Characterization of Copper(II) and Zinc(II) Complexes of Peptides Mimicking the CuZnSOD Enzyme
Abstract
1. Introduction
2. Experimental
2.1. Chemicals
2.2. High-Performance Liquid Chromatography
2.3. Potentiometric Measurements
2.4. Spectroscopic Studies
2.5. Superoxide Dismutase Activity
3. Results and Discussion
3.1. Protonation Equilibria of the Peptides
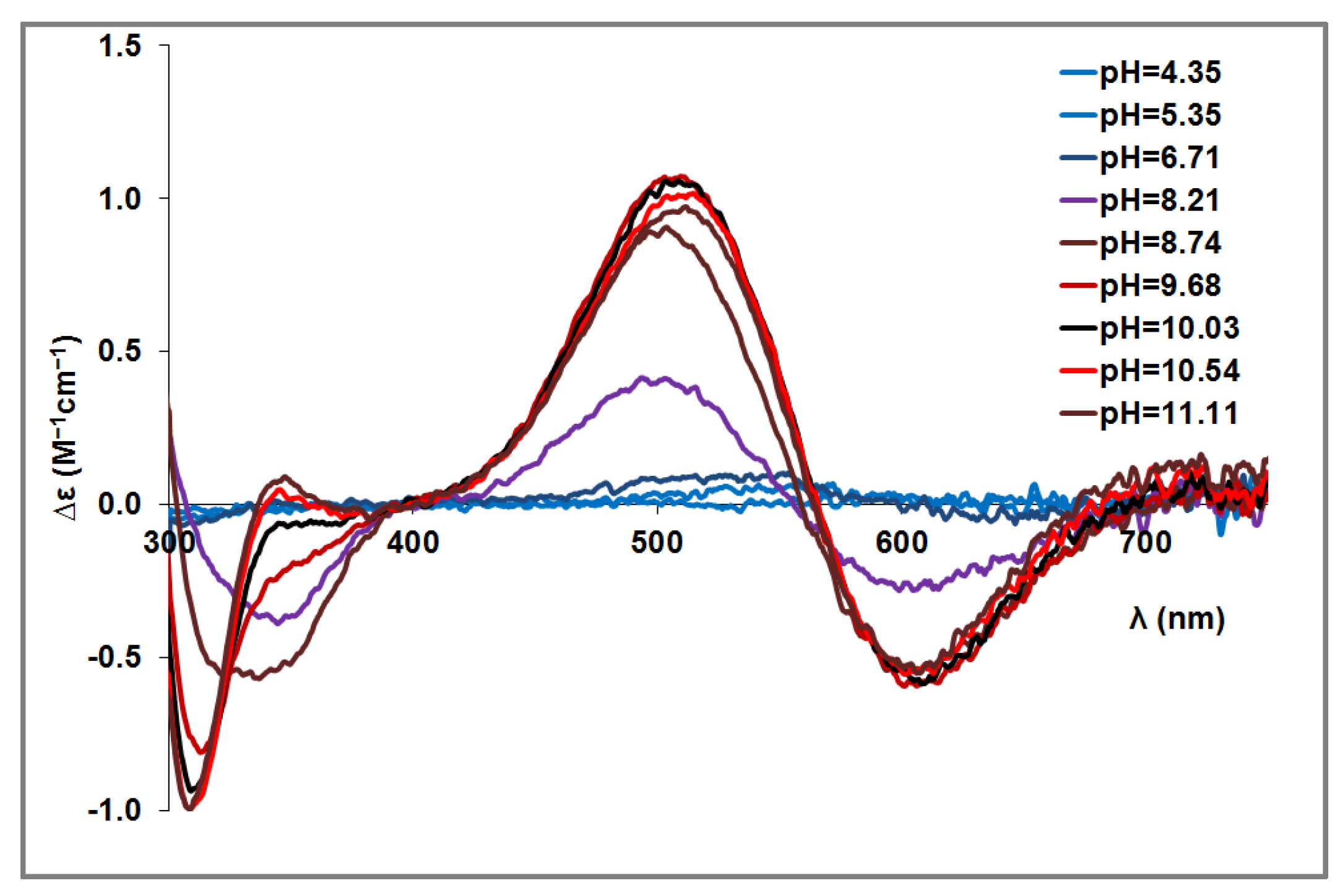
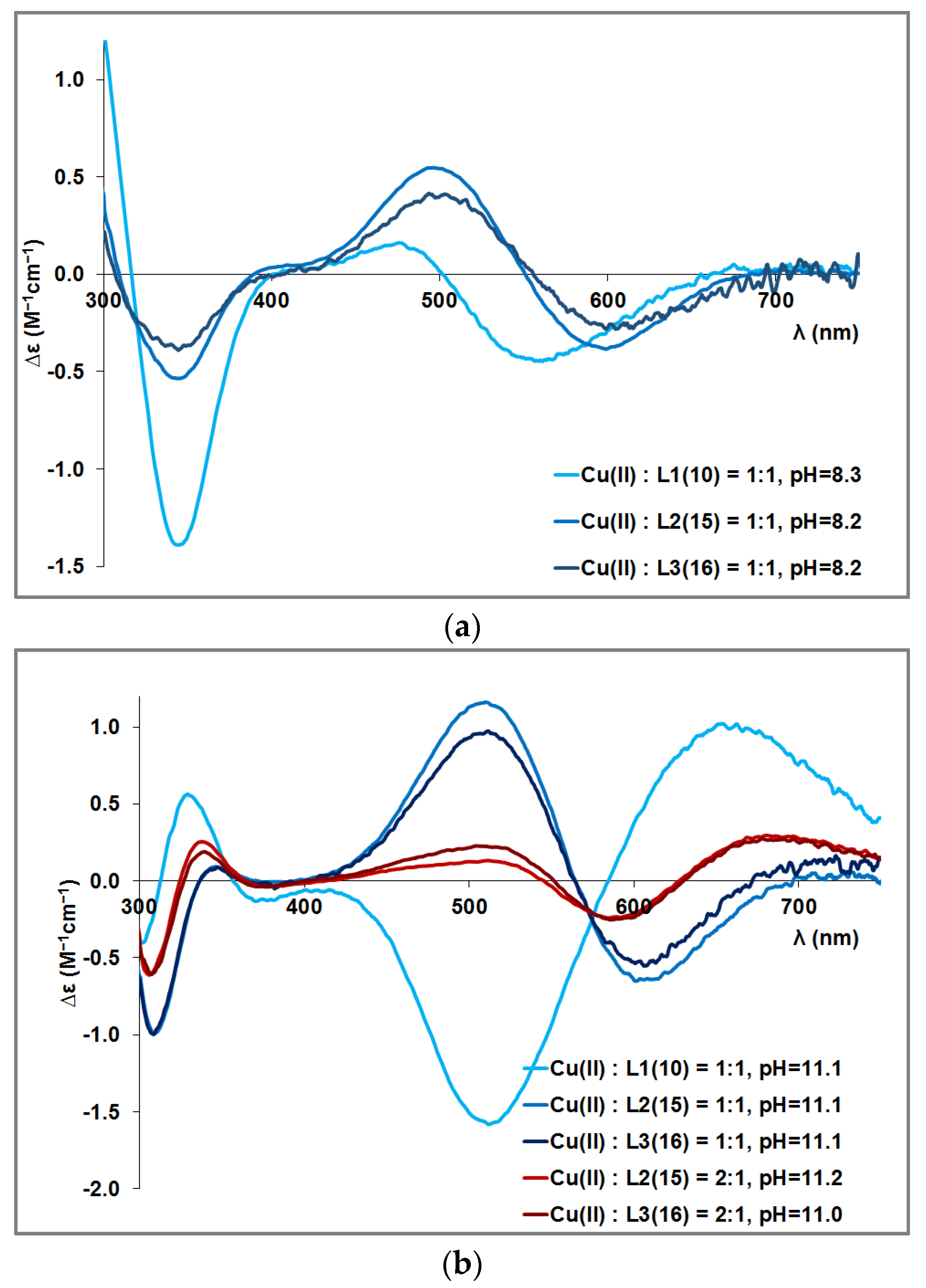
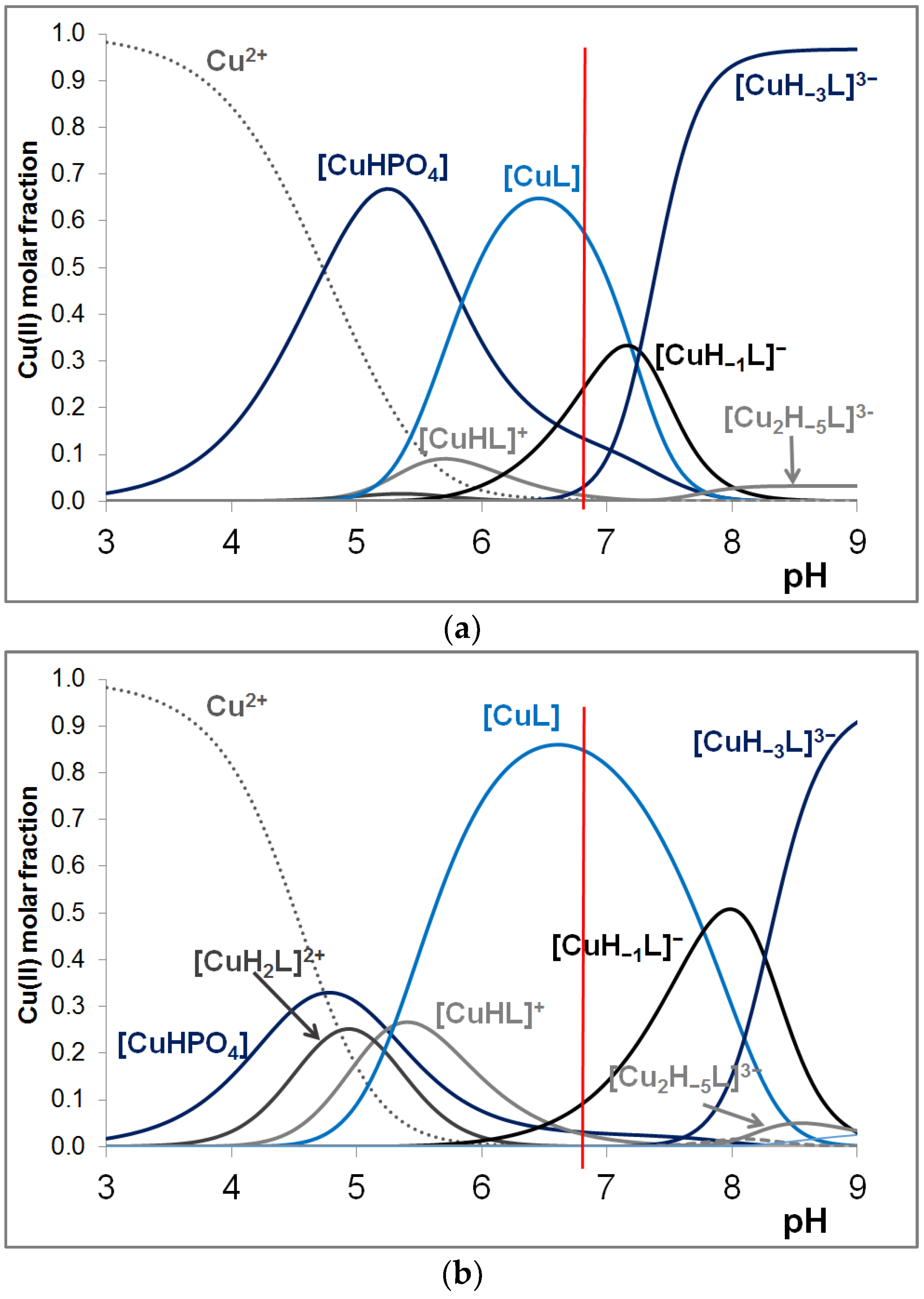
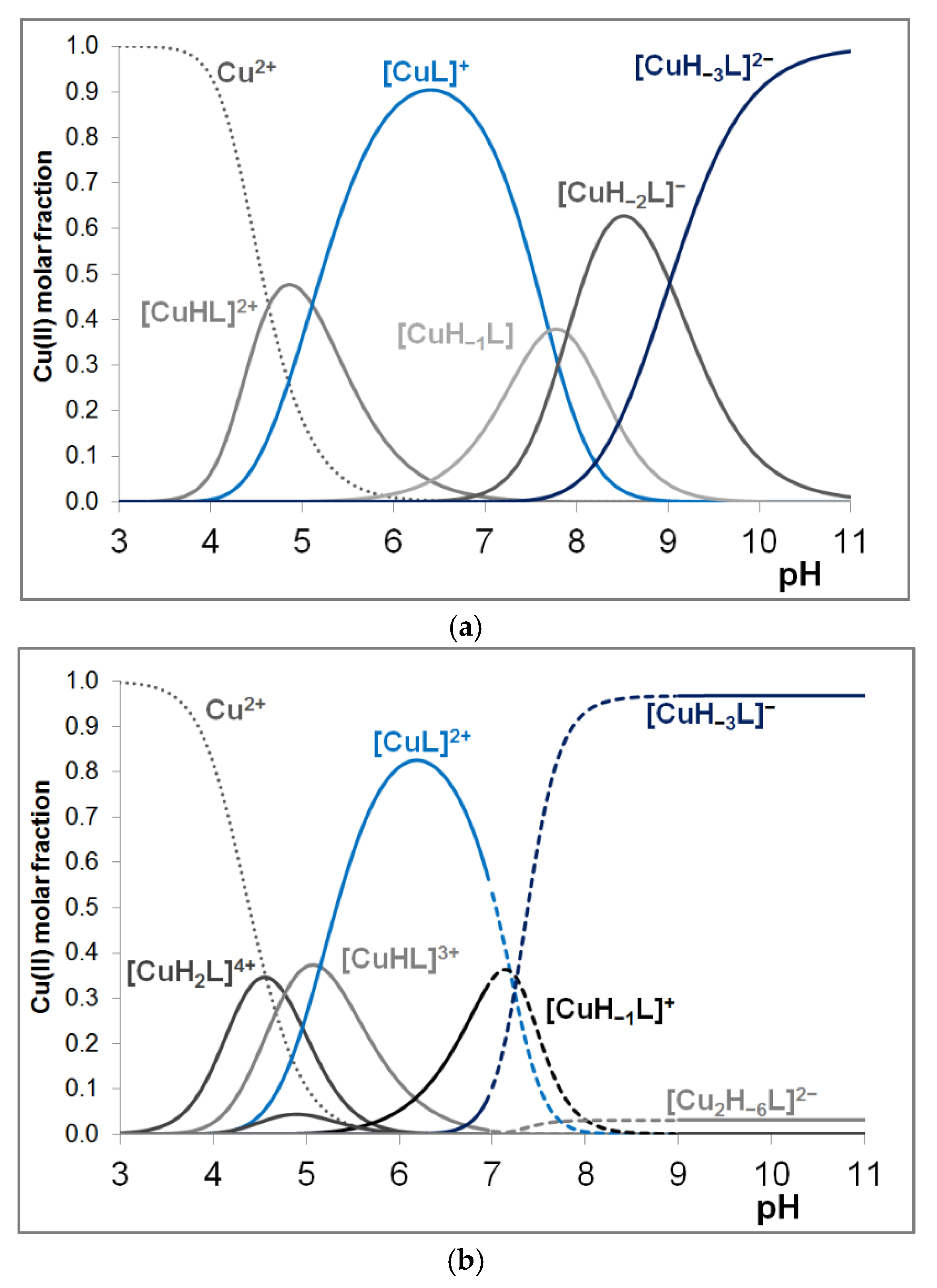
3.2. Copper(II) Complexes of the Peptides
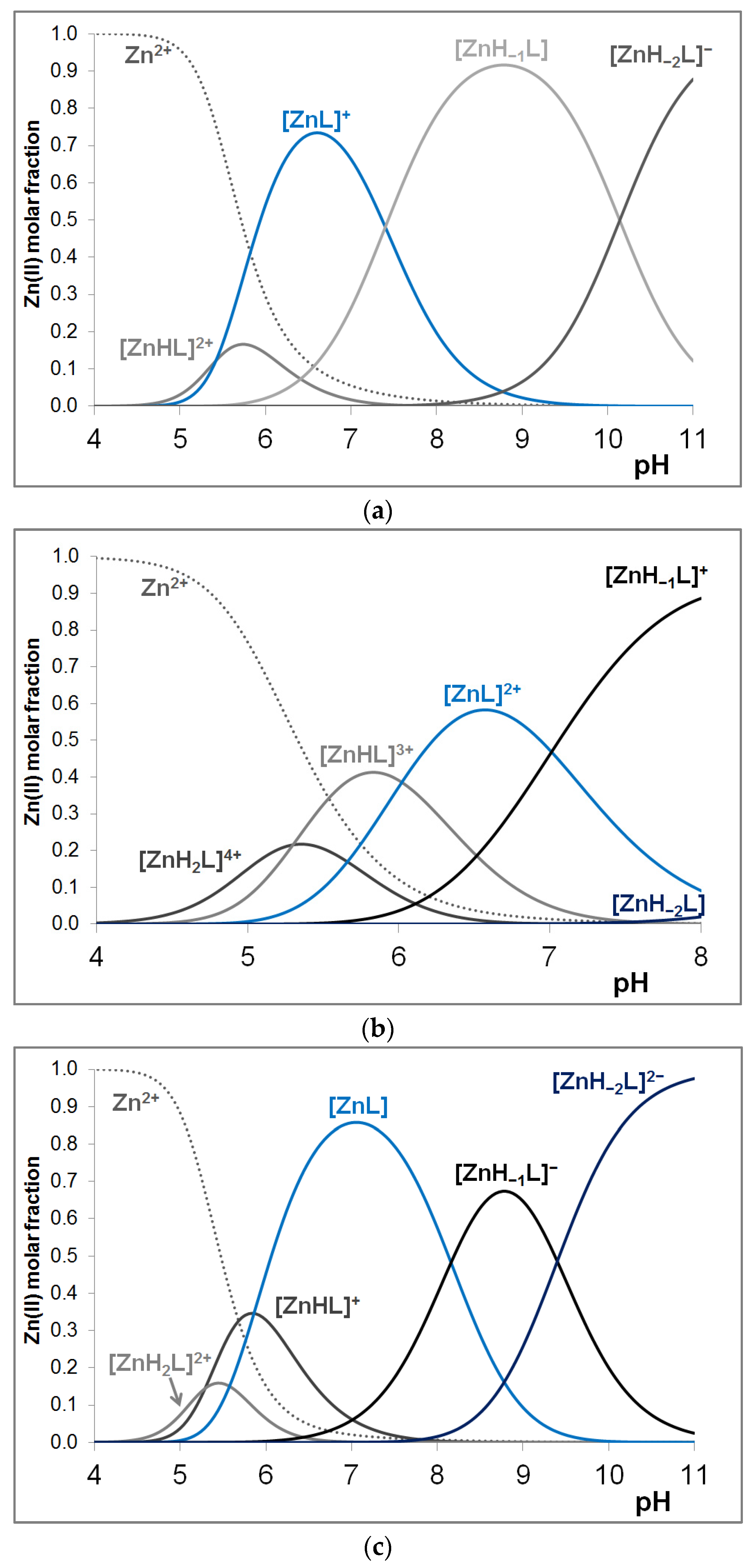
3.3. Zinc(II) Complexes of the Peptides
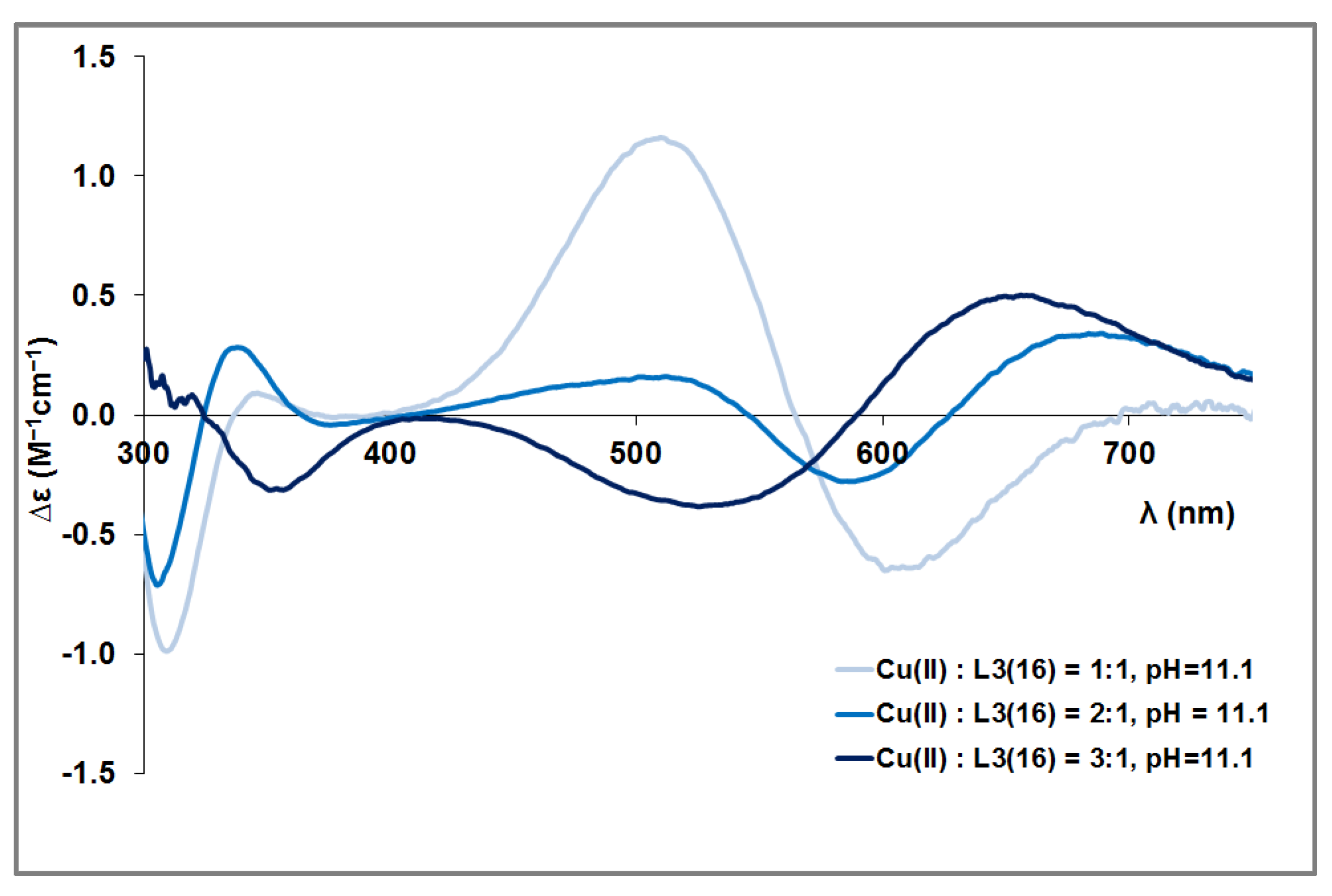
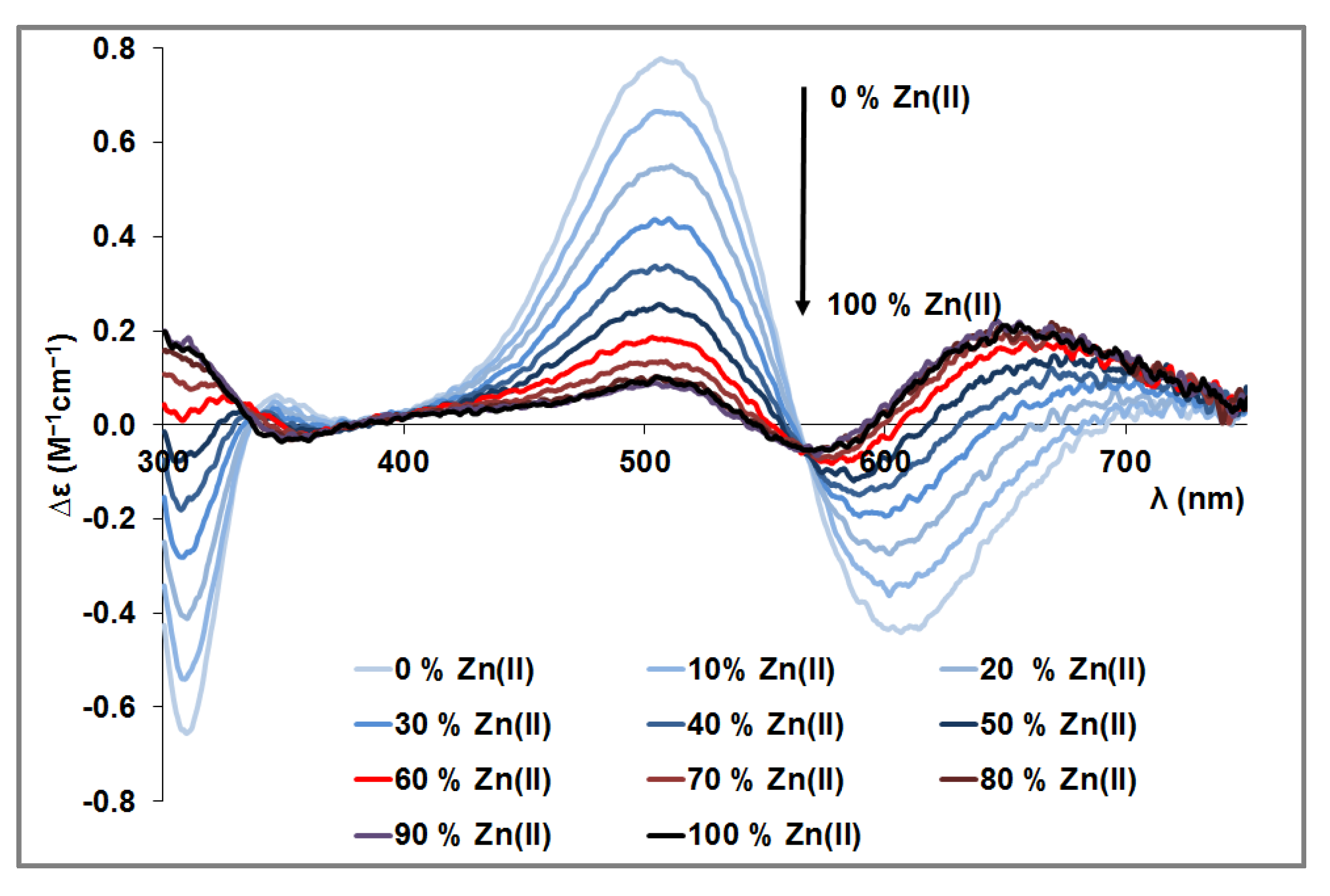
3.4. Mixed Cu(II)/Zn(II) Complexes of the Peptides
3.5. Superoxide Dismutase Activity Measurements:
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Ac2O | acetic anhydride |
| AcOH | acetic acid |
| ACN | acetonitrile |
| A, Ala | alanine, alanyl |
| CD | circular dichroism spectroscopy |
| DCM | dichloromethane |
| DIPEA | N,N-Diisopropylethylamine |
| DMF | N,N-Dimethylmethanamide |
| DODT | 2,2′-(Ethylenedioxy)diethanethiol |
| D, Asp | aspartic acid, aspartyl |
| Et2O | diethyl ether |
| E, Glu | glutamine, glutamyl |
| Fmoc | fluorenylmethyloxycarbonyl |
| F, Phe | phenylalanine, phenylalanyl |
| G, Gly | glycine, glycyl |
| H, His | histidine, histidinyl |
| HOBt | 1H-Benzotriazol-1-ol |
| HPLC | High-Performance Liquid Chromatography |
| IC50 | the concentration that causes 50% inhibition of NBT reduction |
| M, mM, μM | mol·dm−3, mmol·dm−3, μmol·dm−3 |
| NBT | nitro blue tetrazolium chloride |
| NMP | N-methyl-pyrrolidone |
| N, Asn | asparagine, asparaginyl |
| OtBu | 5-tert-Butyl |
| P, Pro | proline, prolyl |
| RP-HPLC | Reversed-Phase High-Performance Liquid Chromatography |
| SOD | superoxide dismutase |
| TBTU | 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium tetrafluoroborate |
| TFA | trifluoroacetic acid |
| TIS | triisopropylsilane |
| Trt | trityl |
| UV-vis | UV-visible spectroscopy |
| V, Val | valine, valyl |
| [X] | equilibrium concentration of X |
| βpqr | stability constants of MpHqLr species |
| the change in absorbance per minute at 560 nm in the absence of complex | |
| the change in absorbance per minute in the presence of X μM of the complex |
Appendix A
| Species | Composition | m/z (Measured) | m/z (Calculated) |
|---|---|---|---|
| L1(10) | |||
| L- | |||
| H2L+ | C59H77N18O14+ | 1261.629 | 1261.586 |
| NaHL+ | C59H76N18NaO14+ | 1283.620 | 1283.568 |
| KHL+ | C59H76KN18O14+ | 1299.592 | 1299.542 |
| Na2L+ | C59H76N18Na2O14+ | 1305.599 | 1305.599 |
| L2(15) | |||
| L | |||
| HL+ | C74H100N25O17+ | 1610.768 | 1610.772 |
| H2L2+ | C74H101N25O172+ | 805.888 | 805.890 |
| H3L3+ | C74H102N25O173+ | 537.595 | 537.596 |
| L3(16) | |||
| L2- | |||
| H3L+ | C80H107N26O22+ | 1783.78 | 1783.8048 |
| NaH2L+ | C80H106N26NaO22+ | 1805.77 | 1805.7867 |
| KH2L+ | C80H106KN26O22+ | 1821.74 | 1821.7606 |
| Na2HL+ | C80H105N26Na2O22+ | 1827.75 | 1827.7687 |
| KNaHL+ | C80H105KN26NaO22+ | 1843.71 | 1843.7426 |
