCI)—60 Cell Line Evaluation
4.3.7. General Instrumental Analysis Procedures
Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC)
LC-MS
NMR and FTIR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyle, S.A.; Reeder, D.R. Colorado Sagebrush: A Conservation Assessment and Strategy. Colorado Division of Wildlife: Grand Junction, CO, USA, 2005. Available online: https://cpw.state.co.us/Documents/WildlifeSpecies/Sagebrush/CHAPTER2overviewsagebrushecosystems.pdf (accessed on 10 October 2023).
- Briggs, G.M. Inanimate Life. 2021. Available online: https://milnepublishing.geneseo.edu/botany/chapter/sagebrush/ (accessed on 12 October 2023).
- Anibogwu, R.; Jesus, K.D.; Pradhan, S.; Pashikanti, S.; Mateen, S.; Sharma, K. Extraction, isolation and characterization of bioactive compounds from Artemisia and their biological significance: A review. Molecules 2021, 26, 6995. [Google Scholar] [CrossRef]
- Bora, K.S.; Sharma, A. The genus Artemisia: A comprehensive review. Pharm. Biol. 2011, 49, 101–109. [Google Scholar] [CrossRef]
- Bisht, D.; Kumar, D.; Kumar, D.; Dua, K.; Chellappan, D.K. Phytochemistry and pharmacological activity of the genus Artemisia. Arch. Pharm. Res. 2021, 44, 439–474. [Google Scholar] [CrossRef]
- Cui, Z.; Li, S.; Chang, J.; Zang, E.; Liu, Q.; Zhou, B.; Li, C.; Li, M.; Huang, X.; Zhang, Z.; et al. The pharmacophylogenetic relationships of two edible medicinal plants in the genus Artemisia. Front. Plant Sci. 2022, 13, 949743. [Google Scholar] [CrossRef]
- West, N.E. Western Intermountain sagebrush steppe. In Temperate Deserts and Semi-Deserts; West, N.E., Ed.; Elsevier Science: Amsterdam, The Netherlands, 1983; pp. 351–374. [Google Scholar]
- Hussain, M.; Thakur, R.K.; Khazir, J.; Ahmed, S.; Khan, M.I.; Rahi, P.; Peer, L.A.; Pragadheesh, V.; Kaur, S.; Raina, S.N.; et al. Traditional uses, phytochemistry, pharmacology, and toxicology of the genus Artemisia, L. (Asteraceae): A high-value medicinal plant. Curr. Top. Med. Chem. 2023, 23. [Google Scholar] [CrossRef] [PubMed]
- Erdogrul, O.T. Antibacterial activities of some plant extracts used in folk medicine. Pharm. Biol. 2002, 40, 269. [Google Scholar] [CrossRef]
- Jelodar, N.B.; Bhatt, A.; Mohamed, K.; Keng, C.L. New cultivation approaches of Artemisia annua L. for a sustainable production of the antimalarial drug artemisinin. J. Med. Plant Resour. 2014, 8, 441–447. [Google Scholar]
- Trendafilova, A.; Moujir, L.; Sousa, P.M.C.; Seca, A.M.L. Research advances on health effects of edible Artemisia species and some sesquiterpene lactones constituents. Foods 2020, 10, 65. [Google Scholar] [CrossRef]
- Kinney, C.R.; Sugihara, J. Constituents of Artemisia tridentate (American Sage brush) II. J. Org. Chem. 1943, 8, 290–294. [Google Scholar] [CrossRef]
- Allen, G. The Herbalist in the Kitchen; University of Illinois Press: Champaign, IL, USA, 2010; ISBN 025209039X. [Google Scholar]
- Yanovsky, E. Food Plants of the North American Indians; U.S. Department of Agriculture: Washington, DC, USA, 1936. [Google Scholar]
- Moerman, D.E. Native American Food Plants: An Ethnobotanical Dictionary; Timber Press Inc.: Portland, OR, USA, 2010; ISBN 9781604691894. [Google Scholar]
