Abstract
Until now, Z-form RNAs were believed to only adopt a left-handed double-helix structure. In this study, we describe the first observation of a right-handed Z-form RNA in NMR solution formed by L-nucleic acid RNA and present the first resolution of structure of the complex between a right-handed Z-form RNA and a curaxin ligand. These results provide a platform for the design of topology specific to Z-form-targeting compounds and are valuable for the development of new potent anticancer drugs.
1. Introduction
Nucleic acids have exquisitely evolved to store genetic information, primarily composed of D-nucleic acids, folding into the most common secondary structure with right-handed double-helical conformational features, fundamentally recognized as A-RNA or B-DNA (Scheme 1, left column). The right-handed A-form RNA plays a crucial role in cellular processes and functions, such as responding to viral infections, post-transcriptional gene silencing, RNA silencing, and RNA interference [1,2,3,4,5,6].
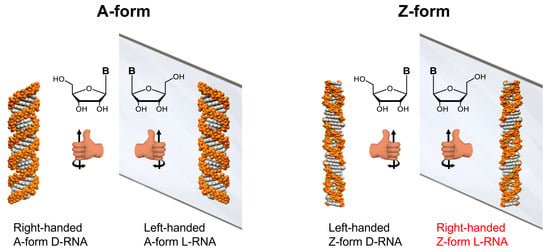
Scheme 1.
Illustration of mirror-image RNA. The left column displays a right-handed helix A-form D-RNA and its mirror-image, a left-handed A-form L-RNA consisting of L-nucleic acids. The right column shows the left-handed helix Z-form D-RNA and its mirror-image with a right-handed Z-form L-RNA consisting of L-nucleic acids (The words in red highlight the typical structure, which is the main focus of this study).
A mirror-image of native D-nucleic acids, L-nucleic acids in a left-handed A-form (Scheme 1, left column), offers superior stability in biological environments due to its resistance to cellular nucleases [7,8]. The unique folding scaffold that L-RNA consists of is used to recognize a variety of small molecules, peptides, and proteins [9,10]. Several aptamers are now under clinical investigation [11]. Simultaneously, chimeras of L-oligonucleotide containing nanotechnology materials are employed for drug delivery by enhancing stability [12].
The left-handed Z-form double-helix structure, including Z-RNA and Z-DNA, has been shown to play an important role in multiple physiological events, including the regulation of gene expression, nucleosome positioning, and genetic instability associated with nucleic acid damage and repair (Scheme 1, right column) [13,14,15,16]. We recently suggested a relationship between this Z-form structure and several diseases, such as cancer and inflammation [3,13,14,17,18]. Double-stranded Z-RNA consists of nucleotides arranged in a pattern where the nucleobases alternate between syn and anti conformations [19]. For the typical purine/pyrimidine repeats, cytidine residues adopt the anti conformation, while guanosine residues display the syn conformation, and the phosphate backbone follows a zig–zag course in a discontinuous manner [20]. Some chemical modifications favor the syn conformation of guanosine, leading to a stabilized Z form [21]. We have demonstrated that trifluoromethyl introduction at the C8 position of guanosine, with the lowest energetic level, allowed guanosine to adopt the syn conformation to stabilize the Z form [22].
Tremendous efforts have been devoted to identifying small molecules for interaction with the Z form. For instance, metal molecules recognize Z-form DNA structures, and our previous report showed that (P)-Helicene displays chiral selection in binding to Z-DNA [23]. We recently identified a small molecule, curaxin CBL0137, which promotes the Z-form transition of DNA and RNA in the nucleus [17]. This transition activates protein Z-DNA-binding protein 1 (ZBP1)-mediated apoptosis and necroptosis, contributing to its antitumor effects.
While Z-form nucleic acid duplexes have been widely studied, all reported structures consistently adopt a left-handed helix. None of the studies have reported on a right-handed Z-form RNA structure and its interaction with ligands. This gap in our understanding motivated us to explore whether a mirror-image, right-handed Z-form nucleic acid could exist and what unique topological features it might present. Such a discovery holds the potential to reveal new biological and medical functions, especially in understanding how nucleic acid molecules fold into complex, high-order structures and participate in vital processes and pathogenesis. Furthermore, studying the interactions between right-handed Z-form RNA and ligands could facilitate the development of novel nucleic acid-based nanomaterials, which have promising applications in drug delivery and targeted therapies. Herein, we present the first NMR structure of right-handed Z-form RNA in solution and investigate its interaction with the curaxin ligand, CBL0137.
2. Results
2.1. Synthesis of Right-Handed Z-Form RNA and Characterization by CD and 19F NMR
To uncover the structural features of right-handed Z-form RNA, we designed and synthesized a novel 8-FG-modified L-guanosine analog (FG), in which a trifluoromethyl (CF3) group is introduced into the C8 position of L-guanosine according to a previously reported method, and incorporated it into an RNA sequence L-r(CGCFGCG)2 (Figure 1 and Supplemental Data S1–S20) [24]. As reported in our previous research, a CF3 group at the C8 position greatly stabilizes the Z-form nucleic acids and acts as a 19F sensor, which can be used to study DNA and RNA structures using 19F NMR [22]. We used circular dichroism spectroscopy (CD) to monitor the conformational state at various NaClO4 concentrations. We observed that L-r(CGCFGCG)2 greatly stabilized the Z-RNA, showing the A–Z transition in the presence of only 50 mM NaClO4 (Figure 2a), while the transition requires 6000 mM NaClO4 for unmodified L-r(CGCGCG)2 (Figure 2b). This result is completely consistent with 19F NMR, which has been successfully used to distinguish RNA structures in our previous studies (Figure 2c) [24]. In 19F NMR, a strong-intensity peak appeared as a Z form (−61.32 ppm) accompanied by a small peak (−60.20 ppm) at a NaClO4 concentration of 100 mM, indicating the major Z-form RNA and a minor A-form RNA. With increasing NaClO4 concentrations, only the peak of the Z-form RNA remained with a strong intensity, whereas the peak of the A-form completely disappeared.