| Na2HL+ | C80H104N26Na3O22+ | 1849.73 | 1849.7506 |
| Species | Binding Mode | λmax (nm) (ε (M−1 cm−1)) | ||
|---|---|---|---|---|
| L1(10) | L2(15) | L3(16) | ||
| [CuH−3L] | [N-, N-, N-, Im] | 659 (+0.989) 510 (−1.563) 410 (−0.068) 372 (−0.128) 332 (+0.547) 300 (−0.378) 261 (+6.341) w | 748 (+0.064) w 606 (−0.684) 511 (+1.191) 348 (+0.097) 309 (−1.012) 278 (+1.309) 256 (+0.677) | 718 (+0.127) 606 (−0.551) 513 (+0.966) 350 (+0.083) 309 (−0.997) 276 (+1.590) 259 (+1.059) |
| [Cu2H−6L] | 2 × [N-, N-, N-, Im] | – | 686 (+0.328) w 586 (−0.271) 516 (+0.144) 379 (−0.042) 339 (+0.286) 307 (−0.686) 266 (+3.060) 253 (+2.850) | 678 (+0.538) 576 (−0.235) 496 (−0.112) 446 (+0.024) 374 (−0.085) 336 (+0.262) 306 (−0.792) 263 (+4.195) |



References
- Fridovich, I. Fundamental aspects of reactive oxygen species, or what’s the matter with oxygen. Ann. N. Y. Acad. Sci. 1999, 893, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Parkes, T.L.; Elia, A.J.; Dickinson, D.; Hilliker, A.J.; Phillips, J.P.; Boulianne, G.L. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat. Genet. 1998, 19, 171–174. [Google Scholar] [CrossRef] [PubMed]
- McCord, J.M.; Fridovich, I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Lyons, T.J.; Nersissian, A.; Huang, H.; Yeom, H.; Nishida, C.R.; Graden, J.A.; Butler Gralla, E.; Selverstone Valentine, J. The metal binding properties of the zinc site of yeast copper-zinc superoxide dismutase: Implications for amyotrophic lateral sclerosis. J. Biol. Inorg. Chem. 2000, 5, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.J.; King, G.W.; LoBuglio, A.F. Superoxide generation by human monocytes and macrophages. Am. J. Hematol. 1978, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Debets-Ossenkopp, Y.; Verbrugh, H.A.; Verhoef, J. Initiation of the respiratory burst of human neutrophils by influenza virus. Infect. Immun. 1981, 32, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Klug, D.; Rabani, J.; Fridovich, I. A direct demonstration of the catalytic action of superoxide dismutase through the use of pulse radiolysis. J. Biol. Chem. 1972, 247, 4839–4842. [Google Scholar] [CrossRef]
- Schmidt, P.J.; Rae, T.D.; Pufahl, R.A.; Hamma, T.; Strain, J.; O’Halloran, T.V.; Culotta, V.C. Multiple protein domains contribute to the action of the copper chaperone for superoxide dismutase. J. Biol. Chem. 1999, 274, 23719–23725. [Google Scholar] [CrossRef]
- Hall, L.T.; Sanchez, R.J.; Holloway, S.P.; Zhu, H.; Stine, J.E.; Lyons, T.J.; Demeler, B.; Schirf, V.; Hansen, J.C.; Nersissian, A.M.; et al. X-ray Crystallographic and analytical ultracentrifugation analyses of truncated and full-length yeast copper chaperones for SOD (LYS7): A dimer−dimer model of LYS7−SOD association and copper delivery. Biochemistry 2000, 39, 3611–3623. [Google Scholar] [CrossRef]
- Torres, A.S.; Petri, V.; Rae, T.D.; O’Halloran, T.V. Copper stabilizes a heterodimer of the yCCS metallochaperone and its target superoxide dismutase. J. Biol. Chem. 2001, 276, 38410–38416. [Google Scholar] [CrossRef]
