Development of a Two-Dimensional Liquid Chromatographic Method for Analysis of Urea Cycle Amino Acids
Abstract
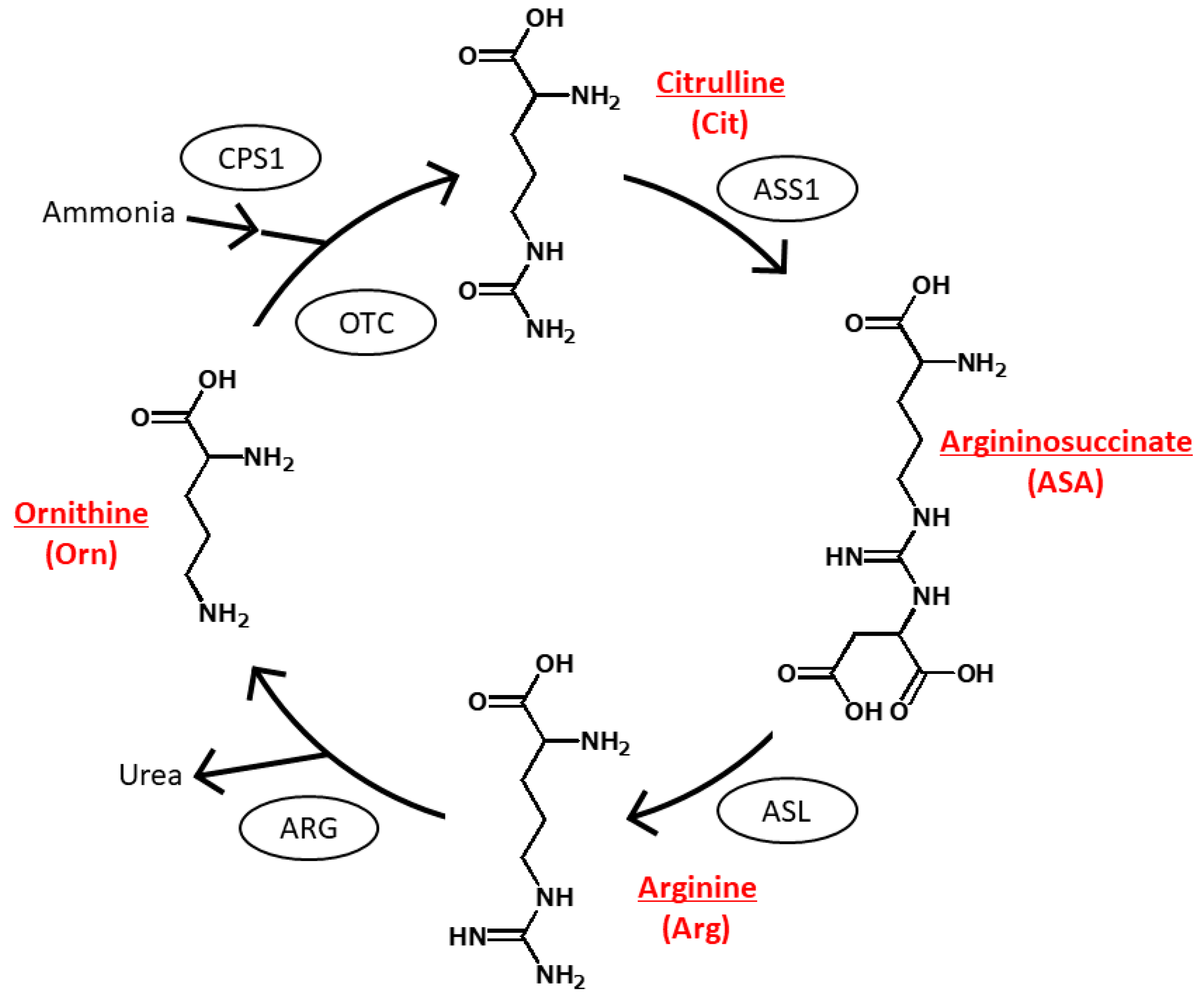
1. Introduction
2. Results and Discussion
2.1. Optimization of the Separation of NBD-Urea Cycle Amino Acids on Reversed-Phase Column
2.2. Retention of NBD-Urea Cycle Amino Acids on Ion-Exchange Column
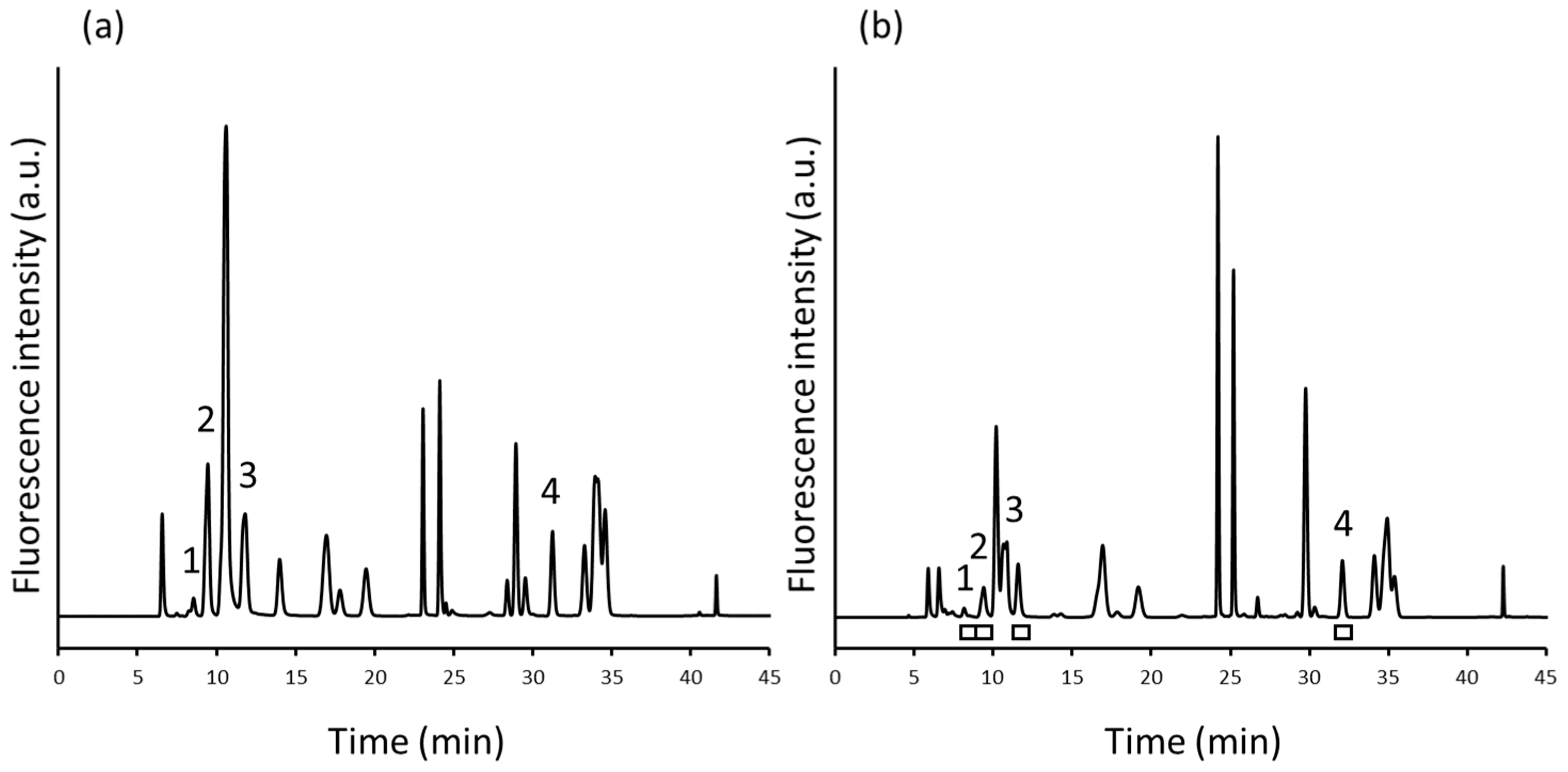
2.3. 2D-LC Separation of NBD-Urea Cycle Amino Acids in Human Plasma
2.4. Method Validation
3. Materials and Methods
3.1. Chemicals
3.2. Pre-Treatment of Human Plasma Samples
3.3. Fluorescence Derivatisation
3.4. Apparatus and Chromatographic Setup of 2D-LC
3.5. Method Validation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matsumoto, S.; Häberle, J.; Kido, H.; Mitsubuchi, F.; Endo, F.; Nakamura, K. Urea cycle disorders-update. J. Hum. Genet. 2019, 64, 833–847. [Google Scholar] [CrossRef]
- Morris, S.M., Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 2002, 22, 87–105. [Google Scholar] [CrossRef]
- Bigot, A.; Tchan, M.C.; Thoreau, B.; Blasco, H.; Maillot, F. Liver involvement in urea cycle disorders: A review of the literature. J. Inherit. Metab. Dis. 2017, 40, 757–769. [Google Scholar] [CrossRef]
- Häberle, J.; Burlina, A.; Chakrapani, A.; Dixon, M.; Karall, D.; Lindner, M.; Mandel, H.; Martinelli, D.; Pintos-Morell, G.; Santer, R.; et al. Suggested guidelines for the diagnosis and management of urea cycle disorders: First revision. J. Inherit. Metab. Dis. 2019, 42, 1192–1230. [Google Scholar] [CrossRef] [PubMed]
- Keshet, R.; Szlosarek, P.; Carracedo, A.; Erez, A. Rewiring urea cycle metabolism in cancer to support anabolism. Nat. Rev. Cancer 2018, 18, 634–645. [Google Scholar] [CrossRef]
- Li, L.; Mao, Y.; Zhao, L.; Li, L.; Wu, J.; Zhao, M.; Du, W.; Yu, L.; Jiang, P. p53 regulation of ammonia metabolism through urea cycle controls polyamine biosynthesis. Nature 2019, 567, 253–256. [Google Scholar] [CrossRef]