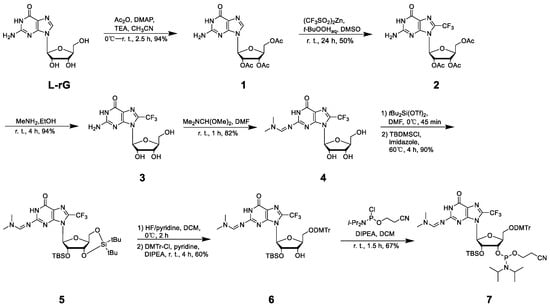
Figure 1.
Synthetic scheme of FG and relative phosphoramidite compound.
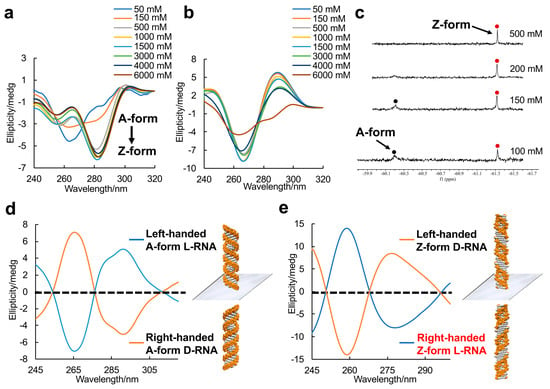
Figure 2.
Enantiomers of A-form and Z-form RNA duplex structures. (a) CD spectra of the A–Z transition of L-RNA r(CGCFGCG)2 at various NaClO4 concentrations at 10 °C. A negative Cotton effect appears around 285 nm with increasing NaClO4 concentration. (b) CD spectra of L-RNA r(CGCGCG)2 at various NaClO4 concentrations at 10 °C. (c) 19F NMR spectra of L-RNA r(CGCFGCG)2 in 5 mM Na-PO4 buffer (pH 7.0) and various NaClO4 concentrations. Red and black spots indicated Z-form and A-form RNA, respectively. (d) CD spectra of A-form RNA mirror enantiomers: left-handed L-RNA r(CGCGCG)2 and right-handed D-RNA r(CGCGCG)2 in presence of 5 mM Na-PO4 buffer (pH 7.0) and 1.5 M NaClO4 at 10 °C. (e) CD spectra of Z-form RNA mirror enantiomers: left-handed D-RNA r(CGCFGCG)2 and right-handed L-RNA r(CGCFGCG)2 (The words in red color label highlights the typical structure which is major studying target in this study) in presence of 5 mM Na-PO4 buffer (pH 7.0) and 1.5 M NaClO4 at 10 °C.
CD was performed to distinguish the RNA mirror enantiomer. The right-handed A-form D-r(CGCGCG)2 exhibited a negative band around 290 nm and a positive band at 266 nm, which are typical A-form signals (Figure 2d). A left-handed A-form L-r(CGCGCG)2 as the enantiomer was completely antithetical. Meanwhile, the right-handed Z-form RNA L-r(CGCFGCG)2 and the left-handed Z-form RNA D-r(CGCFGCG)2 stabilized by CF3-modified D-guanosine showed absolutely mirror-imaged spectral signals (Figure 2e).
2.2. NMR Solution Structure of Right-Handed Z-Form RNA
To reveal the detailed structure of right-handed Z-form RNA, we performed 2D NMR experiments (Figure 3), in which the condition with 2 M NaClO4 was used to ensure complete deletion of NOEs from other structures. A complete list of 1H chemical shifts is shown in Supplemental Table S1. The NOE-restrained refinement provided an unequivocal demonstration that the structure of L-RNA r(CGCFGCG)2 is Z-RNA. Sequential assignments of C1 to C3 and C5 to G6 for the Z-form can complete the pathway: C1(H6/H5′′)-G2(H8/H1′)-C3(H6/H5′′), C5(H6/H5′′)-G6(H8/H1′) (Figure 3a,c,d), indicating a sequence-specific connectivity for right-handed helices. All H5′′ of cytidine were found to shift upfield by 2.85 (C1), 2.70 (C3), and 2.66 (C5) ppm; these alterations only could be observed in the Z-form nucleic acid duplex (Figure 3a). The clear cross-peaks of the imino proton of FG4 (~13.48 ppm) and G2 (~13.36 ppm) with the amino proton of C3 (~6.49 ppm) and C5 (~8.74 ppm) collectively suggested Watson–Crick base pairs (Figure 3b). Moreover, the H8/H1′ cross-peak region of the 2D NOESY spectrum showed only strong intranucleotide G2(H8/H1′) and G6(H8/H1′) cross-peaks (Figure 3c), which indicated the syn conformation of rG residues. Strong intra-residue cross peaks of C1(H6/H5′′), C3(H6/H5′′), and C5(H6/H5′′) indicate anti conformation of all cytidines in the Z form (Figure 3a). We note that the C1H5 proton has cross-peaks to the C5 amino proton (C1H5/C5NH22), and the C5H5 proton has cross-peaks to the C1 amino proton (C5H5/C1NH22) (Figure 3d). Such cross-peaks can only happen between the two interstrand cytosines between C1 in one strand and C5 in another strand of the Z-RNA because of their specific base pair stacking pattern. Additionally, the NOE G2H3′/G2H4′ was observed and indicated that rG residues preserve the C3′-endo sugar pucker while those signals, including C1H1′/C1H2′, C3H1′/C3H2′, and C5H1′/C5H2′, demonstrated rC residues in the C2′-endo sugar pucker (Supplemental Figure S1).
