Bioassay-Guided Isolation and Identification of Xanthine Oxidase Inhibitory Constituents from the Fruits of Chaenomeles speciosa (Sweet) Nakai
Abstract
1. Introduction
2. Results
2.1. In Vitro Inhibition of XO by AP and Three Extracts of C. speciosa
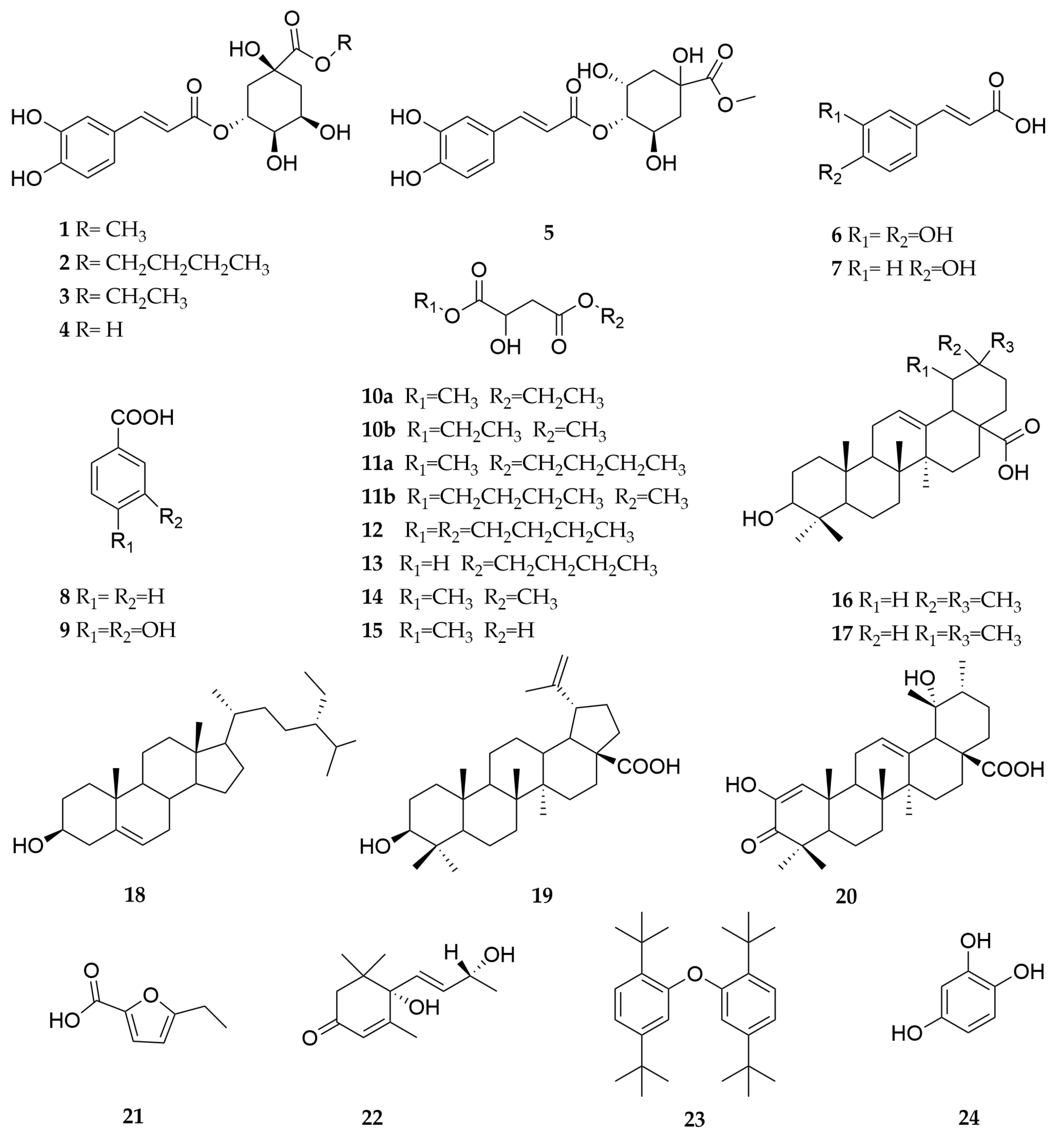
2.2. Monomer Compounds of C. speciosa and Its Inhibitory Effect on XO
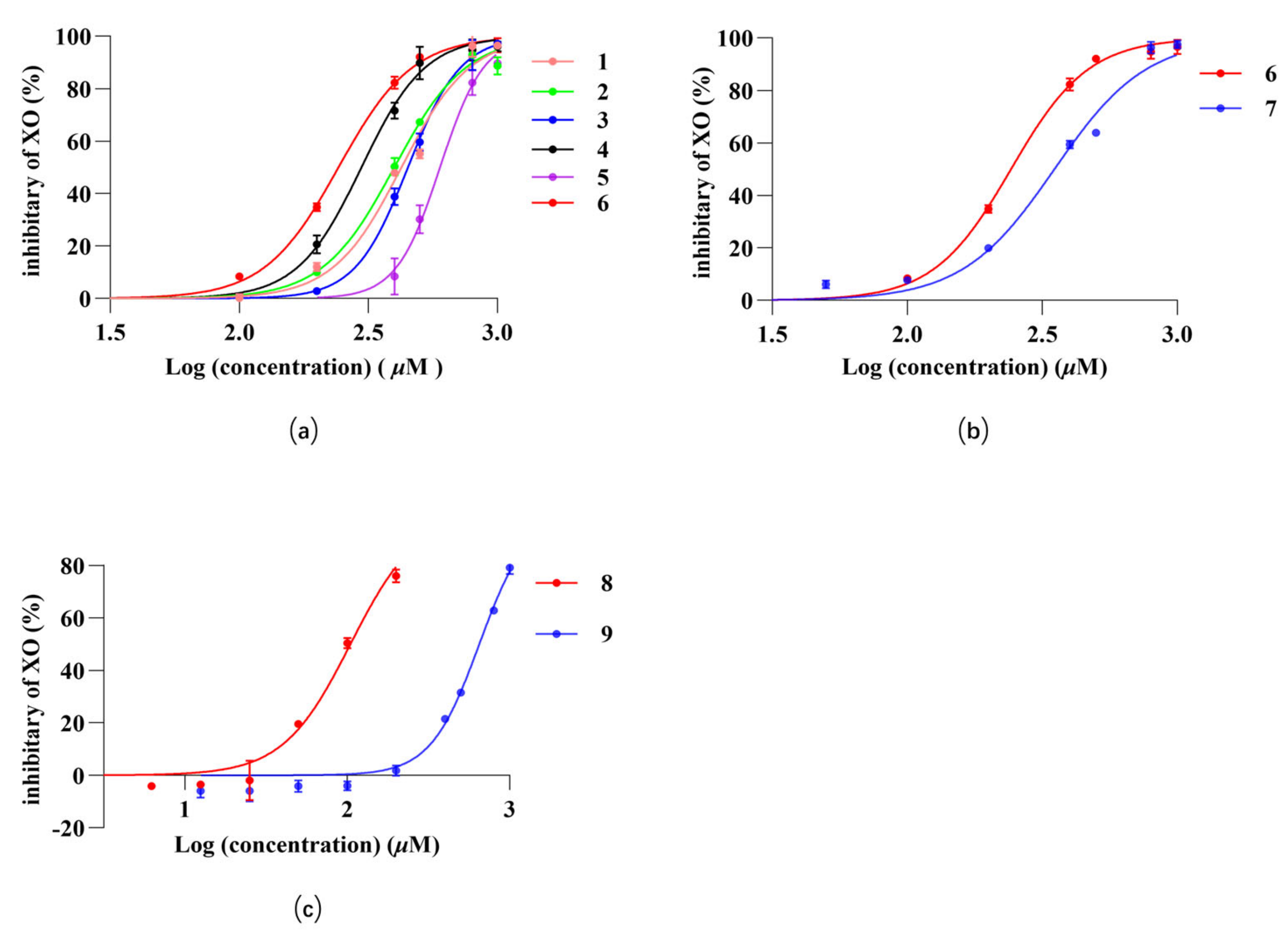
2.3. Inhibition of XO by Monomeric Compounds in C. speciosa
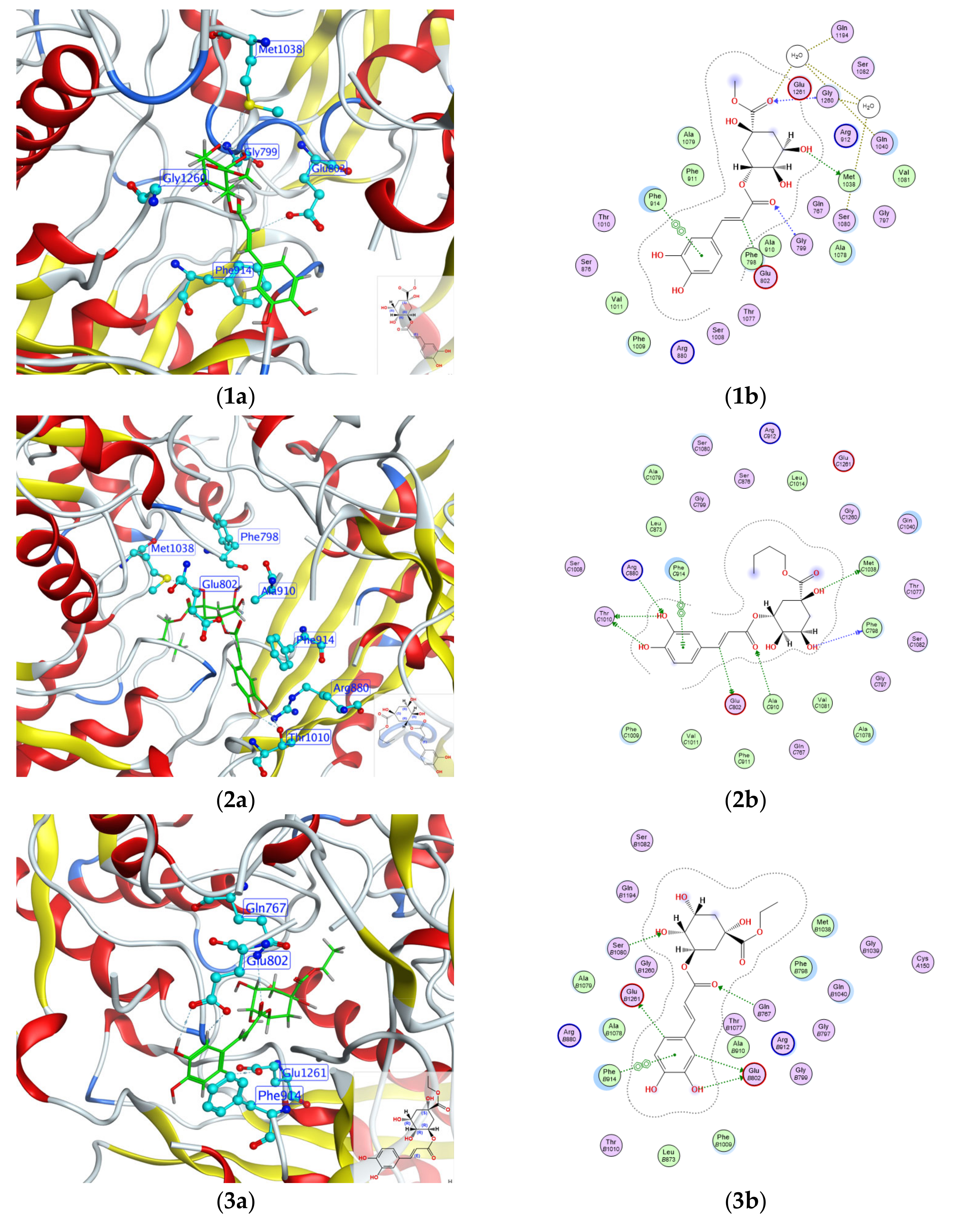
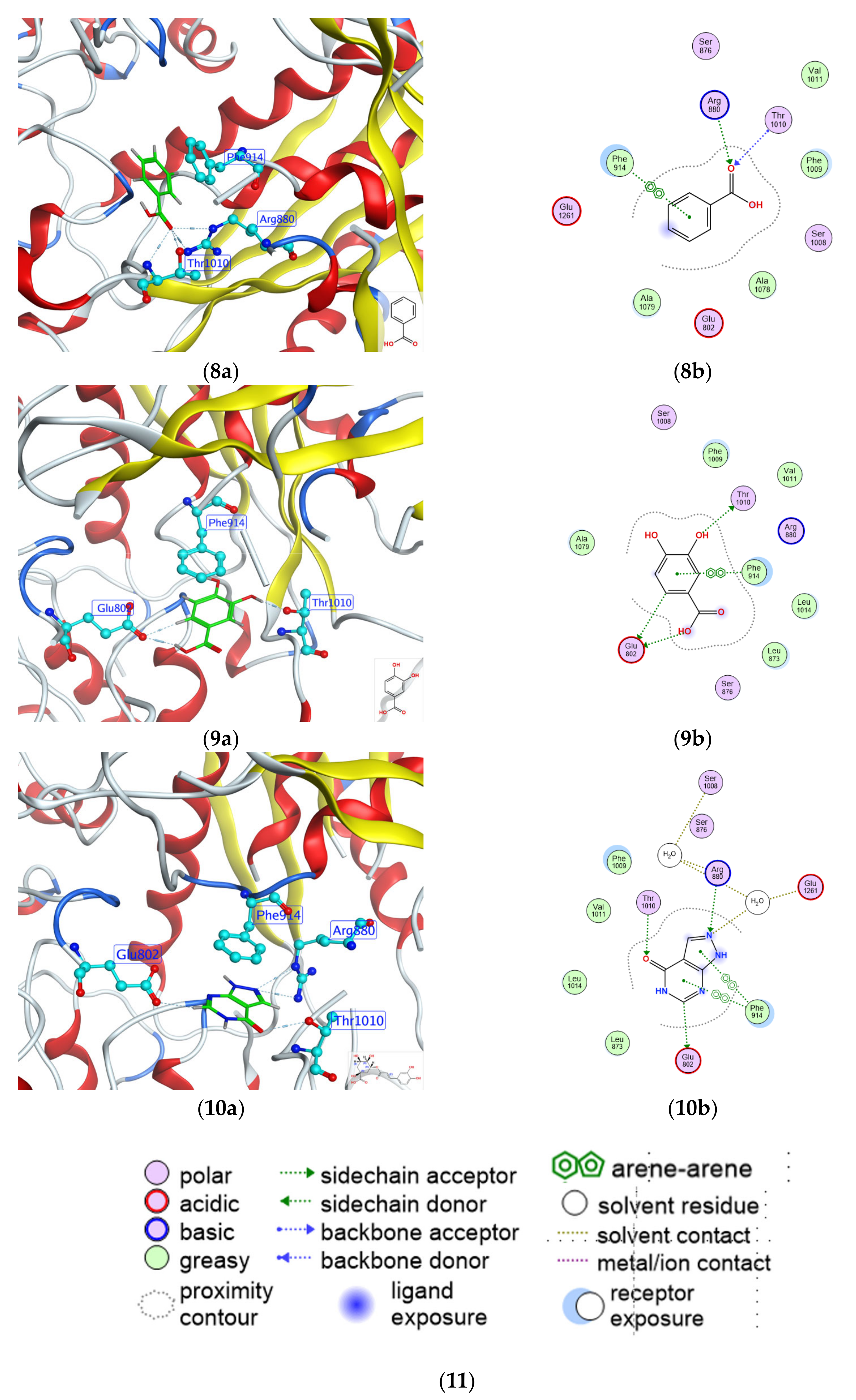
2.4. Molecular Docking Analysis
3. Discussion
4. Materials and Methods
4.1. Instrumentation and General Experimental Techniques
4.2. Plant Materials
4.3. Extraction, Isolation and Structure Identification
4.4. Inhibitory Assays of XO by Different Extracts of C. speciosa Fruits In Vitro
4.5. Extraction and Separation of Monomer Compound
4.6. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, Z.; Wu, K.; Ullah, I.; Zhu, H. Recent advances in xanthine oxidase inhibitors. Mini Rev. Med. Chem. 2024, 24, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Pauff, J.M.; Hille, R. Inhibition Studies of Bovine Xanthine Oxidase by Luteolin, Silibinin, Quercetin, and Curcumin. J. Nat. Prod. 2009, 72, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Qian, J.; Li, Y.; Shen, Y.; Chen, Y.; Fu, G.; Xie, M. Inhibitory mechanism of xanthine oxidase activity by caffeoylquinic acids in vitro. Int. J. Biol. Macromol. 2021, 184, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Fang, Y.; Wu, T.; Liang, F.; Cheng, Y.; Salah, M.; Pan, S.; Xu, X. Insights from multispectral and molecular docking investigation on the xanthine oxidase inhibition by 1,4-dicaffeoylquinic acid. J. Mol. Struct. 2020, 1219, 128475. [Google Scholar] [CrossRef]
- Zhang, G.; Zhu, M.; Liao, Y.; Gong, D.; Hu, X. Action mechanisms of two key xanthine oxidase inhibitors in tea polyphenols and their combined effect with allopurinol. J. Sci. Food Agric. 2022, 102, 7195–7208. [Google Scholar] [CrossRef]