- Bateman, L.A.; Ku, W.-M.; Heslin, M.J.; Contreras, C.M.; Skibola, C.F.; Nomura, D.K. Argininosuccinate Synthase 1 is a Metabolic Regulator of Colorectal Cancer Pathogenicity. ACS Chem. Biol. 2017, 12, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Delage, B.; Fennell, D.A.; Nicholson, L.; McNeish, I.; Lemoine, N.R.; Crook, T.; Szlosarek, P.W. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int. J. Cancer 2010, 126, 2762–2772. [Google Scholar] [CrossRef]
- Liu, Q.; Stewart, J.; Wang, H.; Rashid, A.; Zhao, J.; Katz, M.H.; Lee, J.E.; Fleming, J.B.; Maitra, A.; Wolff, R.A.; et al. Reduced expression of argininosuccinate synthetase 1 has a negative prognostic impact in patients with pancreatic ductal adenocarcinoma. PLoS ONE 2017, 12, E0171985. [Google Scholar] [CrossRef]
- Wu, L.; Li, L.; Meng, S.; Qi, R.; Mao, Z.; Lin, M. Expression of argininosuccinate synthetase in patients with hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2013, 28, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Dai, J.; Zhang, Z.; Yu, H.; Zhang, J.; Zhu, X.; Qin, Y.; Zhang, L.; Zhang, P. ASS1-Mediated Reductive Carboxylation of Cytosolic Glutamine Confers Ferroptosis Resistance in Cancer Cells. Cancer Res. 2023, 83, 1646–1665. [Google Scholar] [CrossRef]
- Yang, H.; Zhai, G.; Ji, X.; Su, J.; Lin, M. Reduced Expression of Argininosuccinate Lyase is Closely Associated With Postresectional Survival in Hepatocellular Carcinoma An Immunohistochemistry Study of 61 Cases. Mol. Morphol. 2012, 20, 602–606. [Google Scholar]
- Cañadas-Garre, M.; Anderson, K.; McGoldrick, J.; Maxwell, A.P.; McKnight, A.J. Proteomic and metabolomic approaches in the search for biomarkers in chronic kidney disease. J. Proteom. 2019, 193, 93–122. [Google Scholar] [CrossRef]
- Yamano, E.; Sugimoto, M.; Hirayama, A.; Kume, S.; Yamato, M.; Jin, G.; Tajima, S.; Goda, N.; Iwai, K.; Fukuda, S.; et al. Index markers of chronic fatigue syndrome with dysfunction of TCA and urea cycles. Sci. Rep. 2016, 6, 34990. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, J.; Sandqvist, A.; Hedeland, M.; Bondesson, U.; Wikström, G.; Rädegran, G. Metabolomic Insights into Cardiovascular Health: A Focus on Coronary Artery Disease. Scand. Cardiovasc. J. 2018, 52, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Soria, L.R.; Mew, N.A.; Brunetti-Pierri, N. Progress and challenges in development of new therapies for urea cycle disorders. Hum. Mol. Genet. 2019, 28, R42–R48. [Google Scholar] [CrossRef] [PubMed]
- Foschi, F.G.; Morelli, M.C.; Sara Savini, S.; Dall’Aglio, A.C.; Lanzi, A.; Cescon, M.; Ercolani, G.; Cucchetti, A.; Pinna, A.D.; Stefanini, G.F. Urea cycle disorders: A case report of a successful treatment with liver transplant and a literature review. World J. Gastroenterol. 2015, 21, 4063–4068. [Google Scholar]
- Martens-Lobenhoffer, J.; Bode-Böger, S.M. Mass spectrometric quantification of L-arginine and its pathway related substances in biofluids: The road to maturity. J. Chromatogr. B 2014, 964, 89–102. [Google Scholar] [CrossRef]
- Jourdan, M.; Nair, K.S.; Carter, R.E.; Schimke, J.; Ford, G.C.; Marc, J.; Aussel, C.; Cynober, L. Citrulline stimulates muscle protein synthesis in the post-absorptive state in healthy people fed a low-protein diet—A pilot study. Clin. Nutr. 2015, 34, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Kaysheva, A.L.; Kopylov, A.T.; Stepanov, A.A.; Malsagova, K.A.; Izotov, A.A.; Shurubor, Y.I.; Krasnikov, B.F. Cite Share Chromatomass-Spectrometric Method for the Quantitative Determination of Amino- and Carboxylic Acids in Biological Samples. Metabolites 2022, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Li, X.; Wang, H.; Cai, Z. Simultaneous determination of methionine cycle metabolites, urea cycle intermediates and polyamines in serum, urine and intestinal tissue by using UHPLC-MS/MS. Talanta 2021, 224, 121868. [Google Scholar] [CrossRef] [PubMed]