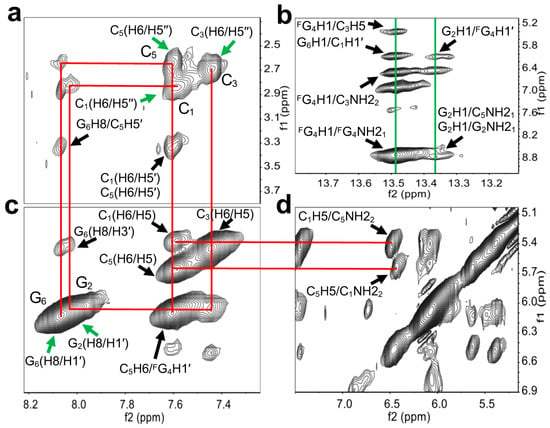
Figure 3.
Structural determination of right-handed Z-form RNA r(C1G2C3FG4C5G6)2. (a,c,d) show wide-ranging connectivity paths (red lines) [C1(H6/H5′′)-G2(H8/H1′)-C3(H6/H5′′), C5(H6/H5′′)-G6(H8/H1′)] in the RNA strand of right-handed Z helices in a contour plot of the 2D-NOESY spectrum. (a) C1(H6/H5′′), C3(H6/H5′′), and C5(H6/H5′′) were found to upshift 2.85, 2.70, and 2.66 ppm (green arrows). Strong intra-residue cross-peaks of C1(H6/H5′′), C3(H6/H5′′), and C5(H6/H5′′) were observed. Additional anomeric–aromatic proton interactions were assigned as C1(H6/H5′), C5(H6/H5′), and G6H8/C5H5′, as marked by black arrows. (b) The cross-peaks of the imino proton of G2 and amino proton of C5, as well as the imino proton of FG4 and the amino proton of C3 associated with G6H1/C1H1′, FG4H1/C3H5, and G2H1/FG4H1′ were observed (green lines). Intraresidue NOE cross-peaks of the imino and amino protons of G2 and FG4 are shown. (c) Strong H8–H1′ cross-peaks were observed (green arrows). C1(H6/H5), C3(H6/H5), C5(H6/H5), G6(H8/H3′), and C5H6/FG4H1′ are labeled by black arrows. (d) The Z-RNA structure-specific cross-peaks were observed between the C5 amino proton and C1H5, as well as between the C1 amino proton and C5H5 from inter-stranded interactions. These signals are linked to C1(H6/H5) and C5(H6/H5) in (c) by red lines.
The structural model of L-r(CGCFGCG)2 is constructed based on the reported Z-form structure and NOE-constrained refining method (Supplemental Tables S2 and S3). The molecular dynamics simulation was carried out in BIOVIA Discovery Studio 4.5 (Accelrys, San Diego, CA, USA) through a standard dynamic cascade with some modifications. The lowest energy conformation was selected as shown in Figure 4a and Supplemental Figure S2. The Watson–Crick scheme of base pairing is observed throughout the duplex, in which rG residues are characterized by a syn conformation in C3′-endo and rC with an anti conformation in C2′-endo (Figure 4b and Supplemental Figures S3 and S4). For both the CpG and GpC steps, base stacking interactions are observed but evidently with no inter-strand base stacking (Figure 4c and Supplemental Figure S5); these results were in high agreement with the landmark of Z helices.
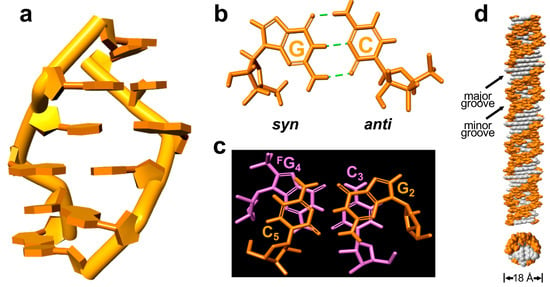
Figure 4.
The structural model of right-handed Z-form RNA r(C1G2C3FG4C5G6)2. (a) Structural right-handed Z-form RNA r(CGCFGCG)2 viewed from the major groove. (b) rG with synconformation and rC with anti-conformation form Watson–Crick base pairs through three hydrogen bonds in Z-form RNA. (c) The stacking pattern within the GpC step in Z-RNA as viewed along the helix z-axis, showing only intra- but not inter-strand stacking. (d) An extended model of the right-handed Z-form RNA helix from Z-form RNA r(CGCFGCG)2 visualized in side (up) and top (bottom) views to show their base pair locations and groove architecture, in which white represent base pairs and orange indicates phosphate–ribose backbones. The duplex diameter is shown, and the major or minor groove is indicated.
To further clearly observe the right-handed Z-form RNA, we generated the model of a full-turn Z-RNA helix by excluding both terminal base pairs of the 6-mer structure (Figure 4d). The right-handed RNA helix has the well-marked zig–zag shape of the phosphorsugar backbone and 2′-OH groups of rG exposed to the outer helix surface (Figure 4d and Supplemental Figure S6). This might be considered a result of the fact that the Z-RNA duplex structure requires higher salt concentration compared to Z-DNA. Another feature of the RNA helix is that both grooves are well defined, possessing a very engraved, deep and much narrower minor groove, and a shallow major groove, which is the same as its mirror image left-handed Z-form D-RNA (Supplemental Figure S6).