- Lamb, A.L.; Torres, A.S.; O’Halloran, T.V.; Rosenzweig, A.C. Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol. 2001, 8, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Teoh, M.L.T.; Walasek, P.J.; Evans, H.D. Leporipoxvirus Cu,Zn-superoxide dismutase (SOD) homologs are catalytically inert decoy proteins that bind copper chaperone for SOD. J. Biol. Chem. 2003, 278, 33175–33184. [Google Scholar] [CrossRef] [PubMed]
- Yount, N.Y.; Bayer, A.S.; Xiong, Y.Q.; Yeaman, M.R. Advances in antimicrobial peptide immunobiology. Biopolymers 2006, 84, 435–458. [Google Scholar] [CrossRef]
- Pasupuleti, M.; Davoudi, M.; Malmsten, M.; Schmidtchen, A. Antimicrobial activity of a C-terminal peptide from human extracellular superoxide dismutase. BMC Res. Notes 2009, 2, 136. [Google Scholar] [CrossRef] [PubMed]
- Steiner, H.; Hultmark, D.; Engstrom, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef]
- Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–54453. [Google Scholar] [CrossRef]
- Gudmundsson, G.H.; Lidholm, D.A.; Asling, B.; Gan, R.; Boman, H.G. The cecropin locus. Cloning and expression of a gene cluster encoding three antibacterial peptides in Hyalophora cecropia. J. Biol. Chem. 1991, 266, 11510–11517. [Google Scholar] [CrossRef]
- Agerberth, B.; Gunne, H.; Odeberg, J.; Kogner, P.; Boman, H.G.; Gudmundsson, G.H. FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc. Natl. Acad. Sci. USA 1995, 92, 195–199. [Google Scholar] [CrossRef]
- Cowland, J.B.; Johnsen, A.H. Borregaard, hCAP-18, a cathelin/pro-bactenecin-like protein of human neutrophil specific granules. FEBS Lett. 1995, 368, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.E.; Gallant, J.W.; Gong, Z.; Hew, C. Cloning and developmental expression of a family of pleurocidin-like antimicrobial peptides from winter flounder, Pleuronectes americanus (Walbaum). Dev. Comp. Immunol. 2001, 25, 137–147. [Google Scholar] [CrossRef]
- Wieprecht, T.; Beyermann, M.; Seelig, J. Thermodynamics of the coil-alpha-helix transition of amphipathic peptides in a membrane environment: The role of vesicle curvature. Biophys. Chem. 2002, 96, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Boman, A.; Boman, H.G. Ascaris nematodes from pig and human make three antibacterial peptides: Isolation of cecropin P1 and two ASABF peptides. Cell. Mol. Life Sci. 2003, 60, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Lequin, O.; Bruston, F.; Convert, O.; Chassaing, G.; Nicolas, P. Helical structure of dermaseptin B2 in a membrane-mimetic environment. Biochemistry 2003, 42, 10311–10323. [Google Scholar] [CrossRef]
- Pukala, T.L.; Brinkworth, C.S.; Carver, J.A.; Bowie, J.H. Investigating the importance of the flexible hinge in caerin 1.1: Solution structures and activity of two synthetically modified caerin peptides. Biochemistry 2004, 43, 937–944. [Google Scholar] [CrossRef]
- Syvitski, R.T.; Burton, I.; Mattatall, N.R.; Douglas, S.E.; Jakeman, D.L. Structural characterization of the antimicrobial peptide pleurocidin from winter flounder. Biochemistry 2005, 44, 7282–7293. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.Q.; Yuan, J.; Osapay, G.; Osapay, K.; Tran, D.; Miller, C.J.; Ouellette, A.J.; Selsted, M.E. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science 1999, 286, 498–502. [Google Scholar] [CrossRef]