- Santucci, L.; Lomuscio, S.; Primiano, A.; Calvani, R.; Persichilli, S.; Iavarone, F.; Picca, A.; Canu, F.; Urbani, A.; Gervasoni, J. Development of a novel Ultra Performance Liquid Chromatography Tandem-Mass Spectrometry (UPLC-MS/MS) method to measure l-arginine metabolites in plasma. Clin. Chim. Acta 2023, 543, 117306. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Kline, J.A.; Wang, M. Development, validation, and comparison of four methods to simultaneously quantify l-arginine, citrulline, and ornithine in human plasma using hydrophilic interaction liquid chromatography and electrospray tandem mass spectrometry. J. Chromatogr. B 2015, 1005, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Funatsu, T.; Tsunoda, M. Rapid determination of amino acids in biological samples using a monolithic silica column. Amino Acids 2012, 42, 1897–1902. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Funatsu, T.; Tsunoda, M. Amino acid analysis using core-shell particle column. J. Chromatogr. B 2013, 927, 214–217. [Google Scholar] [CrossRef]
- Song, Y.; Takatsuki, K.; Sekiguchi, T.; Funatsu, T.; Shoji, S.; Tsunoda, M. Rapid quantitative method for the detection of phenylalanine and tyrosine in human plasma using pillar array columns and gradient elution. Amino Acids 2016, 48, 1731–1735. [Google Scholar] [CrossRef]
- Stoll, D.R.; Carr, P.W. Two-Dimensional Liquid Chromatography: A State of the Art Tutorial. Anal. Chem. 2016, 89, 519–531. [Google Scholar] [CrossRef]
- Pirok, B.W.J.; Stoll, D.R.; Schoenmakers, P.J.J. Recent Developments in Two-Dimensional Liquid Chromatography: Fundamental Improvements for Practical Applications. Anal. Chem. 2018, 91, 240–263. [Google Scholar] [CrossRef]
- Ohtawa, T.; Hattori, A.; Isokawa, M.; Harada, M.; Funatsu, T.; Ito, T.; Tsunoda, M. Determination of intracellular 2-hydroxyglutarate enantiomers using two-dimensional liquid chromatography. J. Chromatogr. Open 2021, 1, 100005. [Google Scholar] [CrossRef]
- Ohtawa, T.; Tsunoda, M. An on-line heart-cutting two-dimensional liquid chromatography method for intracellular 2-hydroxyglutarate enantiomers. Anal. Methods 2023, 15, 2833–2838. [Google Scholar] [CrossRef]