2.3. Complex Structure of Right-Handed Z-Form RNA and Ligand CBL0137
We have identified a small molecule, curaxin CBL0137 (Figure 5a), which is capable of inducing Z-form formation to activate ZBP1-mediated cell death. This encouraged us to explore its binding affinity with the right-handed Z-form RNA model of right-r(CGCFGCG)2. In CD, we observed the appearance of a negative band around 280 nm with the addition of equimolar (1:1) amounts of CBL0137 (Figure 5b). This effect was even more pronounced at a higher ratio (2:1) of CBL0137 to L-r(CGCFGCG)2. These results suggest that CBL0137 effectively converts the A-RNA conformation to Z-RNA.
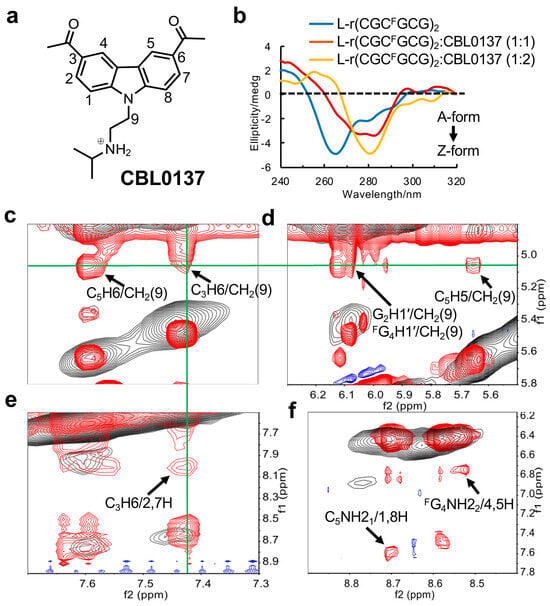
Figure 5.
Study of the CBL0137 ligand binding to Z-form RNA. (a) The structure of CBL0137, comprising a carbazole moiety with a positive charged N-side chain. (b) The CD titration of Z-form RNA r(CGCFGCG)2 with increasing concentrations of CBL0137 at 10 °C in 5 mM Na-PO4 buffer (pH 7.0). The ratio of CBL0137 to RNA is indicated at the top. (c-f) The overlay of NOE spectra. The signals in red and blue from the complex of CBL0137 and RNA. The signals in black from Z-form RNA as a control. (c–e) display strong intermolecular NOEs between RNA and CBL0137. C5H6/CH2(9), C3H6/CH2(9), G2H1′/CH2(9), FG4H1′/CH2(9), C5H5/CH2(9), and C3H6/2,7H are marked by black arrows. The signals derived from CH2(9) or C3H6 are connected by green lines. (f) The black arrows indicate the NOEs between amino protons C5NH21 and the aromatic protons (1H, 8H) of CBL0137, and the NOEs between amino protons FG4NH22 and the aromatic protons (4H, 5H) of CBL0137.
Furthermore, to resolve the complex structure, 1D proton NMR experiments were constructed. As an increasing amount of CBL0137 was titrated into RNA, we observed the emergence of a new set of peaks for the complex after the titration (Supplemental Figure S7). This indicated that the binding of CBL0137 to RNA can be observed in a slow exchange regime on the NMR timescale. The complete lists of 1H chemical shifts of complex CBL0137 and RNA are shown in Supplemental Table S4–S6. As the NMR signals preferred to be stable after the addition of double concentrations of CBL0137 into RNA, we deduced that only a dominant conformation of the 1:2 RNA–ligand complex was present. This is consistent with observations from the CD titration experiments, which showed a clear inflection point around 1:2 for the complex formation (Supplemental Figure S8). Moreover, we found that the addition of the ligand induced only a single peak of phosphate in C3 of the GpC step with lower intensity than the 31P NMR spectrum (Supplemental Figure S9), as well as a resonance shift of FG4 in 19F NMR (Supplemental Figure S10).
The overlay of 2D spectra derived from the complex of CBL0137 and RNA allowed us to obtain more precise NOE interactions between the RNA and ligand (Figure 5c–f). For example, a series of cross-peaks showed the strong intermolecular interactions between CH2(9) in the ligand and aromatic protons such as C5H6, C3H6, and C5H5, as well as the sugar G2H1′ and FG4H1′ protons of the RNA. Similarly, the NOEs between the aromatic protons of CBL0137 and the amino protons or aromatic protons of the RNA were detected. These results suggested that the ligand may insert into G2:C5 and C3:FG4 base pairs via the stacking effect.
The solution structure of the CBL0137-RNA complex was computed using NMR restraints (Figure 6a and Supplemental Figure S11). Two ligands were observed to symmetrically insert into the G2:C5 and C3:FG4 base pairs at the GpC step through π stacking. An expanded view shows clearly the π-π stacking interactions between the carbazole moiety of CBL0137 and the base pairs (Figure 6b and Supplemental Figure S12). The cationic side chain of the ligand approached the phosphate backbone of C3 through intense electrostatic interactions between the positively charged nitrogen atom of the ligand and the negatively charged phosphate backbone of C3 (Figure 6c), while the acetyl groups at the positions 3 and 7 of carbazole protruded into the major groove (Supplemental Figure S13). This is in agreement with the observed NOE cross-peak between the phosphate of C3 and hydrogen of CBL0137 isopropyl from 31P–1H HOESY (heteronuclear Overhauser effect spectroscopy) NMR (Supplemental Figure S14).