- Amorim, M.H.R.; Gilda Costa, R.M.; Lopes, C.; Bastos, M.M.S.M. Sesquiterpene lactones: Adverse health effects and toxicity mechanisms. Crit. Rev. Toxicol. 2013, 43, 559–579. [Google Scholar] [CrossRef]
- Ivanescu, B.; Miron, A.; Corciova, A. Sesquiterpene lactones from Artemisia Genus: Biological activities and methods of analysis. J. Anal. Methods Chem. 2015, 2015, 247685. [Google Scholar] [CrossRef]
- Kawasaki, B.T.; Kalathur, M.; Ana, M. Effects of the sesquiterpene lactone parthenolide on prostate tumor-initiating cells: An integrated molecular profiling approach. Prostate 2009, 69, 827–837. [Google Scholar] [CrossRef]
- Ullah, A.; Munir, S.; Badshah, S.L.; Khan, N.; Ghani, L.; Jaremko, M.; Emwas, A. Important flavonoids and their role as a therapeutic agent. Molecules 2020, 25, 5243. [Google Scholar] [CrossRef] [PubMed]
- Lh, Y.; Ym, J.; Shi, J.; Fa, T.; Datta, N.; Singanusong, R.; Ss, C. Flavonoids in food and their health benefits. Plant Foods Hum. Nutr. 2004, 59, 113–122. [Google Scholar] [CrossRef]
- Khan, A.N.; Dilshad, E. Enhanced Antioxidant and Anticancer Potential of Artemisia carvifolia Buch Transformed with rol A Gene. Metabolites 2023, 13, 351. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.; Luthria, D.L.; Sasaki, T.; Heyerick, A. Flavonoids from Artemisia annua L. as antioxidants and their potential synergism with Artemisinin against malaria and cancer. Molecules 2010, 15, 3135–3170. [Google Scholar] [CrossRef] [PubMed]
- Matvieieva, N.; Drobot, K.; Duplij, V.; Ratushniak, Y.; Shakhovsky, A.M.; Kyrpa-Nesmiian, T.; Mickevičius, S.; Brindza, J. Flavonoid content and antioxidant activity of Artemisia vulgaris L. “hairy” roots. Prep. Biochem. Biotechnol. 2019, 49, 82–87. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Swor, K.; Satyal, P.; Timsina, S.; Setzer, W.N. Chemical Composition and Terpenoid Enantiomeric Distribution of the Essential oil of Artemisia tridentata Subsp. tridentata From Southwestern Idaho. Nat. Prod. Commun. 2022, 17, 1934578X2211174. [Google Scholar] [CrossRef]
- Shafizadeh, F.; Bhadane, N.; Morris, M.S.; Kelsey, R.G.; Khanna, S.N. Sesquiterpene lactones of big sagebrush. Phytochemistry 1971, 10, 2745–2754. [Google Scholar] [CrossRef]
- Geisssman, T.A.; Stewart, T.; Irwin, M.A. Sesquiterpene lactones of Artemisia species-II. Phytochemistry 1967, 6, 901. [Google Scholar] [CrossRef]
- Dadabay, C.Y.; Spaulding, P.B.; Valenzuela, E.; Turner, M.; Eckert, K.E.; Julkunen-Tiitto, R.; Noblit, N.; Mansfield, D.H. Polyphenols from the sagebrush Artemisia tridentata ssp. tridentata affect the redox state of cultured hepatocytes by direct and indirect mechanisms. Curr. Top. Phytochem. 2019, 15, 15–25. [Google Scholar]
- Martinez, M.V.; Munnoz-Zamora, A.; Joseph-Nathan, P. Conformational analysis of achillin and leukodin. J. Nat. Prod. 1988, 51, 221–228. [Google Scholar] [CrossRef]
- Zhanzhaxina, A.; Seiilgazy, M.; Jalmakhanbetova, R.I.; Ishmuratova, M.; Seilkhanov, T.M.; Oyama, M.; Sarmurzina, Z.; Tekebayeva, Z.B.; Suleimen, E.M. flavonoids from Pulicaria vulgaris and their Antimicrobial Activity. Chem. Nat. Compd. 2020, 56, 915–917. [Google Scholar] [CrossRef]
