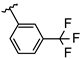
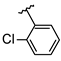
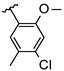
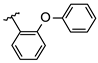
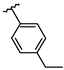
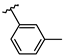
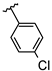
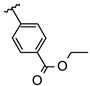
Abstract
The Caf1/CNOT7 nuclease is a catalytic component of the Ccr4-Not deadenylase complex, which is a key regulator of post-transcriptional gene regulation. In addition to providing catalytic activity, Caf1/CNOT7 and its paralogue Caf1/CNOT8 also contribute a structural function by mediating interactions between the large, non-catalytic subunit CNOT1, which forms the backbone of the Ccr4-Not complex and the second nuclease subunit Ccr4 (CNOT6/CNOT6L). To facilitate investigations into the role of Caf1/CNOT7 in gene regulation, we aimed to discover and develop non-nucleoside inhibitors of the enzyme. Here, we disclose that the tri-substituted 2-pyridone compound 5-(5-bromo-2-hydroxy-benzoyl)-1-(4-chloro-2-methoxy-5-methyl-phenyl)-2-oxo-pyridine-3-carbonitrile is an inhibitor of the Caf1/CNOT7 nuclease. Using a fluorescence-based nuclease assay, the activity of 16 structural analogues was determined, which predominantly explored substituents on the 1-phenyl group. While no compound with higher potency was identified among this set of structural analogues, the lowest potency was observed with the analogue lacking substituents on the 1-phenyl group. This indicates that substituents on the 1-phenyl group contribute significantly to binding. To identify possible binding modes of the inhibitors, molecular docking was carried out. This analysis suggested that the binding modes of the five most potent inhibitors may display similar conformations upon binding active site residues. Possible interactions include π-π interactions with His225, hydrogen bonding with the backbone of Phe43 and Van der Waals interactions with His225, Leu209, Leu112 and Leu115.
1. Introduction
The human Caf1/CNOT7 nuclease is a member of the DEDDh subgroup of the RNAse D family of exonucleases [1,2,3,4,5,6,7]. The enzyme removes the 3′ terminal nucleotide in a distributive manner and selectively recognises poly(A) RNA substrates. As is typical for enzymes of the RNAse D family, enzyme activity is dependent on two Mg2+ ions that are coordinated by three Asp (D40, D161 and D230) and a Glu (E42) residue in the active site [4,5,6,7,8,9]. Caf1/CNOT7 is a catalytic component of the human Ccr4-Not complex [10,11,12,13], which is involved in the degradation of cytoplasmic mRNA by the shortening and removal of the poly(A) tail [14,15,16,17,18]. This initiates the removal of the 5′ cap structure and the exposure of the 5′ end of mRNA to degradation by the 5′-3′ exoribonuclease Xrn1 [2,3,4,5].
In addition to their catalytic roles, Caf1/CNOT7 and its paralogue Caf1/CNOT8 also have a structural role in the Ccr4-Not complex by mediating the interaction between the large subunit of the complex, CNOT1, and the second nuclease subunit, Ccr4 (CNOT6/CNOT6L) [3,4,5,10,12,19]. The contribution of the two nuclease subunits is not well understood: it is currently not clear whether the nuclease activities of the Caf1 and Ccr4 subunits are collaborative and inhibition of one subunit impacts on the second [13,20,21,22,23,24], or whether the Caf1 and Ccr4 nucleases have specialised roles [25,26].
In higher eukaryotes, Caf1 is encoded by two paralogous genes: CNOT7 and CNOT8. In human and mouse cells, CNOT7 and CNOT8 have overlapping functions and can partially compensate for loss-of-function of the other paralogue [27,28,29,30]. Both paralogues are required for efficient cell proliferation of breast and gastric cancer cells [27,31]. However, the paralogues also have unique roles. In mice, Cnot7 is a repressor of osteoblast function and deletion of Cnot7 improves bone density [32,33]. Moreover, genome-wide association studies using recombinant inbred mice identified the involvement of Cnot7 gene variants in bone biology [34]. The use of recombinant inbred mouse strains also led to the identification of Ccr4-Not components as modulators of cancer metastasis [35]. Specifically, Caf1/Cnot7 promotes metastatic disease in a murine model of breast cancer [36]. In this case, the role of Cnot7 in promoting metastasis is dependent on its catalytic activity suggesting that inhibitors of Caf1/CNOT7 may reduce the incidence of breast cancer metastasis [36].