- Harada, E.; Nishiyori, A.; Tokunaga, Y.; Watanabe, Y.; Kuriya, N.; Kumashiro, R.; Kuno, T.; Kuromaru, R.; Hirose, S.; Ichikawa, K.; et al. Late-onset ornithine transcarbamylase deficiency in male patients: Prognostic factors and characteristics of plasma amino acid profile. Pediatr. Int. 2006, 48, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Santra, S.; Cameron, J.M.; Shyr, C.; Zhang, L.; Drögemöller, B.; Ross, C.J.; Wasserman, W.W.; Wevers, R.A.; Rodenburg, R.J.; Gupte, G.; et al. Cytosolic phosphoenolpyruvate carboxykinase deficiency presenting with acute liver failure following gastroenteritis. Mol. Genet. Metab. 2016, 118, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The Human Serum Metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed]


| Limit of Detection [nM] | Limit of Quantification [nM] | Calibration Range [nM] | r2 | Intraday Precision (%) (n = 5) | Interday Precision (%) (n = 4–5) | Recovery (%) (n = 3) | |
|---|---|---|---|---|---|---|---|
| Arg | 2.6 | 8.5 | 15–600 | 0.9985 | 4.0 | 3.8 | 95.4 |
| Orn | 0.96 | 3.2 | 15–600 | 0.9998 | 7.9 | 15 | 109 |
| Cit | 0.43 | 1.4 | 15–600 | 0.9988 | 0.98 | 7.7 | 99.9 |
| ASA | 0.53 | 1.8 | 3–600 | 0.9969 | 7.9 | 6.9 | 86.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sumida, Y.; Tsunoda, M. Development of a Two-Dimensional Liquid Chromatographic Method for Analysis of Urea Cycle Amino Acids. Molecules 2024, 29, 700. https://doi.org/10.3390/molecules29030700
Sumida Y, Tsunoda M. Development of a Two-Dimensional Liquid Chromatographic Method for Analysis of Urea Cycle Amino Acids. Molecules. 2024; 29(3):700. https://doi.org/10.3390/molecules29030700
Chicago/Turabian StyleSumida, Yuko, and Makoto Tsunoda. 2024. "Development of a Two-Dimensional Liquid Chromatographic Method for Analysis of Urea Cycle Amino Acids" Molecules 29, no. 3: 700. https://doi.org/10.3390/molecules29030700
APA StyleSumida, Y., & Tsunoda, M. (2024). Development of a Two-Dimensional Liquid Chromatographic Method for Analysis of Urea Cycle Amino Acids. Molecules, 29(3), 700. https://doi.org/10.3390/molecules29030700