- Yount, N.Y.; Yeaman, M.R. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 7363–7368. [Google Scholar] [CrossRef]
- Ganz, T. Defensins and other antimicrobial peptides: A historical perspective and an update. Comb. Chem. High. Throughput Screen 2005, 8, 209–217. [Google Scholar] [CrossRef]
- Wieprecht, T.; Apostolov, O.; Beyermann, M.; Seelig, J. Thermodynamics of the α-helix-coil transition of amphipathic peptides in a membrane environment: Implications for the peptide-membrane binding equilibrium. J. Mol. Biol. 1999, 294, 785–794. [Google Scholar] [CrossRef]
- Marshall, S.H. Antimicrobial peptides: A natural alternative to chemical antibiotics and a potential for applied biotechnology. Electron. J. Biotechnol. 2003, 6, 3. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Andersson, E.; Rydengård, V.; Sonesson, A.; Mörgelin, M.; Björck, L.; Schmidtchen, A. Antimicrobial activities of heparin-binding peptides. Eur. J. Biochem. 2004, 271, 1219–1226. [Google Scholar] [CrossRef]
- Nordahl, E.A.; Rydengård, V.; Nyberg, P.; Nitsche, D.P.; Mörgelin, M.; Malmsten, M.; Björck, L.; Schmidtchen, A. Activation of the complement system generates antibacterial peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 16879–16884. [Google Scholar] [CrossRef] [PubMed]
- Pasupuleti, M.; Walse, B.; Nordahl, E.A.; Morgelin, M.; Malmsten, M.; Schmidtchen, A. Preservation of antimicrobial properties of complement peptide C3a, from invertebrates to humans. J. Biol. Chem. 2007, 282, 2520–2528. [Google Scholar] [CrossRef] [PubMed]
- Nordahl, E.A.; Rydengård, V.; Mörgelin, M.; Schmidtchen, A. Domain 5 of high molecular weight kininogen is antibacterial. J. Biol. Chem. 2005, 280, 34832–34839. [Google Scholar] [CrossRef] [PubMed]
- Malmsten, M.; Davoudi, M.; Schmidtchen, A. Bacterial killing by heparin-binding peptides from PRELP and thrombospondin. Matrix Biol. 2006, 25, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Malmsten, M.; Davoudi, M.; Walse, B.; Rydengard, V.; Pasupuleti, M.; Morgelin, M.; Schmidtchen, A. Antimicrobial peptides derived from growth factors. Growth Factors 2007, 25, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, F.G.; Xu, T.; McMillian, F.M.; Levitz, S.M.; Diamond, R.D.; Offner, G.D.; Troxler, R.F. Histatins, a novel family of histidine-rich proteins in human parotid secretion.Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J. Biol. Chem. 1988, 263, 7472–7477. [Google Scholar] [CrossRef]
- Frank, R.W.; Gennaro, R.; Schneider, K.; Przybylski, M.; Romeo, D. Amino acid sequences of two proline-rich bactenecins. Antimicrobial peptides of bovine neutrophils. J. Biol. Chem. 1990, 265, 18871–18874. [Google Scholar] [CrossRef] [PubMed]
- Selsted, M.E.; Novotny, M.J.; Morris, W.L.; Tang, Y.Q.; Smith, W.; Cullor, J.S. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J. Biol. Chem. 1992, 267, 4292–4295. [Google Scholar] [CrossRef] [PubMed]