Inhibitors of Caf1/CNOT7 offer an alternative approach to genetic methods and can provide valuable starting points for possible subsequent drug discovery programmes. Therefore, to facilitate the discovery and identification of drug-like inhibitors of the Caf1/CNOT7 nuclease, we previously developed a quantitative, fluorescence-based assay using purified components [20,37]. This assay was used to develop Caf1/CNOT7 inhibitors based on the presence of a N-hydroxy-imide moiety [38], which was proposed to coordinate two Mg2+ ions in the active sites of the FEN-1 and HIV RNase H ribonucleases [39,40,41,42].
We also used the fluorescence-based assay to identify inhibitors of the Caf1/CNOT7 nuclease by screening a library of compounds [37]. Here, we disclose the structure of the most potent ‘hit’ compound of the screen for drug-like inhibitors, a tri-substituted 2-pyridone, and report the results of a preliminary structure–activity relationship analysis. We also report plausible binding interactions between the most potent analogues and the binding pocket of human Caf1/CNOT7 using molecular docking.
2. Results and Discussion
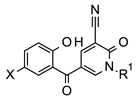
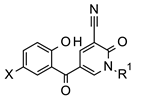
Previously, we carried out a virtual screen (83,086 compounds) to identify inhibitors of the Caf1/CNOT7 nuclease and assayed the top 1440 ranked compounds using a fluorescence-based assay [37]. Here, we disclose the structure of the most potent compound of the screen, 5-(5-bromo-2-hydroxybenzoyl)-1-(4-chloro-2-methoxy-5-methylphenyl)-2-oxo-1,2-dihydropyridine-3-carbonitrile (1, Figure 1). The activity of the compound was confirmed in a gel-based assay and shown to be selective to Caf1/CNOT7 [37].
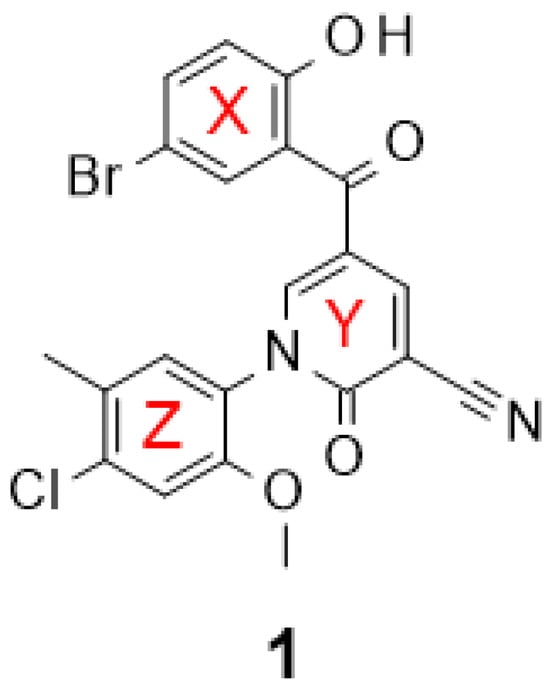
Figure 1.
Structure of 5-(5-bromo-2-hydroxybenzoyl)-1-(4-chloro-2-methoxy-5-methylphenyl)-2-oxo-1,2-dihydropyridine-3-carbonitrile, an inhibitor of the human Caf1/CNOT7 nuclease. The reported IC50 value is 14.6 ± 3.1 μM [37].
To confirm the activity of 1, we re-purchased the compound and confirmed inhibition of the Caf1/CNOT7 enzyme. The obtained IC50 (24.1 ± 3.3 µM, n = 3) conformed with the originally determined value (14.6 ± 3.1 µM, n = 3) (p = 0.104, two-tailed, unpaired t-test) (Table 1).

Table 1.
The inhibitory activity of 5-benzoyl-2-oxo-1-phenyl-1,2-dihydropyridine-3-carbonitrile analogues versus Caf1/CNOT7 a.
To obtain insight into the probable binding mode of 1, we used molecular docking using the X-ray structure of human Caf1/CNOT7 in complex with the MIF4G domain of CNOT1 and Ccr4/CNOT6L (PDB 7VOI) [12]. Because the active site Mg2+ ions were not resolved in this structure, the metal ions were transposed from the structure of the homologous Schizosaccharomyces pombe Pop2 protein, which displays a highly similar conformation of the coordinating side chains of amino acids corresponding to Asp40, Glu42, Asp161 and Asp230 (Figure 2A). Next, the position of the RNA substrate was modelled by superimposing the structures of Caf1/CNOT7 and the DEDDh-type Schizosaccharomyces pombe Pan2 ribonuclease in complex with oligo(A) RNA (PDB 6R9J) [45] (Figure 2B, Supplementary Figure S2A). To assess the effectiveness of the docking approach, we initially redocked a native ligand, poly(A) RNA, into the active site of human Caf1/CNOT7. To this end, the ligand from the crystal structure was retrieved and redocked resulting in a binding orientation closely matching the original pose (docking score, −8.4 kcal/mol, RMSD 1.6Å) (Supplementary Figure S2B).