- Ojha, R.; Singh, J.; Ojha, A.; Singh, H.; Sharma, S.; Nepali, K. An updated patent review: Xanthine oxidase inhibitors for the treatment of hyperuricemia and gout (2011–2015). Expert. Opin. Ther. Pat. 2016, 27, 311–345. [Google Scholar] [CrossRef]
- Guo, X.; Gao, Y.; Yang, Y.; Zhu, Q.; Guan, H.; He, X.; Zhang, C.; Wang, Y.; Xu, G.; Zou, S.; et al. Amelioration effects of α-viniferin on hyperuricemia and hyperuricemia-induced kidney injury in mice. Phytomedicine 2023, 116, 154868–154882. [Google Scholar] [CrossRef]
- White, W.; Saag, K.; Becker, M.; Borer, J.; Gorelick, P.; Whelton, A.; Hunt, B.; Castillo, M.; Gunawardhana, L.; Investigators, C. Cardiovascular safety of febuxostat or allopurinol in patients with gout. N. Engl. J. Med. 2018, 378, 1200–1210. [Google Scholar] [CrossRef]
- Cao, P.; Huang, Y.; Zong, M.; Xu, Z. De novo assembly and comparative analysis of the complete mitochondrial genome of Chaenomeles speciosa (Sweet) Nakai revealed the existence of two structural isomers. Genes 2023, 14, 526–541. [Google Scholar] [CrossRef]
- Xu, R.; Kuang, M.; Li, N. Phytochemistry and pharmacology of plants in the genus Chaenomeles. Arch. Pharm. Res. 2023, 46, 825–854. [Google Scholar] [CrossRef]
- Xu, R.; Deng, P.; Ma, Y.; Li, K.; Ren, F.; Li, N. Anti-hyperuricemic effects of extracts from Chaenomeles speciosa (Sweet) Nakai Fruits on hyperuricemic rats. Metabolites 2024, 14, 117–128. [Google Scholar] [CrossRef]
- Zeller, W. Synthesis of 1-O-methylchlorogenic acid: Reassignment of structure for MCGA3 isolated from bamboo (Phyllostachys edulis) leaves. J. Agric. Food Chem. 2014, 62, 1860–1865. [Google Scholar] [CrossRef] [PubMed]