- Cuong, D.T.D.; Dat, H.T.; Duan, N.T.; Thuong, P.D.; Phat, N.T.; Tri, M.D.; Dang, V.; Hoa, N.T.; Tuyen, P.N.K.; Phụng, N.K.P. Isolation and characterization of six flavonoids from the leaves of Sterculia foetida Linn. Vietnam. J. Chem. 2019, 57, 438–442. [Google Scholar] [CrossRef]
- Batra, P.; Sharma, A.K. Anti-cancer potential of flavonoids: Recent trends and future perspectives. Biotech 2013, 3, 439–459. [Google Scholar] [CrossRef]
- Yuan, H.; Lu, X.; Ma, Q.; Li, D.; Xu, G.; Piao, G. Flavonoids from Artemisia sacrorum Ledeb. and their cytotoxic activities against human cancer cell lines. Exp. Ther. Med. 2016, 12, 1873–1878. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, H.; Tang, C.; Yao, S.; Ke, C.; Xu, C.; Ye, Y. Cytotoxic sesquiterpene lactones from Artemisia anomala. Phytochem. Lett. 2017, 20, 177–180. [Google Scholar] [CrossRef]
- Gunawardena, K.; Rivera, S.B.; Epstein, W.W. The monoterpenes of Artemisia tridentata ssp. vaseyana, Artemisia cana ssp. viscidula and Artemisia tridentata ssp. spiciformis. Phytochemistry 2002, 59, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.H.; Bhat, K.A.; Khuroo, M.A. Arglabin: From isolation to antitumor evaluation. Chem.-Biol. Interact. 2015, 240, 180–198. [Google Scholar] [CrossRef] [PubMed]
- Chadwick, M.; Trewin, H.; Gawthrop, F.; Wagstaff, C. Sesquiterpenoids lactones: Benefits to plants and people. Int. J. Mol. Sci. 2013, 14, 12780–12805. [Google Scholar] [CrossRef]
- Kalidindi, S.; Jeong, W.B.; Schall, A.; Bandichhor, R.; Nosse, B.; Reiser, O. Enantioselective synthesis of arglabin. Angew. Chem. Int. Ed. 2007, 46, 6361–6363. [Google Scholar] [CrossRef] [PubMed]
- Burits, M.; Asres, K.; Bucar, F. The antioxidant activity of the essential oils of Artemisia afra, Artemisia abyssinica and Juniperus procera. Phytother. Res. 2001, 15, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Benzie, I.F.; Strain, S. The ferric reducing ability of plasma (FRAP) as a measure of “Antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- NCI-60 Screening Methodology|NCI-60 Human Tumor Cell Lines Screen|Discovery & Development Services|Developmental Therapeutics Program (DTP). Available online: https://dtp.cancer.gov/discovery_development/nci-60/methodology.htm (accessed on 19 March 2021).
| Protons | Chemical Shift (ppm) | Integration | Multiplicity | Coupling Constants (Hz) |
|---|---|---|---|---|
| H3 | 2.458 | 1H | dq | J3,3α =11.2 Hz; J3,10 =6.9 Hz |
| H3α | 2.347 | 1H | m | |
| H4 | 4.830 | 1H | td | J4,3α/J4,5α = 10.7 Hz; J4,5β = 2.1 Hz |
| H5α | 2.726 | 1H | dd | J5α,5β = 13.4 Hz; J5α,4 = 11.0 Hz |
| H5β | 2.375 | 1H | dd | J5α,5β = 13.4 Hz; J5β,4 = 2.1 Hz |
| H8 | 6.179 | 1H | m | J6,12 = 1.8 Hz |
| H9α | 3.416 | 1H | d | J9α,9β = 10.0 Hz |
| H9β | 3.728 | 1H | dd | J9β,3α = 10.3 Hz; J9β,9α = 10.0 Hz |
| H10 | 1.340 | 3H | d | J10,3 = 6.9 Hz |
| H11 | 2.413 | 3H | bs | |
| H12 | 2.349 | 3H | bs | |
| H14 | 2.117 | 3H | s |
| 13C NMR: CDCl3 | Chemical Shift (ppm) | BioRad Spectra Base Comparison |
|---|---|---|
| C2 | 176.85 | 176.6 |
| C3 | 40.73 | 40.7 |
| C3α | 59.10 | 59.1 |
| C4 | 70.38 | 70.4 |
| C5 | 44.75 | 44.7 |
| C6 | 145.15 | 145.0 |
| C6α | 133.28 | 133.3 |
| C7 | 195.27 | 195.1 |