- Cabiaux, V.; Agerberth, B.; Johansson, J.; Homble, F.; Goormaghtigh, E.; Ruysschaert, J.M. Secondary structure and membrane interaction of PR-39, a Pro+ Arg-rich antibacterial peptide. Eur. J. Biochem. 1994, 224, 1019–1027. [Google Scholar] [CrossRef]
- Gudmundsson, G.H.; Magnusson, K.P.; Chowdhary, B.P.; Johansson, M.; Andersson, L.; Boman, H.G. Structure of the gene for porcine peptide antibiotic PR-39, a cathelin gene family member: Comparative mapping of the locus for the human peptide antibiotic FALL-39. Proc. Natl. Acad. Sci. USA 1995, 92, 7085–7089. [Google Scholar] [CrossRef] [PubMed]
- Schibli, D.J.; Hwang, P.M.; Vogel, H.J. Structure of the antimicrobial peptide tritrpticin bound to micelles: A distinct membrane-bound peptide fold. Biochemistry 1999, 38, 16749–16755. [Google Scholar] [CrossRef]
- Rozek, A.; Friedrich, C.L.; Hancock, R.E. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry 2000, 39, 15765–15774. [Google Scholar] [CrossRef]
- Rydengard, V.; Olsson, A.K.; Morgelin, M.; Schmidtchen, A. Histidinerich glycoprotein exerts antibacterial activity. FEBS J. 2007, 274, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Rydengard, V.; Shannon, O.; Lundqvist, K.; Kacprzyk, L.; Chalupka, A.; Olsson, A.K.; Morgelin, M.; Jahnen-Dechent, W.; Malmsten, M.; Schmidtchen, A. Histidine-rich glycoprotein protects from systemic Candida infection. PLoS Pathog. 2008, 4, e1000116. [Google Scholar] [CrossRef]
- Frueh, J.; Gai, M.; Yang, Z.; He, Q. Influence of Polyelectrolyte Multilayer Coating on the Degree and Type of Biofouling in Freshwater Environment. J. Nanosci. Nanotechnol. 2014, 14, 4341–4350. [Google Scholar] [CrossRef]
- Badaraev, A.D.; Lerner, M.I.; Bakina, O.V.; Sidelev, D.V.; Tran, T.-H.; Krinitcyn, M.G.; Malashicheva, A.B.; Cherempey, E.G.; Slepchenko, G.B.; Kozelskaya, A.I.; et al. Antibacterial Activity and Cytocompatibility of Electrospun PLGA Scaffolds Surface-Modified by Pulsed DC Magnetron Co-Sputtering of Copper and Titanium. Pharmaceutics 2023, 15, 939. [Google Scholar] [CrossRef]
- Suksrichavalit, T.; Prachayasittikul, S.; Nantasenamat, C.; Isarankura-NaAyudhya, C.; Prachayasittikul, V. Copper complexes of pyridine derivatives with superoxide scavenging and antimicrobial activities. Eur. J. Med. Chem. 2009, 44, 3259–3265. [Google Scholar] [CrossRef]
- Mehta, J.V.; Gajera, S.B.; Patel, M.N. Antimalarial, antimicrobial, cytotoxic, DNA interaction and SOD like activities of tetrahedral copper(II) complexes. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 136, 1881–1892. [Google Scholar] [CrossRef]
- Joseph, J.; Boomadevi Janaki, G.; Nagashri, K.; Selwin Joseyphus, R. Antimicrobial, antioxidant and SOD activities of copper(II) complexes derived from 2-aminobenzothiazole derivatives. J. Coord. Chem. 2017, 70, 242–260. [Google Scholar] [CrossRef]