- Kirmizibekmez, H.; Bassarello, C.; Piacente, S.; Celep, E.; Atay, I.; Mercanoglu, G.; Yesilada, E. Phenolic compounds from hypericum calycinum and their antioxidant activity. Nat. Prod. Commun. 2009, 4, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Dong, X.; Wang, W.; Ju, P.; Luo, S. Phenolic compounds from Viburnum cylindricum. Helv. Chim. Acta 2005, 88, 339–342. [Google Scholar] [CrossRef]
- Tong, Y.; Li, G.; Shi, X.; Wang, L.; Zhou, J.; Chu, M.; Wang, Z.; Abd El-Aty, A.M.; Dang, J. Protection against myocardial ischemia/reperfusion injury in mice by 3-caffeoylquinic acid isomers isolated from Saxifraga tangutica. RSC Adv. 2024, 14, 6642–6655. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chang, F.; Lin, Y.; Wang, L.; Chen, J.; Wu, Y.; Wu, M. Identification of phenolic antioxidants from Sword Brake fern (Pteris ensiformis Burm.). Food Chem. 2007, 105, 48–56. [Google Scholar] [CrossRef]
- Rho, T.; Yoon, K. Chemical constituents of nelumbo nucifera seeds. Nat. Prod. Sci. 2017, 23, 253–257. [Google Scholar] [CrossRef]
- Lü, S.; Zhao, S.; Zhao, M.; Guo, Y.; Li, G.; Yang, B.; Wang, Q.; Kuang, H. Systematic screening and characterization of prototype constituents and metabolites of triterpenoid saponins of Caulopphyllum robustum Maxim using UPLC-LTQ Orbitrap MS after oral administration in rats. J. Pharm. Biomed. Anal. 2019, 168, 75–82. [Google Scholar] [CrossRef]
- Wang, W.; Yang, C.R.; Zhang, Y.J. Phenolic constituents from the fruits of Amomum tsaoko (Zingiberaceae). Acta Botanica Yunnanica 2009, 31, 284–288. [Google Scholar] [CrossRef]
- Dias, C.; Dias, M.; Borges, C.; Almoster Ferreira, M.A.; Paulo, A.; Nascimento, J. Structural elucidation of natural 2-hydroxy di- and tricarboxylic acids and esters, phenylpropanoid esters and a flavonoid from Autonoë madeirensis using gas chromatographic/electron ionization, electrospray ionization and tandem mass spectrometric techniques. J. Mass. Spectrom. 2003, 38, 1240–1244. [Google Scholar] [CrossRef]