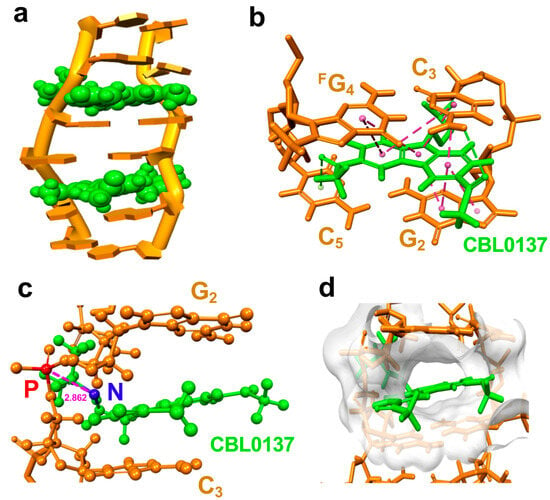
Figure 6.
The molecular dynamic model of CBL0137 ligand binding to Z-form right-hand RNA. (a) Solution structure of the CBL0137-RNA complex with cartoon representation. CBL0137 (in green CPK representation) symmetrically binding with Z-form RNA (orange) in a 2:1 molecular ratio. (b) An expanded view of CBL0137 (in green stick representation) and base pairs indicates the π-π stacking formation between the carbazole moiety and base pairs of G2:C5 and C3:FG4 as pink dashed lines connected by each aromatic ring’s center. (c) A side view showing the close proximity between the cationic N-side chain (shown as a blue sphere) of CBL0137 and the anionic phosphate group (shown as a red sphere) of C3 (~2.862 Å in distance). (d) The solvent hydrophobicity representation of the surface of RNA (white) shows CBL0137 (in green stick representation) positioned in the cleft pocket.
We noted that π-π stacking between the planar carbazole moiety and base pairs of RNA produces a hydrophobic cavity as a cleft pocket in RNA (Figure 6d and Supplemental Figure S15). This suggests that the CBL0137 binding to Z-form RNA may result from a preference that Z-form RNA provides such a cleft pocket, whereas A-form RNA lacks the space for CBL0137 binding. This is consistent with the higher binding constant K = 1.63 × 105 M−1 of CBL0137 and the right-handed Z-form RNA r(CGCFGCG)2 complex compared to that of K = 4.31 × 104 M−1 of CBL0137 and the right-handed A-form RNA r(CGCGCG)2 complex (Supplemental Figure S16).
3. Discussion
The present data clearly show the right-handed Z-form L-RNA are the exact mirror images of left-handed helix Z-form D-RNA; except for chirality, both possess the same conformation. Such a fine structure provides an essential piece of information in the higher order structures of RNAs, completing the puzzle of mirror images of RNA A-form and Z-form enantiomers.
Previous studies revealed that Z-RNA plays critical roles in antiviral defense and cancer therapy, which has generated significant research interest. Additionally, L-nucleic acids are highly resistant to enzymatic degradation, making them ideal candidates for nanomaterial applications in biosensing and drug delivery. These results consistently demonstrate that Z-form L-RNA possesses potentially applicable values in the biological and medical fields. Moreover, the incorporated 8-FG-modified L-guanosine could serve as a sensitive tool for 19F NMR studies, enabling the observation and investigation of the biological functions of Z-form L-RNA within cells.
We demonstrated that curaxin CBL0137 interacts with right-handed Z-form RNA predominantly through π stacking and electrostatic interactions. The conjugated aromatic ring of CBL0137 adopts a centrosymmetric orientation, facilitating π-π stacking interactions with the base pairs of the upper G2 and lower C3. However, the N-side chain of CBL0137 does not localize symmetrically but shifts towards the phosphate backbone of the C3 residue. This shift enables strong electrostatic interactions between the positively charged nitrogen of CBL0137 and the negatively charged phosphate backbone of C3 [25]. From the resolved structure of the complex, we infer that enhancing the binding affinity or selectivity of small molecules towards specific Z-form RNA structures can be achieved by optimizing the steric properties of the planar stacking and the cationic properties of the side chain.
Previous studies indicated that the Z-form DNA comprising 2′-deoxy-L-ribose adopts a right-hand helix and could bind with small molecules [26,27]. These results, corroborated with the findings in this study, are perfectly in line with our recent report, which showed that CBL0137 triggers Z-form formation and contributes to its antitumor effects. Advanced NMR studies on the resulting complex of left-handed helix Z-form D-RNA and CBL0137 and the implications of their formation are currently underway.
4. Materials and Methods
4.1. RNA Sample Preparation
By using an automatic solid-phase phosphoramidite chemistry and DNA/RNA synthesizer, the d-, l-, and trifluoromethyl-labeled RNAs were synthesized at a ratio of 1.0 µmol. Following the RNAs being cleaved from the column and deprotected by using Ammonium Hydroxide/40% aqueous methylamine 1:1 v/v (AMA) at room temperature for 20 min and at 65 °C for 10 min, respectively. Tert-butyldimethylsilyl (TBDMS) protections were removed by treatment with triethylamine trihydrofluoride followed by filtration through an ion exchange cartridge. The oligomers were further purified by high-performance liquid chromatography (HPLC) in a linear gradient of 50 mM ammonium formate in 1:1 acetonitrile/H2O and 50 mM ammonium formate in H2O. The oligomers were desalted through a NAP 10 column (disposable column, GE Healthcare) and identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) on an Autoflex III smart beam mass spectrometer (negative mode) (Supplemental Data S21–S24).