- Csire, G.; Timári, S.; Asztalos, J.; Király, J.M.; Kiss, M.; Várnagy, K. Coordination, redox properties and SOD activity of Cu(II) complexes of multihistidine peptides. J. Inorg. Biochem. 2017, 177, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Grenács, Á.; Kaluha, A.; Kállay, C.; Jószai, V.; Sanna, D.; Sóvágó, I. Binary and ternary mixed metal complexes of terminally free peptides containing two different histidyl binding sites. J. Inorg. Biochem. 2013, 128, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Raics, M.; Sanna, D.; Sóvágó, I.; Kállay, C. Copper(II), nickel(II) and zinc(II) complexes of hexapeptides containing separate aspartyl and histidyl residues. Inorg. Chim. Acta 2015, 426, 99–106. [Google Scholar] [CrossRef]
- Csire, G.; Nagy, L.; Várnagy, K.; Kállay, C. Copper(II) interaction with the Human Prion 103-112 fragment-Coordination and oxidation. J. Inorg. Biochem. 2017, 170, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Irving, H.M.; Miles, M.G.; Pettit, L.D. A study of some problems in determining the stoicheiometric proton dissociation constants of complexes by potentiometric titrations using a glass electrode. Anal. Chim. Acta 1967, 38, 475–488. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. SUPERQUAD: An improved general program for computation of formation constants from potentiometric data. J. Chem. Soc. Dalton Trans. 1985, 6, 1195–1200. [Google Scholar] [CrossRef]
- Zékány, L.; Nagypál, I. Computational Methods for the Determination of Formation Constants; Leggett, D.J., Ed.; Plenum Press: New York, NY, USA, 1985; pp. 291–353. [Google Scholar]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1974, 44, 276–287. [Google Scholar] [CrossRef]
- Goldstein, S.; Michel, C.; Bors, W.; Saran, M.; Czapski, G. A critical reevaluation of some assay methods for superoxide dismutase activity. Free Radic. Biol. Med. 1988, 4, 295–303. [Google Scholar] [CrossRef]
- Durot, S.; Policar, C.; Cisnetti, F.; Lambert, F.; Renault Pelosi, G.; Blain, G.; Korri-Youssoufi, H.; Mahy, J.-P. Series of Mn complexes based on N-centered ligands and superoxide−reactivity in an anhydrous medium and SOD-like activity in an aqueous medium correlated to Mn II/Mn III redox potentials. Eur. J. Inorg. Chem. 2005, 2005, 3513–3523. [Google Scholar] [CrossRef]
- Székely, E.; Csire, G.; Balogh, B.D.; Erdei, J.Z.; Király, J.M.; Kocsi, J.; Pinkóczy, J.; Várnagy, K. The role of side chains in the fine-tuning of the metal-binding ability of multihistidine peptides. Molecules 2022, 27, 3435. [Google Scholar] [CrossRef]
- Kállay, C.; Várnagy, K.; Malandrinos, G.; Hadjiliadis, N.; Sanna, D.; Sóvágó, I. Thermodynamic and structural characterization of the macrochelates formed in the reactions of copper(II) and zinc(II) ions with peptides of histidine. Inorg. Chim. Acta 2009, 362, 935–945. [Google Scholar] [CrossRef]
- Lukács, M.; Szunyog, G.; Grenács, Á.; Lihi, N.; Kállay, C.; Di Natale, G.; Campagna, T.; Lanza, V.; Tabbi, G.; Pappalardo, G.; et al. Copper(II) coordination abilities of the tau protein’s N-terminus peptide fragments: A combined potentiometric, spectroscopic and mass spectrometric study. Chem. Plus Chem. 2019, 84, 1697–1708. [Google Scholar] [CrossRef]
- Billo, E. Copper(II) chromosomes and the rule of average environment. J. Inorg. Nucl. Chem. Lett. 1974, 10, 613–617. [Google Scholar] [CrossRef]