- Samoylenko, V.; Zhao, J.; Dunbar, D.; Khan, I.; Rushing, J.; Muhammad, I. New constituents from Noni (Morinda citrifolia) fruit juice. J. Agric. Food Chem. 2006, 54, 6398–6402. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.; Song, S.; Jang, D. A method for preparation of unnatural (R)-malic acid derivatives with phenylsilanes. Synth. Commun. 2003, 33, 515–519. [Google Scholar] [CrossRef]
- Akihisa, T.; Tochizawa, S.; Takahashi, N.; Yamamoto, A.; Zhang, J.; Kikuchi, T.; Fukatsu, M.; Tokuda, H.; Suzuki, N. Melanogenesis-inhibitory saccharide fatty acid esters and other constituents of the fruits of Morinda citrifolia (Noni). Chem. Biodivers. 2012, 9, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- Xing, R.; Wu, L.; Fei, Z.; Wu, P. Palladium phthalocyaninesulfonate functionalized mesoporous polymer: A highly efficient photocatalyst for degradation of 4-chlorophenol under visible light irradiation. J. Mol. Catal. A Chem. 2013, 371, 15–20. [Google Scholar] [CrossRef]
- Houston, T.; Wilkinson, B.; Blanchfield, J. Boric acid catalyzed chemoselective esterification of α-hydroxycarboxylic acids. Org. Lett. 2004, 6, 679–681. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ge, Z.; Liao, X.; Xue, J.; Wu, L.; Liang, L. α-Glucosidase inhibitory phytochemical components of chinese endemic plant whitfordiodendron filipes var. tomentosum. Plants 2024, 13, 692–703. [Google Scholar] [CrossRef]
- Silva, M.; Vieira, I.; Mendes, F.; Albuquerque, I.; dos Santos, R.; Silva, F.; Morais, S. Variation of ursolic acid content in eight Ocimum species from northeastern Brazil. Molecules 2008, 13, 2482–2487. [Google Scholar] [CrossRef]
- Koizumi, N.; Fujimoto, Y.; Takeshita, T.; Ikekawa, N. Carbon-13 nuclear magnetic resonance of 24-substituted steroids. Chem. Pharm. Bull. 1979, 27, 38–42. [Google Scholar] [CrossRef]