4.2. Circular Dichroism
CD experiments were performed by using a JASCO model J-820 CD spectrophotometer (JASCO Corporation, Tokyo, Japan). The RNA samples were prepared at a 10 μM concentration in the presence of different concentrations of NaClO4, and 5 mM Na-PO4 buffer (pH 7.0). For the CBL0137 and RNA binding assay, RNAs were prepared at 10 µM in 5 mM NaPO4 buffer (pH 7.0). CBL0137 was added into the RNA solution and kept at room temperature for 30 min before measurement. Titrating concentrations for ligands were prepared at [RNA duplex]/[CBL0137] ratios of 1:0, 1:1, and 1:2 in the presence of a fixed concentration (5 μM) of the duplex RNA. The binding constant K was calculated by analyzing CD data at a fixed wavelength versus ligand concentrations according to our previous study [28].
4.3. 1H NMR Experiments
NMR data were recorded on a BRUKER AVANCE 400 and 600 MHz spectrometer (Bruker, Billerica, MA, USA). For spectra recorded in 90% H2O/10% D2O water, the signal was suppressed using the 3-9-19 WATER-GATE (water suppression by gradient-tailored excitation) pulse sequence or excitation sculpting with a gradient pulse. The data were processed with TopSpin 3.0 (Bruker BioSpin Gmbh) software and analyzed with MestReNova software (Mestrelab Research, USC, ES). For 1D NMR measurement (1H and 31P spectrum), RNA samples of a concentration of 0.3 or 2 mM were dissolved in 150 µL of the designed solution containing 10% D2O, 2 M NaClO4 and 10 mM Na-PO4 buffer (pH 7.0). Two-dimensional NOESY spectra in 90% H2O/10% D2O were collected from 512 scans with a 400 ms mixing time at 20 °C. On average, 2048 complex points and 512 FIDs (free induction decays) were collected within the spectral width of 14,097 Hz. The sample solutions were as follows: 2 mM RNA were dissolved in 150 µL of designed solution containing 10% D2O, 2 M NaClO4, and 10 mM Na-PO4 buffer (pH 7.0). Heteronuclear 2D NOESY spectra (1H–31P) were carried out in an identical solution containing 10% D2O, 2 M NaClO4, and 10 mM Na-PO4 buffer (pH 7.0), in which the chemical shift of phosphate from Na-PO4 was referred to as the internal standard at 0 ppm. The samples were prepared by heating the oligonucleotides at 85 °C for 3 min and gradually cooling them to room temperature. To study the RNA duplex binding with the CBL0137 ligand, the samples were prepared by the denaturing and annealing of RNA and followed by incubation at 4 °C overnight before the addition of the ligand. Conditions: 0.15 or 1 mM RNA duplex with 0.3 or 2 mM CBL0137 dissolved in the solution of 2 M NaClO4 and 10 mM Na-PO4 buffer (pH 7.0) containing 10% D2O.
4.4. 19F NMR Experiments
For 19F NMR measurement, RNA samples of a concentration of 100 μM were dissolved in 150 μL of the designed solution containing 10% D2O, in the presence of different concentrations of NaClO4 and 5 mM Na-PO4 buffer (pH 7.0). The samples were prepared by heating the 19F-labeled oligonucleotides at 90 °C for 3 min and gradually cooling them to room temperature. The 19F NMR spectrum was measured on a Bruker AVANCE 400 MHz spectrometer (Bruker, Billerica, MA, USA) at a frequency of 376.05 MHz and referenced to the internal standard CF3COOH (–75.66 ppm). The experimental parameters are recorded as follows: spectral width 89.3 kHz, 19F excitation pulse 15.0 μs, relaxation delay 1.5 s, acquisition time 0.73 s, scan numbers 1000, and temperature 10 °C.
4.5. Structural Determination
All assigned NOESY cross peaks were classified as strong (1.8–3.0 Å), medium (3.0–3.7 Å), weak (3.7–5.5 Å), and very weak (5.5–7.5 Å) inter-proton distance restraints based on the intensity of NOESY. The average of cytosine H5-H6 proton distances was used as a reference (2.42 Å). Distance restraints for the hydrogen bonding in each Watson–Crick base pair were 1.8–3.7 Å. The force constant of hydrogen bonds and NOE restraints were kept between 5 and 50 kcal mol−1 Å−2 throughout the computation. Then, molecular dynamics simulations were performed by the structural solvation and standard dynamics cascade in BIOVIA Discovery Studio 4.5 with modifications. Generally, the structure was heated from 50 K to 300 K over 4 ps and equilibration at 300 K with 100 ps simulation time. The save results interval in the production step was 2 ps during 100 ps simulation time at 300 K. The 10 best conformations generated by simulation were further energy minimized until the gradient of energy was less than 0.1 kcal mol−1. The conformation with the lowest energy was selected as the final presentation. The all-atom RMSD was calculated using Z-form L-RNA in superimposition.
4.6. Molecular Modeling
We manually prepared the initial model using L-ribonucleosides, which are the mirror images of D-ribonucleosides by using the L-RNA model (PDB: 1R3O) with A-form structure served as the template [29]. Next, the structure was modified as the known structural features of Z-form D-RNA and D-DNA with the syn conformation of guanosine and anti conformation of cytidine, which are consistent with the NMR results in this study. All of the above molecular model constructions were accomplished by BIOVIA Discovery Studio 4.5.
4.7. Molecular Docking Simulation
The molecular docking calculations were performed by using the BIOVIA Discovery Studio 4.5. All assigned NOESY cross-peaks in the Z-form L-RNA duplex–CBL0137 complex were used to determine the inter-proton distance restraints. The NOE peaks of H5-H6 from the cytosine bases were used for calibration of the distance measurements. The structure of CBL0137 was optimized before use as a ligand in the binding assay. The Z-form L-RNA duplex was collected from the former NMR structure in the solution in this study. All possible docking sites were provided by the BIOVIA Discovery Studio 4.5 based on the calculation results and were further selected reasonably according to their NOEs intensities. The docking experiment was implemented and provided at least five conformations, in which the proper one was used for the subsequent standard dynamics cascade and energy minimization processes. A total of five conformations were generated and the conformation with lowest energy was selected for the final presentation. The all-atom RMSDs were calculated using the Z-form L-RNA-CBL0137 complex in superimposition.