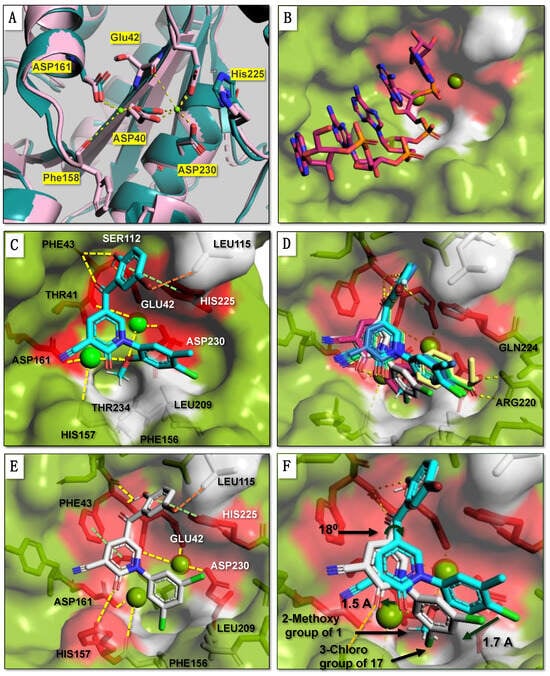
Figure 2.
Molecular docking of inhibitors into the active site of human Caf1/CNOT7. (A) Catalytic site of the Caf1/CNOT7 enzyme. Shown is the position of the residues coordinating two Mg2+ ions (bright green) in the active site of Schizosaccharomyces pombe Pop2 protein (PDB 2P51, slate blue) [9] and the corresponding coordinating residues of human Caf1/CNOT7 (PDB 7VOI, salmon red) [12]. (B) Model of human Caf1/CNOT7 bound to poly(A) RNA. The RNA was obtained by superposition of the structure of human Caf1/CNOT7 (PDB 7VOI) [12] and Schizosaccharomyces pombe Pan2 in complex with poly(A) RNA (PDB 6R9J) [45]. Shown are the surface views of the residues developing; polar interactions (red) and nonpolar interactions (white) with the analogues (1, 8, 9, 11, 15 and 17) in the active site. (C) Molecular docking of 1 (cyan) into the active site of Caf1/CNOT7. (D) Overlay of 1 (cyan) with the plausible conformation of the five most potent analogues 8 (light green), 9 (light yellow), 11 (slate blue), 15 (magenta) and 17 (white). (E) Molecular docking of 17 (white) into the active site of Caf1/CNOT7 (F) Overlay of 1 (cyan) with the plausible conformation of the most potent analogue 17, into the active site of Caf1/CNOT7.
Molecular docking of compound 1 indicated that its probable position in the active site blocks access of the substrate (Figure 2C). Plausible interactions between 1 and the Caf1/CNOT7 protein primarily include polar contacts between the benzoyl group and the residues Glu42, Phe43, His225, Asp161 and Asp230 (Figure 2C, red surface) and nonpolar contacts between the methoxy group and residues Phe156, Leu209 and Thr234 (Figure 2C, grey surface). Here, the backbone amide of Phe43 forms hydrogen bonds with the 2-hydroxyl and 1-carbonyl groups of the benzoyl moiety of 1. The aromatic phenyl (Figure 1, ring X) of the benzoyl moiety develops Van der Waals interactions with the residues Ser112, Leu115 and His225, in addition to Van der Waals interactions between the 5-bromo group and Ser112. His225 also forms T-shaped π-π interactions with both the benzoyl and the 1-phenyl ring structures of 1. In the case of the 2-methoxy substitution on the phenyl (Figure 1, ring Z), the methyl group protrudes to the lipophilic trough between the residues Phe156, Leu209 and Thr234. Moreover, the methoxy oxygen coordinates interactions with two Mg2+ ions that mutually coordinate cohesive ionic bonds with other protein residues, including Thr41-Mg2+(OMe)-Asp230 and Asp161-Mg2+(OMe)-His157.