- Jószai, V.; Nagy, Z.; Ősz, K.; Sanna, D.; Di Natale, G.; La Mendola, D.; Pappalardo, G.; Rizzarelli, E.; Sóvágó, I. Transition metal complexes of terminally protected peptides containing histidyl residues. J. Inorg. Biochem. 2006, 100, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Csire, G.; Turi, I.; Sóvágó, I.; Kárpáti, E.; Kállay, C. Complex formation processes and metal ion catalyzed oxidation of model peptides related to the metal binding site of the human prion protein. J. Inorg. Biochem 2020, 203, 110927. [Google Scholar] [CrossRef]
- Ohtsu, H.; Shimazaki, Y.; Odani, A.; Yamauchi, O.; Mori, W.; Itoh, S.; Fukuzumi, S. Synthesis and characterization of imidazolate-bridged dinuclear complexes as active site models of Cu,Zn-SOD. J. Am. Chem. Soc. 2000, 122, 5733–5741. [Google Scholar] [CrossRef]
- Diószegi, R.; Bonczidai-Kelemen, D.; Bényei, A.C.; May, N.V.; Fábián, I.; Lihi, N. Copper(II) complexes of pyridine-2,6-dicarboxamide ligands with high SOD activity. Inorg. Chem. 2022, 61, 2319–2332. [Google Scholar] [CrossRef] [PubMed]
- Kachadourian, R.; Batinic-Haberle, I.; Fridovich, I. Syntheses and superoxide dismuting activities of partially (1−4) β-chlorinated derivatives of manganese(III) meso-tetrakis(N-ethylpyridinium-2-yl)porphyrin. Inorg. Chem. 1999, 38, 391–396. [Google Scholar] [CrossRef]
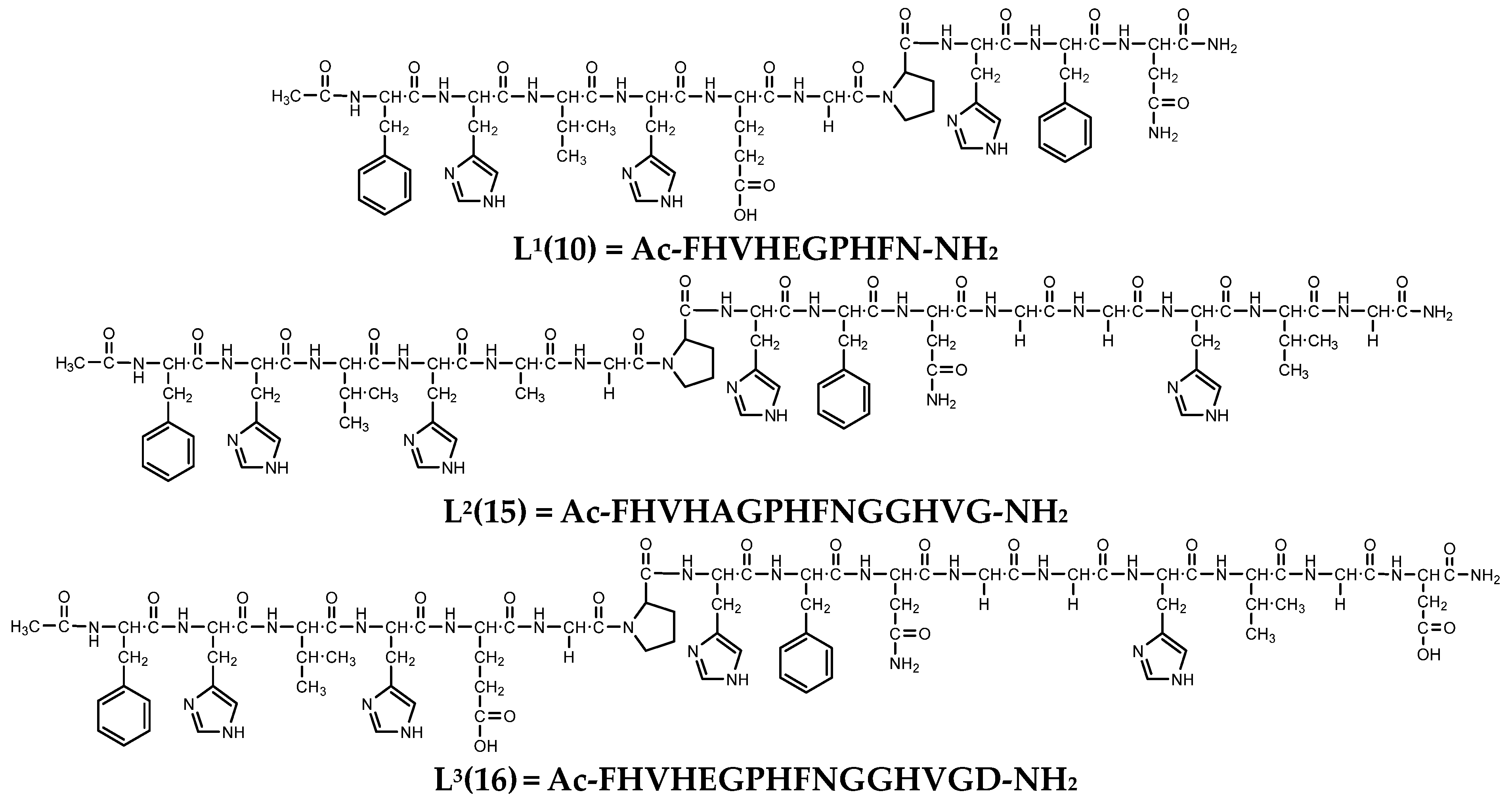

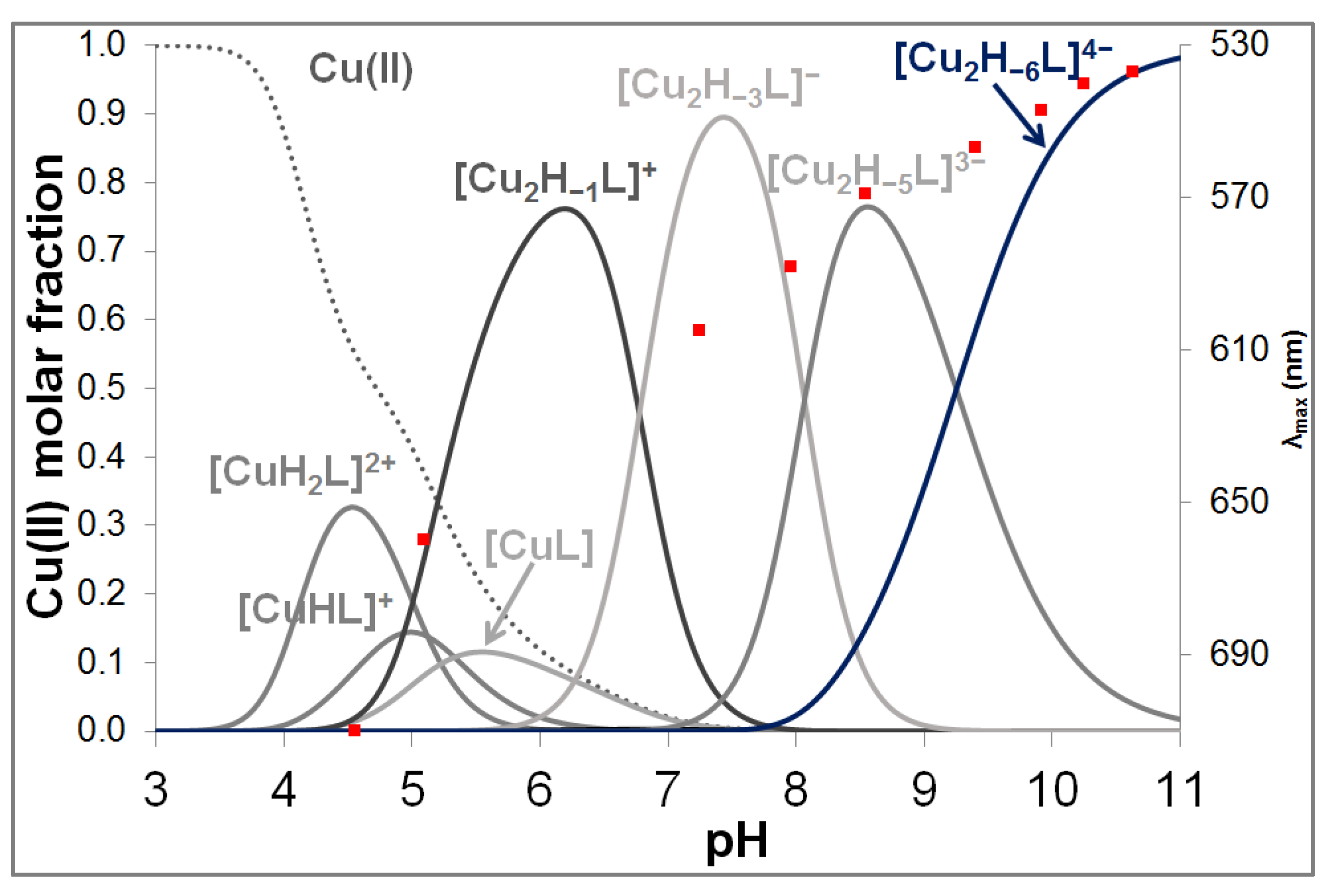
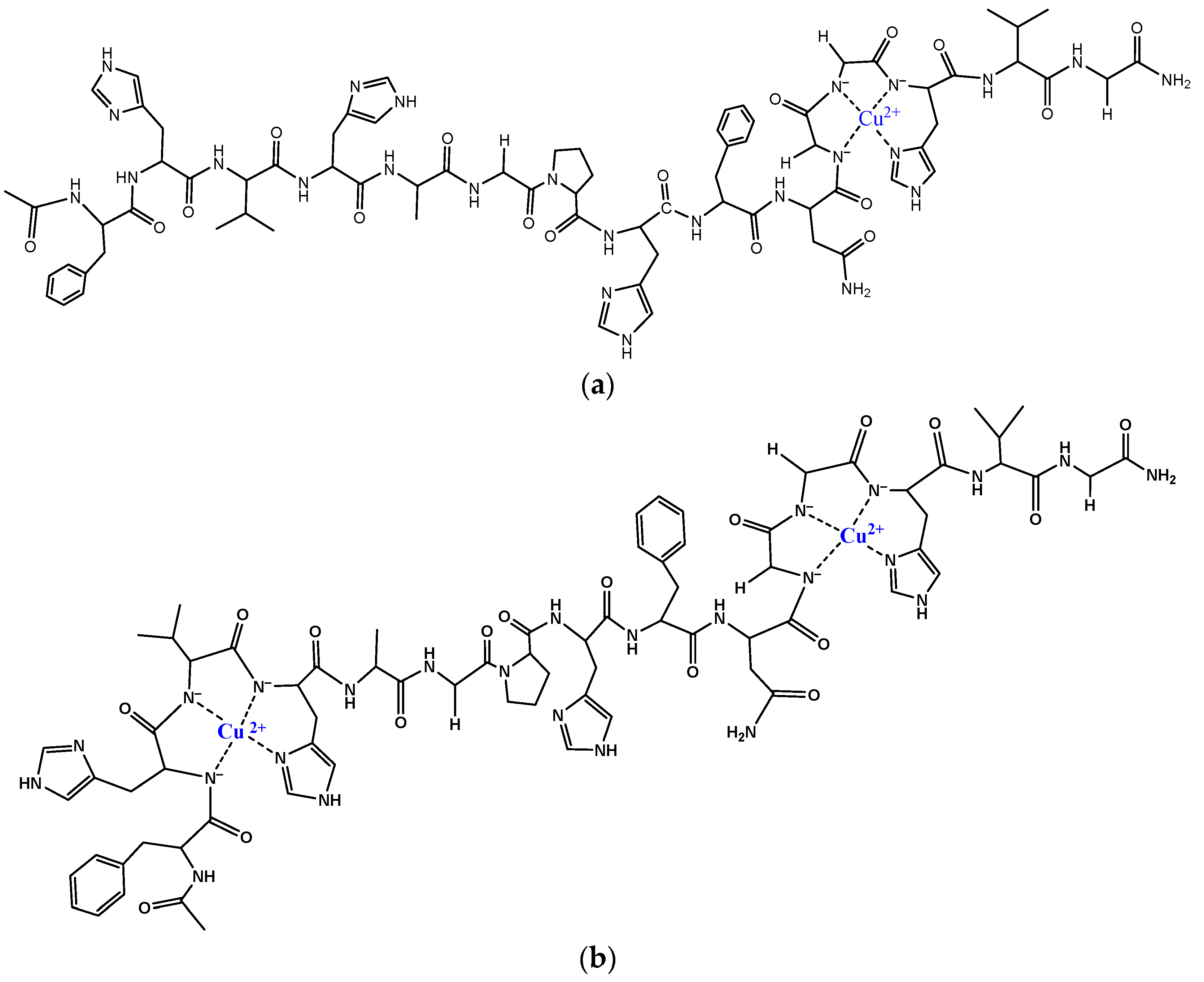
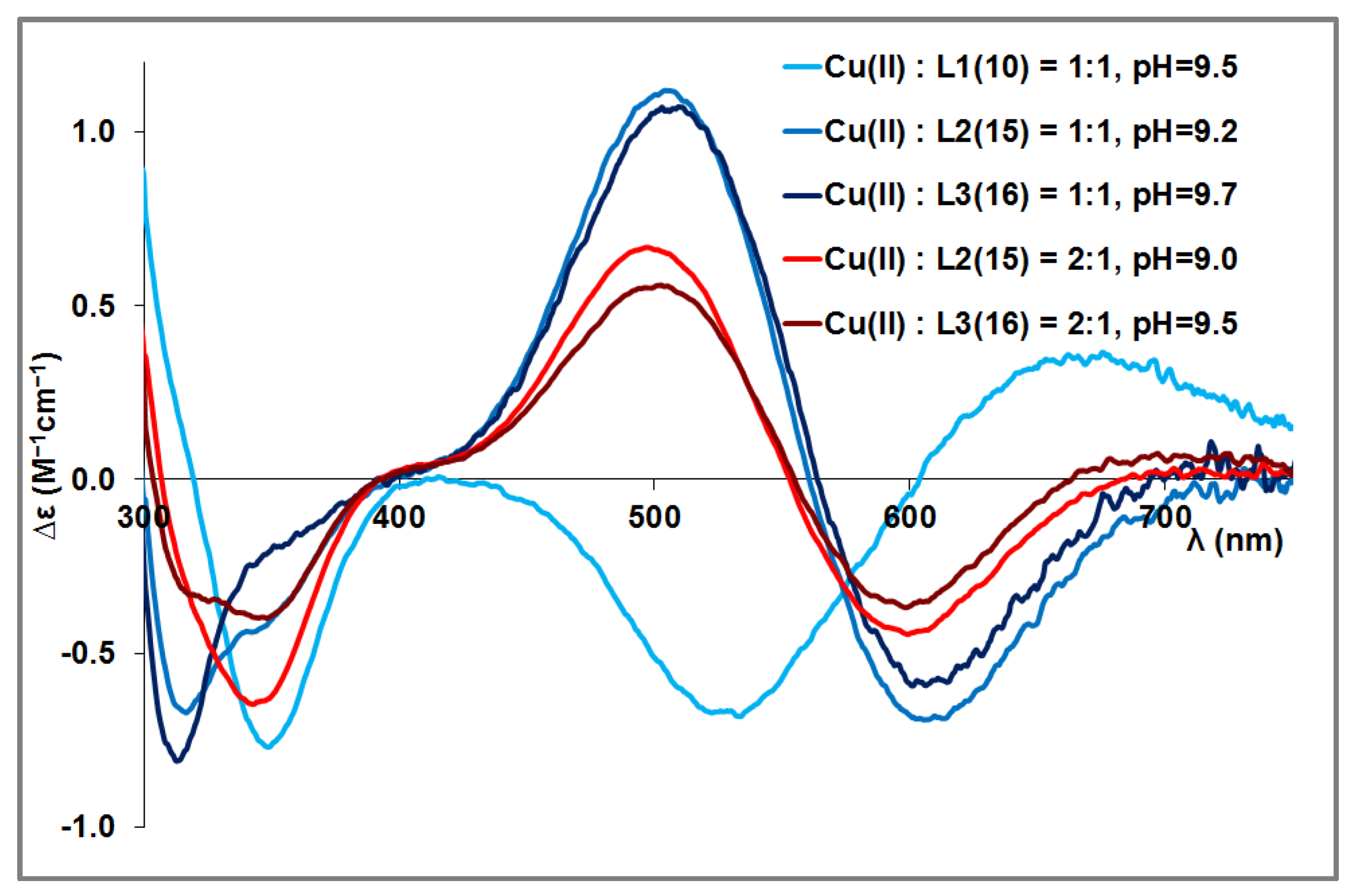

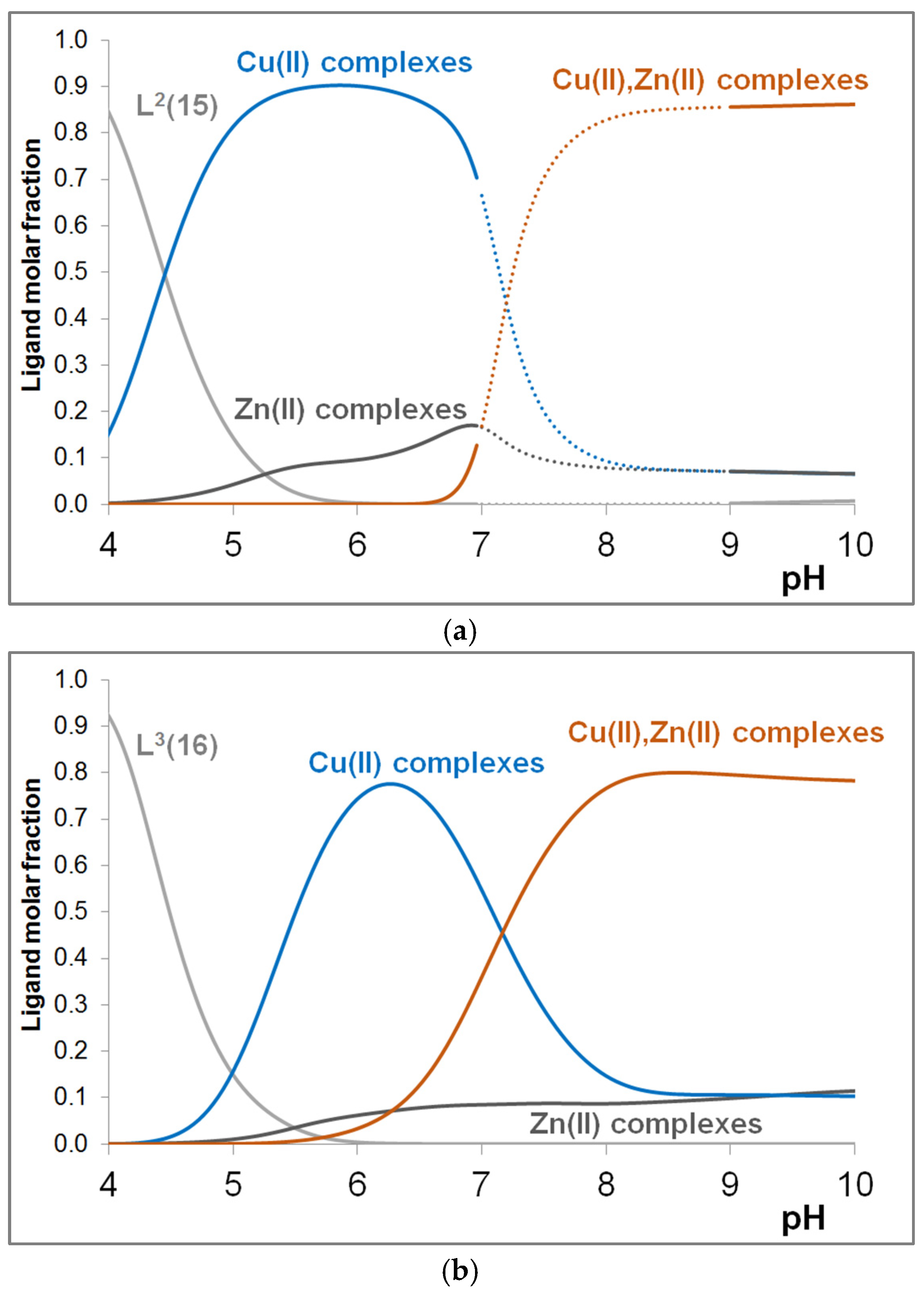
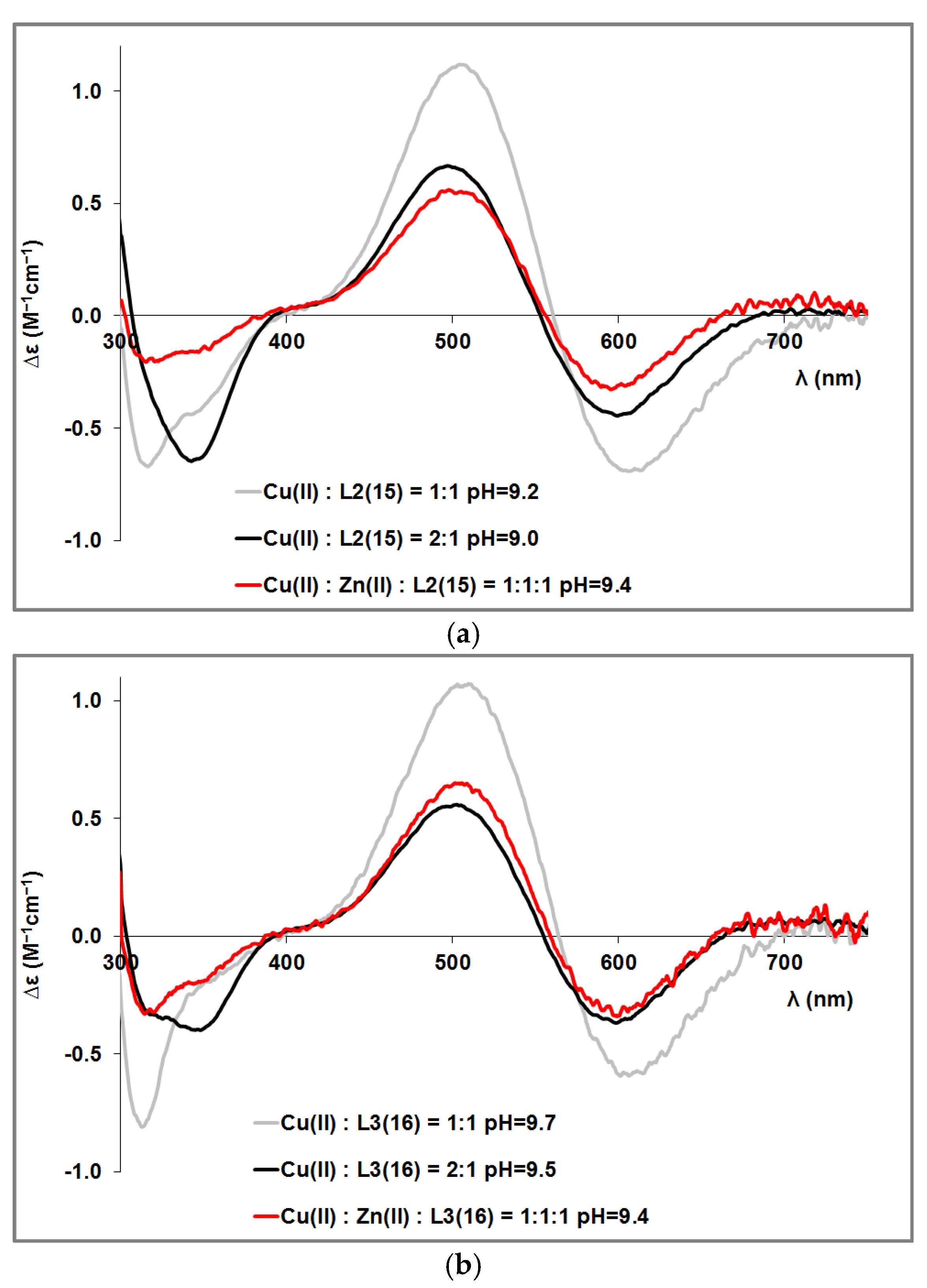
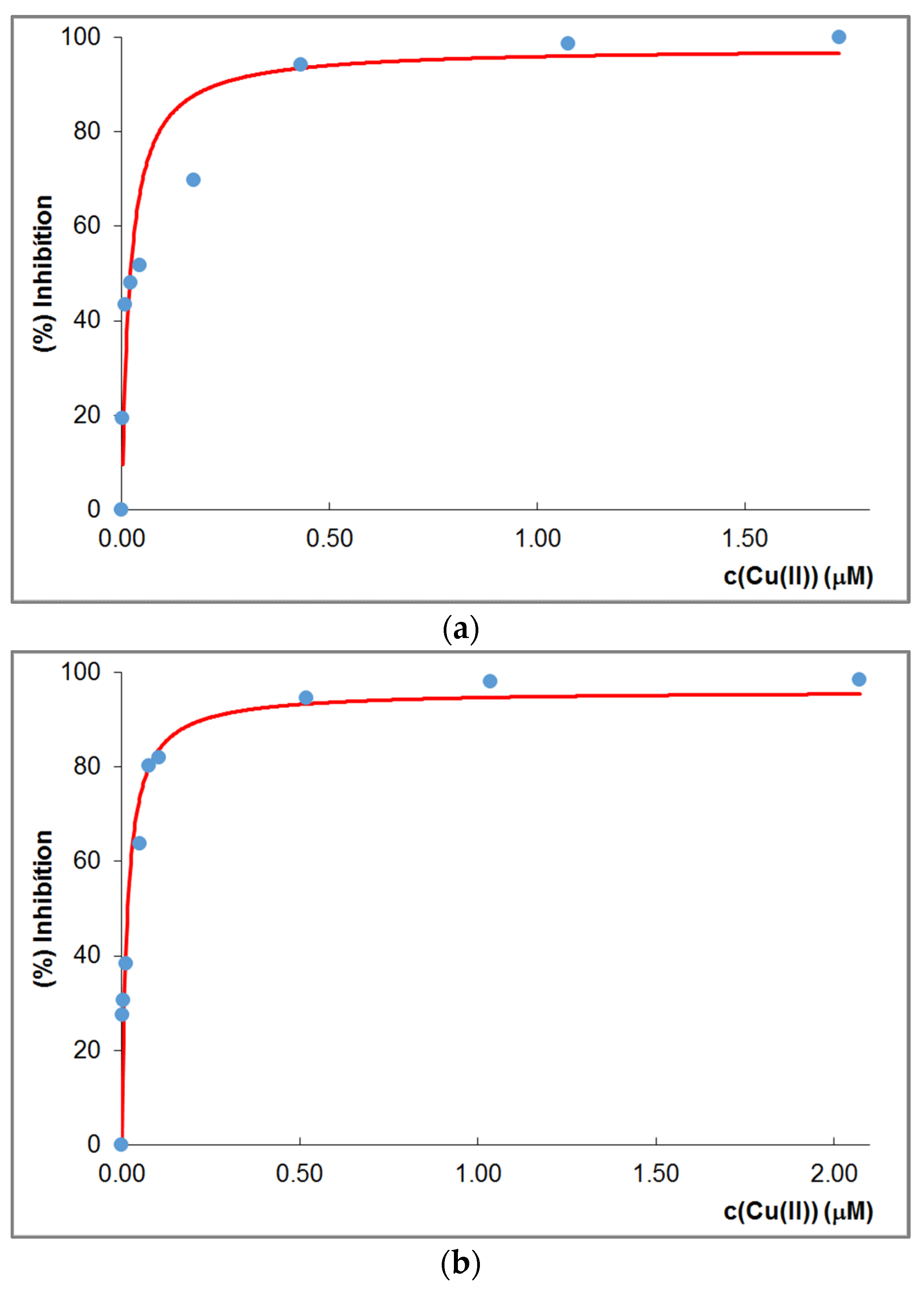
| pK(Asp)1 | pK(Asp)2 | pK(Glu) | pK (His)1 | pK(His)2 | pK(His)3 | pK(His)4 | |
|---|---|---|---|---|---|---|---|
| L1(10) | – | – | 4.15 (3) | 5.97 (2) | 6.32 (1) | – | 7.62 (7) |
| L2(15) | – | – | – | 5.43 (1) | 6.12 (1) | 6.47 (1) | 6.83 (2) |
| L3(16) | 3.49 (1) | 4.27 (1) | 5.60 (1) | 6.36 (1) | 6.53 (1) | 7.85 (3) | |
| Ac-HADHAH-NH2 [63] | 2.83 | – | – | 5.90 | – | 6.48 | 7.11 |
| Ac-HDHAHDH-NH2 [63] | 2.96 | 3.72 | – | 5.75 | 6.38 | 6.66 | 7.17 |
| L1(10) | L2(15) | L3(16) | Ac-HADHAH-NH2 [63] | Ac-HDHAHDH- NH2 [63] | |
|---|---|---|---|---|---|
| [CuH3L] | − | − | − | − | 25.10 |
| [CuH2L] | − | 19.21 (2) | 21.18 (3) | − | − |
| [CuHL] | 14.15 (2) | 14.44 (2) | 16.04 (7) | 13.64 | 16.54 |
| [CuL] | 9.04 (2) | 9.29 (2) | 10.74 (6) | 8.65 | 10.95 |
| [CuH−1L] | 1.34 (12) | 2.10 (7) | 2.97 (12) | − | − |
| [CuH−2L] | −6.56 (12) | − | − | −6.20 | −4.36 |
| [CuH−3L] | −15.58 (9) | −12.42 (18) | −13.55 (9) | −15.70 | −14.61 |
| [Cu2L] | − | 12.18 (8) | − | − | − |
| [Cu2H−1L] | − | − | 8.92 (12) | − | − |
| [Cu2H−2L] | − | 0.64 (3) | − | −1.02 | 3.26 |
| [Cu2H−3L] | − | − | −5.22 (43) | −7.54 | − |
| [Cu2H−4L] | − | − | − | −14.64 | −12.16 |
| [Cu2H−5L] | − | − | −21.32 (33) | −23.64 | −22.59 |
| [Cu2H−6L] | − | −28.39 (12) | −30.45 (26) | −34.54 | −33.84 |
| logK(Cu(II) + 2NIm) | 6.53 | 5.91 | 6.80 | 6.53 | − |
| logK(Cu(II) + 3NIm) | 9.04 | 7.61 | 8.19 | 8.65 | 9.37 |
| logK(Cu(II) + 4NIm) | − | 9.29 | 10.74 | − | 10.95 |
| pK(Im) | 5.11 | 4.77; 5.15 | 5.14; 5.30 | 4.99 | 4.28; 5.59 |
| pK(amide)1 | 7.70 | 7.19 | 7.77 | 7.42 (av.) | 7.65 (av.) |