5. Conclusions
In summary, we described a right-handed Z-form L-RNA, and demonstrated its binding with the CBL0137 ligand at a high resolution. These results indicate that the new nucleic acid structure might produce applicable biofunctions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29204900/s1, Figure S1: Anomeric regions of Z-form RNA in 2D NOESY; Figure S2: Structural model of Z-form RNA L-r(CGCFGCG)2; Figure S3: Inter-strand Watson–Crick base pairs involved in the Z-form RNA duplex; Figure S4: Stick model of Z-form RNA; Figure S5: Stacking pattern within the CpG and GpC steps in Z-form RNA as viewed along the helix z-axis; Figure S6: The refined structure of Z-form RNA L-r(C1G2C3FG4C5G6)2 is a half turn of a right-handed RNA helix; Figure S7: 1D NMR spectra of Z-form L-r(CGCFGCG)2 with CBL0137; Figure S8: Study of the CBL0137 ligand binding to Z-form RNA; Figure S9: 31P spectra of Z-form L-r(CGCFGCG)2/CBL0137 complex; Figure S10: 19F spectra of Z-form L-r(CGCFGCG)2/CBL0137 complex; Figure S11: Structural model of Z-form RNA L-r(CGCFGCG)2 and the CBL0137 complex; Figure S12: An expanded view of the RNA and CBL0137 complex models; Figure S13: Molecular model show the acetyl groups (purple, CPK presentation) at the position of 3 and 7 of carbazole (green, CPK presentation) protruding into the major groove of the Z-form duplex; Figure S14: 2D heteronuclear 31P-1H NMR spectroscopy of the Z-form RNA L-r(CGCFGCG)2 and the CBL0137 complex; Figure S15: Molecular model of Z-form L-r(CGCFGCG)2/CBL0137 complex; Figure S16: Titration data of CBL0137 and RNA derived by monitoring the wavelength at 280 nm of CD. Data S1-S20: Synthesis of 8-FG-modified L-guanosine phosphoramidite; Data S21–S24: MALDI-MS of oligonucleotide in this study; Table S1: 1H chemical shift assignments of Z-form RNA L-r(CGCFGCG)2; Table S2: Statistics for the structure determination of Z-form RNA L-r(CGCFGCG)2; Table S3: A comprehensive list of NMR distance restraints of Z-form RNA L-r(CGCFGCG)2; Table S4: 1H chemical shift assignments of CBL0137; Table S5: 1H chemical shift assignments of Z-form RNA L-r(CGCFGCG)2 in presence of CBL0137; Table S6: 1H chemical shift assignments of CBL0137 in the presence of Z-form RNA L-r(CGCFGCG)2.
Author Contributions
The two authors, Y.S. and S.W., contributed equally. Both authors gave approval to the final version of the manuscript. Conceptualization, Y.X.; methodology, Y.X., Y.S. and S.W.; Software, S.W.; validation, Y.X., Y.S. and S.W.; formula analysis, Y.S.; investigation, Y.S. and S.W.; resources, Y.X., Y.S. and S.W.; curation, Y.X. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by JSPS KAKENHI Grant Numbers 21H02081 (to Y.X.) and 24K01648 (to Y.X.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wan, Y.; Kertesz, M.; Spitale, R.C.; Segal, E.; Chang, H.Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011, 12, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Wang, M.; Liu, J.; Ma, P.; Pang, S.; Liu, W.; Liu, A. Diagnostics and analysis of SARS-CoV-2: Current status, recent advances, challenges and perspectives. Chem. Sci. 2023, 14, 6149–6206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yin, C.; Boyd, D.F.; Quarato, G.; Ingram, J.P.; Shubina, M.; Ragan, K.B.; Ishizuka, T.; Crawford, J.C.; Tummers, B.; et al. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 2020, 180, 1115–1129. [Google Scholar] [CrossRef]
- Ji, D.; Lyu, K.; Zhao, H.; Kwok, C.K. Circular L-RNA aptamer promotes target recognition and controls gene activity. Nucleic Acids Res. 2021, 49, 7280–7291. [Google Scholar] [CrossRef]
- Umar, M.I.; Chan, C.Y.; Kwok, C.K. Development of RNA G-quadruplex (rG4)-targeting L-RNA aptamers by rG4-SELEX. Nat. Protoc. 2022, 17, 1385–1414. [Google Scholar] [CrossRef]
- Pu, F.; Ren, J.; Qu, X. Nucleobases, nucleosides, and nucleotides: Versatile biomolecules for generating functional nanomaterials. Chem. Soc. Rev. 2018, 47, 1285–1306. [Google Scholar] [CrossRef]
- Oberthür, D.; Achenbach, J.; Gabdulkhakov, A.; Buchner, K.; Maasch, C.; Falke, S.; Rehders, D.; Klussmann, S.; Betzel, C. Crystal structure of a mirror-image L-RNA aptamer (Spiegelmer) in complex with the natural L-protein target CCL2. Nat. Commun. 2015, 6, 6923. [Google Scholar] [CrossRef]
- Ji, D.; Yuan, J.H.; Chen, S.B.; Tan, J.H.; Kwok, C.K. Selective targeting of parallel G-quadruplex structure using L-RNA aptamer. Nucleic Acids Res. 2023, 51, 11439–11452. [Google Scholar] [CrossRef]