- Chatterjee, P.; Pezzuto, J.; Kouzi, S. Glucosidation of betulinic acid by Cunninghamell species. J. Nat. Prod. 1999, 62, 761–763. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Q.; Wang, J.; Jang, W.; Hai, P.; Jia, C.; Ren, L.; Wu, X.; Kang, C.; Yang, J.; et al. Triterpenoids in sorbus pohuashanensis suspension cell treated with yeast extract. China J. Chin. Mater. Med. 2024, 49, 130–140. [Google Scholar] [CrossRef]
- Lee, S.; Kim, B.; Cho, S.; Shin, K. Phytochemical constituents from the fruits of Acanthopanax sessiliflorus. Arch. Pharm. Res. 2002, 25, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Lundgren, L. Monoaryl and cyclohexenone glycosides from needles of Pinus sylvestris. Phytochemistry 1988, 27, 559–562. [Google Scholar] [CrossRef]
- Sun, B.; Shen, H.; Wu, H.; Yao, L.; Cheng, Z. Isolation and identification of chemical constituents from Peronia verruculata. Chin. Pharm. J. 2014, 25, 1019–1021. [Google Scholar] [CrossRef]
- Huang, Y.; Zeng, G.; Yang, Y.; Zhao, Y. Chemical constituents from euphorbia royleana. J. Yunnan Univ. Nat. Sci. Ed. 2021, 30, 205–208. [Google Scholar]
- Ghallab, D.; Shawky, E.; Metwally, A.; Celik, I.; Ibrahim, R.; Mohyeldin, M. Integrated in silico—In vitro strategy for the discovery of potential xanthine oxidase inhibitors from Egyptian propolis and their synergistic effect with allopurinol and febuxostat. RSC Adv. 2022, 12, 2843–2872. [Google Scholar] [CrossRef]
- Song, J.; Chen, M.; Meng, F.; Chen, J.; Wang, Z.; Zhang, Y.; Cui, J.; Wang, J.; Shi, D. Studies on the interaction mechanism between xanthine oxidase and osmundacetone: Molecular docking, multi-spectroscopy and dynamical simulation. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 299, 122861. [Google Scholar] [CrossRef]
- Hille, R. Xanthine oxidase-a personal history. Molecules 2023, 28, 1921. [Google Scholar] [CrossRef]
- Wang, R.; Halimulati, M.; Huang, X.; Ma, Y.; Li, L.; Zhang, Z. Sulforaphane-driven reprogramming of gut microbiome and metabolome ameliorates the progression of hyperuricemia. J. Adv. Res. 2023, 52, 19–28. [Google Scholar] [CrossRef]