| pK(amide)2 | 7.90 | 7.26 (av.) | 8.26 (av.) | ||
| pK(amide)3 | 9.02 | 9.50 | 10.25 |
| Species | Binding Mode | λmax (nm) (ε (M−1 cm−1)) | ||||
|---|---|---|---|---|---|---|
| L1(10) | L2(15) | L3(16) | Ac-HADHAH-NH2 [63] | Ac-HDHAHDH-NH2 [63] | ||
| [CuL] | [3 × Im] or [4 × Im] | 624 (52) w | 615 (97) w | 600 (108) w | 615 (85) | 605 (60) |
| [CuH − 3L] | [N-, N-, N-, Im] | 530 (102) | 536 (166) w | 540 (147) w | 532 (132) | 538 (103) |
| [Cu2H − 6L] | 2 × [N-, N-, N-, Im] | – | 545 (191) w | 537 (163) w | – | – |
| L1(10) | L2(15) | L3(16) | Ac-HADHAH-NH2 [63] | Ac-HDHAHDH- NH2 | |
|---|---|---|---|---|---|
| [ZnH2L] | − | 17.38 (6) | 18.51 (16) | − | 18.18 (3) |
| [ZnHL] | 11.54 (8) | 12.08 (2) | 13.22 (6) | 10.91 | 12.80 (2) |
| [ZnL] | 6.12 (2) | 6.06 (2) | 7.36 (4) | 5.63 | 6.97 (2) |
| [ZnH−1L] | −1.28 (6) | −0.95 (2) | −0.80 (7) | −2.40 | −0.72 (3) |
| [ZnH−2L] | −11.42 (8) | −10.60 (3) | −10.20 (7) | −11.23 | − |
| logK(Zn(II) + 3NIm) | 6.12 | 5.25 | 5.37 | 5.63 | 5.63 |
| logK(Zn(II) + 4NIm) | − | 6.06 | 7.36 | − | 6.97 |
| logK(ZnL/ZnH−1L) | 7.40 | 7.01 | 8.16 | 8.03 | 7.69 |
| logK(ZnH−1L/ZnH−2L) | 10.14 | 8.65 | 9.40 | 8.83 | − |
| L2(15) | L3(16) | |
|---|---|---|
| [CuZnH−1L] | – | 6.6 (4) |
| [CuZnH−2L] | – | 0.11 (5) |
| [CuZnH−3L] | – | −7.95 (7) |
| [CuZnH−4L] | – | −17.15 (9) |
| [CuZnH−5L] | −22.7 (5) | – |
| System | pH | IC50 (µM) | Relative Activity % [63] | kcat/106 (M−1s−1) | Ref. |
|---|---|---|---|---|---|
| CuZnSOD | 6.8 | 0.0030 | 100 | 914 | [53] |
| CuZnSOD | 0.010 | 430 | [69] | ||
| Cu(II)-L3(16) | 6.8 | 0.017 ± 0.004 | 17.6 | 161 | this work |
| Cu(II)-Zn(II)-L3(16) | 6.8 | 0.014 ± 0.002 | 21.4 | 196 | this work |
| Cu(II)-L2(15) | 6.8 | 0.021 ± 0.007 | 14.3 | 131 | this work |
| Cu(II)-Zn(II)-L2(15) | 6.8 | 0.019 ± 0.008 | 15.8 | 144 | this work |
| Cu(II)-Ac-HDHAHDH-NH2 | 6.8 | 0.020 ± 0.006 | 15.0 | 137 | this work |
| Cu(II)-Ac-HAAHGH-NH2 | 6.8 | 0.072 | 4.17 | [53] | |
| Cu(II)-Ac-HGGGHGH-NH2 | 6.8 | 0.071 | 4.22 | [53] | |
| Cu(II)-PydiTyr | 0.028 | 96.2 | [70] | ||
| [Mn(PI)2](PF6)(CH3CN) | 7.0 | 0.87 | 6.6 | [62] | |
| [Mn(III)Cl4 TE-2-PyP] | 0.0065 | 400 | [71] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Székely, E.; Molnár, M.; Lihi, N.; Várnagy, K. Characterization of Copper(II) and Zinc(II) Complexes of Peptides Mimicking the CuZnSOD Enzyme. Molecules 2024, 29, 795. https://doi.org/10.3390/molecules29040795
Székely E, Molnár M, Lihi N, Várnagy K. Characterization of Copper(II) and Zinc(II) Complexes of Peptides Mimicking the CuZnSOD Enzyme. Molecules. 2024; 29(4):795. https://doi.org/10.3390/molecules29040795
Chicago/Turabian StyleSzékely, Enikő, Mariann Molnár, Norbert Lihi, and Katalin Várnagy. 2024. "Characterization of Copper(II) and Zinc(II) Complexes of Peptides Mimicking the CuZnSOD Enzyme" Molecules 29, no. 4: 795. https://doi.org/10.3390/molecules29040795
APA StyleSzékely, E., Molnár, M., Lihi, N., & Várnagy, K. (2024). Characterization of Copper(II) and Zinc(II) Complexes of Peptides Mimicking the CuZnSOD Enzyme. Molecules, 29(4), 795. https://doi.org/10.3390/molecules29040795