- Dey, S.; Sczepanski, J.T. In vitro selection of L-DNA aptamers that bind a structured D-RNA molecule. Nucleic Acids Res. 2020, 48, 1669–1680. [Google Scholar] [CrossRef]
- Umar, M.I.; Kwok, C.K. Specific suppression of D-RNA G-quadruplex–protein interaction with an L-RNA aptamer. Nucleic Acids Res. 2020, 48, 10125–10141. [Google Scholar] [CrossRef]
- Zhou, L.Y.; Qin, Z.; Zhu, Y.H.; He, Z.Y.; Xu, T. Current RNA-based therapeutics in clinical trials. Curr. Gene Ther. 2019, 19, 172–196. [Google Scholar] [CrossRef] [PubMed]
- Skaanning, M.K.; Bønnelykke, J.; Nijenhuis, M.A.; Samanta, A.; Smidt, J.M.; Gothelf, K.V. Self-Assembly of Ultrasmall 3D Architectures of (l)-Acyclic Threoninol Nucleic Acids with High Thermal and Serum Stability. J. Am. Chem. Soc. 2024, 146, 20141–20146. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A. Z-DNA and Z-RNA in human disease. Commun. Biol. 2019, 2, 7. [Google Scholar] [CrossRef]
- Jiao, H.; Wachsmuth, L.; Kumari, S.; Schwarzer, R.; Lin, J.; Eren, R.O.; Fisher, A.; Lane, R.; Young, G.R.; Kassiotis, G.; et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 2020, 580, 391–395. [Google Scholar] [CrossRef]
- Mondal, M.; Yang, L.; Cai, Z.; Patra, P.; Gao, Y.Q. perspective on the molecular simulation of DNA from structural and functional aspects. Chem. Sci. 2021, 12, 5390–5409. [Google Scholar] [CrossRef]
- Ha, S.C.; Lowenhaupt, K.; Rich, A.; Kim, Y.G.; Kim, K.K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature 2005, 437, 1183–1186. [Google Scholar] [CrossRef]
- Zhang, T.; Yin, C.; Fedorov, A.; Qiao, L.; Bao, H.; Beknazarov, N.; Wang, S.; Gautam, A.; Williams, R.M.; Crawford, J.C.; et al. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 2022, 606, 594–602. [Google Scholar] [CrossRef]
- Hubbard, N.W.; Ames, J.M.; Maurano, M.; Chu, L.H.; Somfleth, K.Y.; Gokhale, N.S.; Werner, M.; Snyder, J.M.; Lichauco, K.; Savan, R.; et al. ADAR1 mutation causes ZBP1-dependent immunopathology. Nature 2022, 607, 769–775. [Google Scholar] [CrossRef]
- Hall, K.; Cruz, P.; Tinoco, I., Jr.; Jovin, T.M.; Van De Sande, J.H. ‘Z-RNA’—A left-handed RNA double helix. Nature 1984, 311, 584–586. [Google Scholar] [CrossRef]
- Nichols, P.J.; Krall, J.B.; Henen, M.A.; Welty, R.; Macfadden, A.; Vicens, Q.; Vögeli, B. Z-Form Adoption of Nucleic Acid is a Multi-Step Process Which Proceeds through a Melted Intermediate. J. Am. Chem. Soc. 2023, 146, 677–694. [Google Scholar] [CrossRef]
- Xu, Y.; Ikeda, R.; Sugiyama, H. 8-Methylguanosine: A powerful Z-DNA stabilizer. J. Am. Chem. Soc. 2003, 125, 13519–13524. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.L.; Masuzawa, T.; Oyoshi, T.; Xu, Y. Oligonucleotides DNA containing 8-trifluoromethyl-2′-deoxyguanosine for observing Z-DNA structure. Nucleic Acids Res. 2020, 48, 7041–7051. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, Y.X.; Sugiyama, H.; Umano, T.; Osuga, H.; Tanaka, K. (P)-helicene displays chiral selection in binding to Z-DNA. J. Am. Chem. Soc. 2004, 126, 6566–6567. [Google Scholar] [CrossRef]
- Wang, S.; Xu, Y. RNA structure promotes liquid-to-solid phase transition of short RNAs in neuronal dysfunction. Commun. Biol. 2024, 7, 137. [Google Scholar] [CrossRef]
- Dallavalle, S.; Mattio, L.M.; Artali, R.; Musso, L.; Aviñó, A.; Fàbrega, C.; Eritja, R.; Gargallo, R.; Mazzini, S. Exploring the interaction of curaxin CBL0137 with G-quadruplex DNA oligomers. Int. J. Mol. Sci. 2021, 22, 6476. [Google Scholar] [CrossRef]
- Drozdzal, P.; Manszewski, T.; Gilski, M.; Brzezinski, K.; Jaskolski, M. Right-handed Z-DNA at ultrahigh resolution: A tale of two hands and the power of the crystallographic method. Acta Crystallogr. D Struct. Biol. 2023, 79, 133–139. [Google Scholar] [CrossRef]
- Urata, H.; Ogura, E.; Shinohara, K.; Ueda, Y.; Akagi, M. Synthesis and properties of mirror-image DNA. Nucleic Acids Res. 1992, 20, 3325–3332. [Google Scholar] [CrossRef]
- Stootman, F.H.; Fisher, D.M.; Rodger, A.; Aldrich-Wright, J.R. Improved curve fitting procedures to determine equilibrium binding constants. Analyst 2006, 131, 1145–1151. [Google Scholar] [CrossRef]
- Vallazza, M.; Perbandt, M.; Klussmann, S.; Rypniewski, W.; Einspahr, H.; Erdmann, V.; Betzel, C. First look at RNA in L-configuration. Acta. Cryst. 2004, 60, 1–7. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).