- Chen, J.; Xu, L.; Jiang, L.; Wu, Y.; Wei, L.; Wu, X.; Xiao, S.; Liu, Y.; Gao, C.; Cai, J.; et al. Sonneratia apetala seed oil attenuates potassium oxonate/hypoxanthine-induced hyperuricemia and renal injury in mice. Food Funct. 2021, 12, 9416–9431. [Google Scholar] [CrossRef]
- Becker, M.; Schumacher, H.; Wortmann, R.; MacDonald, P.; Eustace, D.; Palo, W.; Streit, J.; Joseph-Ridge, N. Febuxostat compared with allopurinol in patients with hyperuricemia and gout. N. Engl. J. Med. 2005, 353, 2450–2461. [Google Scholar] [CrossRef]
- Bai, Y.; Wu, B.; Gou, L.; Fang, Z.; Xu, T.; Zhang, T.; Li, Y. Cardiovascular safety evaluation of febuxostat and allopurinol: Findings from the FDA adverse event reporting system. J. Clin. Med. 2023, 12, 6089–6101. [Google Scholar] [CrossRef] [PubMed]
- Yong, T.; Liang, D.; Xiao, C.; Huang, L.; Chen, S.; Xie, Y.; Gao, X.; Wu, Q.; Hu, H.; Li, X.; et al. Hypouricemic effect of 2,4-dihydroxybenzoic acid methyl ester in hyperuricemic mice through inhibiting XOD and down-regulating URAT1. Biomed. Pharmacother. 2022, 153, 113303–113313. [Google Scholar] [CrossRef] [PubMed]
- Masuoka, N.; Kubo, I. Characterization of the xanthine oxidase inhibitory activity of alk(en)yl phenols and related compounds. Phytochemistry 2018, 155, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Lee, F.; Chen, C.; Huang, S.; Tsai, S.; Huang, S.; Lin, C. Structure-activity relationship of C6-C3 phenylpropanoids on xanthine oxidase-inhibiting and free radical-scavenging activities. Free Radic. Biol. Med. 2007, 43, 1541–1551. [Google Scholar] [CrossRef] [PubMed]
- Gawlik-Dziki, U.; Dziki, D.; Świeca, M.; Nowak, R. Mechanism of action and interactions between xanthine oxidase inhibitors derived from natural sources of chlorogenic and ferulic acids. Food Chem. 2017, 225, 138–145. [Google Scholar] [CrossRef]