| C8 | 135.95 | 135.8 |
| C9 | 169.73 | 169.6 |
| C9α | 51.59 | 51.6 |
| C9β | 81.12 | 81.1 |
| C10 | 15.06 | 15.0 |
| C11 | 21.44 | 21.1 |
| C12 | 20.01 | 19.0 |
| C13 | 169.82 | 169.7 |
| C14 | 21.23 | 21.3 |
| HETCOR (CDCl3) | Chemical Shift (ppm) | 13C |
|---|---|---|
| H3 | 2.458 | 40.73 |
| H3α | 2.347 | 59.10 |
| H4 | 4.830 | 70.38 |
| H5α | 2.726 | 44.75 |
| H5β | 2.375 | 44.75 |
| H8 | 6.179 | 135.95 |
| H9α | 3.416 | 51.59 |
| H9b | 3.728 | 81.12 |
| H10 | 1.340 | 15.06 |
| H11 | 2.413 | 21.44 |
| H12 | 2.349 | 20.01 |
| H14 | 2.117 | 21.23 |
| COSY (CDCl3) | Chemical Shift (ppm) | Correlations | |||
|---|---|---|---|---|---|
| H3 | 2.458 | 2.347 | 1.340 | ||
| H3α | 2.347 | 4.830 | 3.728 | 2.458 | |
| H4 | 4.830 | 2.726 | 2.375 | 2.347 | |
| H5α | 2.726 | 4.830 | 2.726 | ||
| H5β | 2.375 | 4.830 | 2.375 | ||
| H8 | 6.179 | 2.349 | 3.416 (wk) | 2.413 (wk) | |
| H9α | 3.416 | 3.728 | 6.179 (wk) | 2.413 (wk) | 2.347 (wk) |
| H9β | 3.728 | 3.416 | 2.347 | ||
| H10 | 1.340 | 2.458 | |||
| H11 | 2.413 | 6.179 | 3.416 | ||
| H12 | 2.349 | 6.179 | 3.416 | ||
| H14 | 2.117 | ||||
| HMBC (CDCl3) | Chemical Shift (ppm) | Correlations | ||||||
|---|---|---|---|---|---|---|---|---|
| H3 | 2.458 | 176.85 | 59.10 | 15.06 | 70.38 | 20.01 | ||
| H3α | 2.347 | 81.12 | 70.38 | 40.73 | 195.27 | 51.59 | 44.75 | 15.06 |
| H4 | 4.830 | 169.82 | ||||||
| H5α | 2.375 | 145.15 | 70.38 | 133.28 | 59.10 | 21.44 | ||
| H5β | 2.726 | 145.15 | 70.38 | 133.28 | 59.10 | 21.44 | ||
| H8 | 6.179 | 195.27 | 169.73 | 133.28 | 51.59 | 20.01 | ||
| H9α | 3.416 | 169.73 | 133.28 | 81.12 | 145.15 (wk) | 135.95 | 59.10 | |
| H9β | 3.728 | 133.28 | 70.38 | |||||
| H10 | 1.340 | 40.73 | 176.85 | 59.10 | ||||
| H11 | 2.413 | 145.15 | 133.28 | 44.75 | ||||
| H12 | 2.349 | 169.73 | 51.59 | |||||
| H14 | 2.117 | 169.82 | 20.01 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anibogwu, R.; Jesus, K.D.; Pradhan, S.; Leuven, S.V.; Sharma, K. Sesquiterpene Lactones and Flavonoid from the Leaves of Basin Big Sagebrush (Artemisia tridentata subsp. tridentata): Isolation, Characterization and Biological Activities. Molecules 2024, 29, 802. https://doi.org/10.3390/molecules29040802
Anibogwu R, Jesus KD, Pradhan S, Leuven SV, Sharma K. Sesquiterpene Lactones and Flavonoid from the Leaves of Basin Big Sagebrush (Artemisia tridentata subsp. tridentata): Isolation, Characterization and Biological Activities. Molecules. 2024; 29(4):802. https://doi.org/10.3390/molecules29040802
Chicago/Turabian StyleAnibogwu, Rosemary, Karl De Jesus, Samjhana Pradhan, Shanae Van Leuven, and Kavita Sharma. 2024. "Sesquiterpene Lactones and Flavonoid from the Leaves of Basin Big Sagebrush (Artemisia tridentata subsp. tridentata): Isolation, Characterization and Biological Activities" Molecules 29, no. 4: 802. https://doi.org/10.3390/molecules29040802
APA StyleAnibogwu, R., Jesus, K. D., Pradhan, S., Leuven, S. V., & Sharma, K. (2024). Sesquiterpene Lactones and Flavonoid from the Leaves of Basin Big Sagebrush (Artemisia tridentata subsp. tridentata): Isolation, Characterization and Biological Activities. Molecules, 29(4), 802. https://doi.org/10.3390/molecules29040802