- Wang, S.; Chen, C.; Huang, S.; Yu, S.; Lai, Z.; Huang, S.; Lin, C. Hydrophilic ester-bearing chlorogenic acid binds to a novel domain to inhibit xanthine oxidase. Planta Med. 2009, 75, 1237–1240. [Google Scholar] [CrossRef]
- Falodun, A.; Ali, S.; Quadir, I.M.; Choudhary, I.M.I. Phytochemical and biological investigation of chloroform and ethylacetate fractions of Euphorbia heterophylla leaf (Euphorbiaceae). J. Med. Plant Res. 2008, 2, 365–369. [Google Scholar]
- Li, J.; Gong, Y.; Li, J.; Fan, L. In vitro inhibitory effects of polyphenols from Tartary buckwheat on xanthine oxidase: Identification, inhibitory activity, and action mechanism. Food Chem. 2022, 379, 132100–132109. [Google Scholar] [CrossRef]
- Palafox-Carlos, H.; Gil-Chavez, J.; Sotelo-Mundo, R.R.; Namiesnik, J.; Gorinstein, S.; Gonzalez-Aguilar, G.A. Antioxidant interactions between major phenolic compounds found in ‘Ataulfo’ mango pulp: Chlorogenic, gallic, protocatechuic and vanillic acids. Molecules 2012, 17, 12657–12664. [Google Scholar] [CrossRef]
- Shao, Y.; Hu, Z.; Yu, Y.; Mou, R.; Zhu, Z.; Beta, T. Phenolic acids, anthocyanins, proanthocyanidins, antioxidant activity, minerals and their correlations in non-pigmented, red, and black rice. Food Chem. 2018, 239, 733–741. [Google Scholar] [CrossRef]
- Foss, K.; Przybylowicz, K.E.; Sawicki, T. Antioxidant Activity and Profile of Phenolic Compounds in Selected Herbal Plants. Plant Foods Hum. Nutr. 2022, 77, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jin, W.; Zhang, W.; Zheng, G. The inhibitory kinetics and mechanism of quercetin-3-O-rhamnoside and chlorogenic acid derived from Smilax china L. ethyl acetate fraction on xanthine oxidase. Int. J. Biol. Macromol. 2022, 213, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Karhana, S.; Dabral, S.; Garg, A.; Bano, A.; Agarwal, N.; Khan, M.A. Network pharmacology and molecular docking analysis on potential molecular targets and mechanism of action of BRAF inhibitors for application in wound healing. J. Cell Biochem. 2023, 124, 1023–1039. [Google Scholar] [CrossRef] [PubMed]
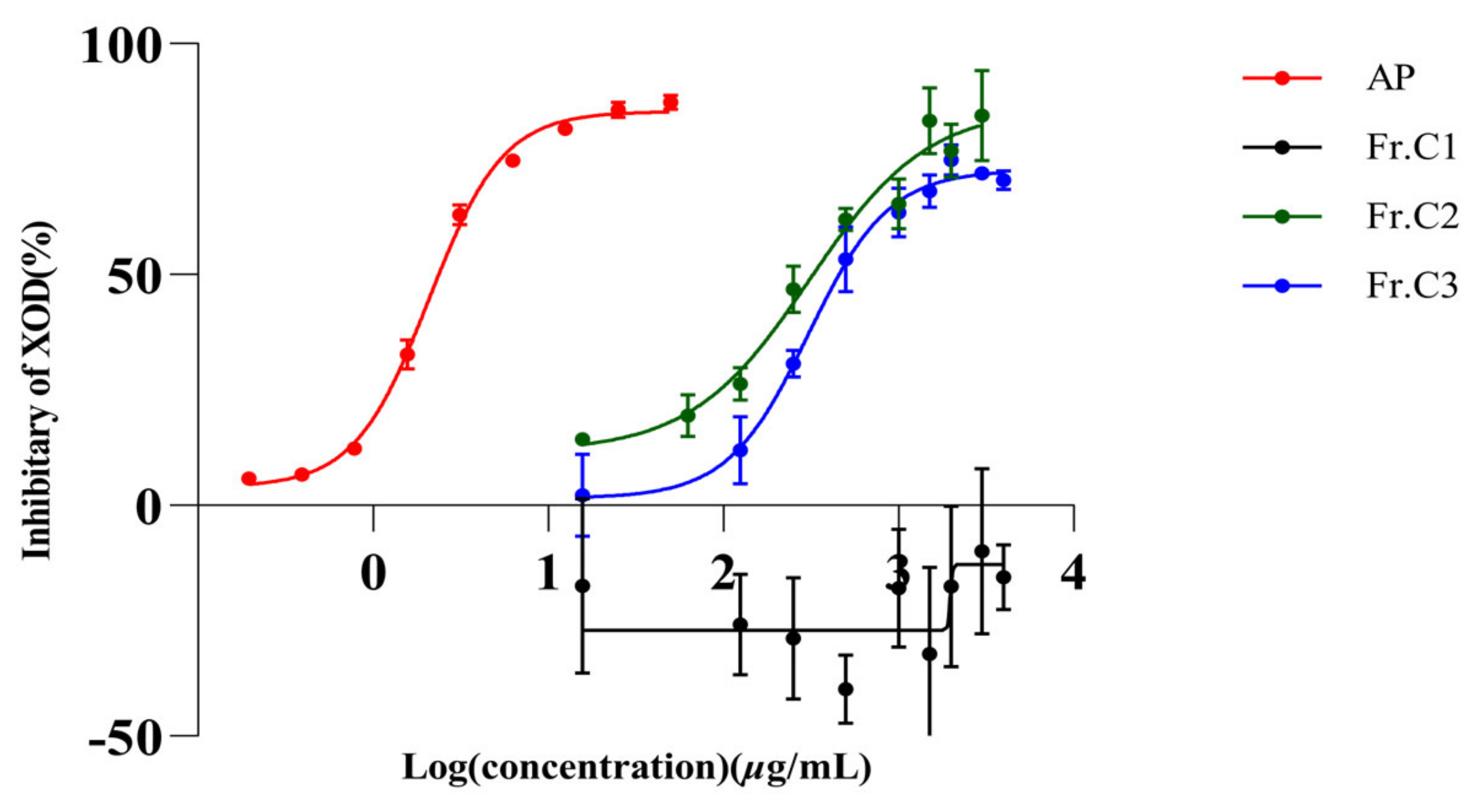

| Extract | IC50 (µg/mL) |
|---|---|
| Fr.C1 | nc |
| Fr.C2 | 341.80 |
| Fr.C3 | 321.10 |
| AP | 2.08 |
| NO. | Compound | Classification | IC50 (µg/mL) |
|---|---|---|---|
| 1 | methyl chlorogenate | penylpropanoids | 156.5 |
| 2 | butyl chlorogenate | penylpropanoids | 163.6 |
| 3 | ethyl chlorogenate | penylpropanoids | 172.2 |
| 4 | chlorogenic acid | penylpropanoids | 105.4 |
| 5 | cryptochlorogenic acid methyl ester | penylpropanoids | 220.9 |
| 6 | caffeic acid | hydroxycinnamic acid | 43.6 |
| 7 | p-coumaric acid | hydroxycinnamic acid | 57.6 |
| 8 | benzoic acid | aromatic carboxylic acids | 12.7 |
| 9 | protocatechuic acid | aromatic carboxylic acids | 100.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Xu, R.; Kuang, M.; Ma, W.; Li, N. Bioassay-Guided Isolation and Identification of Xanthine Oxidase Inhibitory Constituents from the Fruits of Chaenomeles speciosa (Sweet) Nakai. Molecules 2024, 29, 4468. https://doi.org/10.3390/molecules29184468
Li K, Xu R, Kuang M, Ma W, Li N. Bioassay-Guided Isolation and Identification of Xanthine Oxidase Inhibitory Constituents from the Fruits of Chaenomeles speciosa (Sweet) Nakai. Molecules. 2024; 29(18):4468. https://doi.org/10.3390/molecules29184468
Chicago/Turabian StyleLi, Kui, Ruoling Xu, Mengting Kuang, Wei Ma, and Ning Li. 2024. "Bioassay-Guided Isolation and Identification of Xanthine Oxidase Inhibitory Constituents from the Fruits of Chaenomeles speciosa (Sweet) Nakai" Molecules 29, no. 18: 4468. https://doi.org/10.3390/molecules29184468
APA StyleLi, K., Xu, R., Kuang, M., Ma, W., & Li, N. (2024). Bioassay-Guided Isolation and Identification of Xanthine Oxidase Inhibitory Constituents from the Fruits of Chaenomeles speciosa (Sweet) Nakai. Molecules, 29(18), 4468. https://doi.org/10.3390/molecules29184468