Based on the probable interactions between 1 and the Caf1/CNOT7 nuclease, we hypothesised that changing the position of substituents on the 1-phenyl group or the introduction of alternative substituents may lead to additional Van der Waals interactions. We therefore acquired 15 analogues of 1 with different substituents on the 1-phenyl group (Figure 1, ring Z) and determined their Caf1/CNOT7 nuclease inhibitory potential (IC50 values) to establish a preliminary structure–activity relationship (Table 1). All compounds were selected based on their predicted drug-likeness, favourable predicted pharmacokinetic profile, and accessibility to synthetic chemistry (Supplementary Tables S2–S4). The activity of 8 and 15 indicates that the 4-chloro and 3-methyl substituents appear to contribute to binding. The 2-methoxy group alone does not appear to contribute significantly as evident from the reduced potency of 6. Moderate differences in potency were observed for the remaining compounds with one or two substituents. Only compound 17, which contains 3, 5-dichloro substituents on the 1-phenyl group, displayed a lower IC50 value than 1, although the difference was not statistically significant (p = 0.9914). Compound 12, which has no substituents on the 1-phenyl moiety, displayed a 4–5-fold lower potency compared to 1 (p < 0.0001), indicating that substituents make a significant contribution to potency.
In addition to compounds with different substituents in the 1-phenyl group, we also acquired an analogue of 1 containing a 5-fluoro atom instead of a 5-bromo substituent on the benzoyl moiety (11). Compound 11 displayed a modest (<2-fold) reduction in potency compared to 1, which was not statistically significant (p = 0.517). Perhaps the larger atom size of the 5-bromo substituent compared to the 5-fluoro substituent may increase the probability of Van der Waals interactions with adjoining protein residues.
To understand the activity of the analogues, we also evaluated the probable binding mode of the five most potent analogues 8, 9, 11, 15, and 17 using molecular docking (Figure 2D, Supplementary Table S1). In all cases, the probable conformation of the benzoyl group adopts a similar conformation as observed for 1 (Figure 2D). However, some differences in the position of the central pyridone ring (Figure 1, ring Y) and the 1-phenyl group (Figure 1, ring Z) were observed. The probable interactions and conformations of 8 and 11 were very similar to that of 1. In case of 9, however, the oxygen and carbonyl of the 4-ethoxycarbonyl group forms possible hydrogen bonds with the side chains of Arg220 and Gln224, respectively.
More significant differences were observed for the probable interactions of 15 and 17 with active site residues. In these cases, His225 makes T-shaped π-π interactions with the benzoyl ring while Phe43 forms π-π interactions with the central pyridone ring (Figure 1, ring Y) in addition to a probable hydrogen bond between the Phe43 backbone amide-NH and the 2-hydroxyl and 1-carbonyl groups of the benzoyl group. The benzoyl ring (Figure 1, ring X) forms Van der Waals interactions with the residues Phe43, Ser112, Leu115 and His225, and in case of 17, also with Phe156 (Figure 2E).
When compared to 1 (Figure 2E,F), the central pyridone ring (Figure 1, ring Y) of 17 may swing clockwise by 18° thereby shifting the 3,5-dichlorophenyl ring (Figure 1, ring Z) by 1.7 Å so that the 3-chloro group takes the position of 2-methoxy group of 1. This orientation also moves 2-ketone group of the pyridone ring (Figure 1, ring Y) of 17 by 1.5 Å towards His157 to form a polar interaction with its carbamide oxygen. The docking scores of the selected analogues showed a moderate correlation with experimental activities (IC50) (Supplementary Figure S2C).
In conclusion, we disclose that 5-(5-bromo-2-hydroxy-benzoyl)-1-(4-chloro-2-methoxy-5-methyl-phenyl)-2-oxo-pyridine-3-carbonitrile is an inhibitor of the Caf1/CNOT7 nuclease. The activity of 16 structural analogues was determined. Substituents on the 1-phenyl moiety contributed to the interaction with the Caf1/CNOT7, but no compound was identified with a statistically significant increase in inhibitory activity towards the Caf1/CNOT7 nuclease.
3. Materials and Methods
Compounds (purity > 95%) were obtained from Enamine. Purity of the compounds was verified by LC-MS. The inhibitory activity of the compounds was assessed using a fluorescence-based activity assay described before [37,46]. Briefly, purified Caf1/CNOT7 was pre-incubated in the presence of compound or vehicle only (DMSO) before addition of a Fluorescein-labelled RNA oligonucleotide substrate. The composition of the final reaction mixture (10 µL) was: 0.4 µM Caf1/CNOT7, 1 µM Flc-RNA substrate, 20 mM Tris–HCl pH 7.9, 50 mM NaCl, 2 mM MgCl2, 10% glycerol, 1 mM β-mercaptoethanol and 5% DMSO. After incubation at 30 °C for 60 min, 10 µL probe mix (5 µM TAMRA-DNA probe, 1% SDS and 20 mM Tris–HCl pH 8.0, 0.5 mM EDTA) was added. Fluorescence intensity was measured using a BioTek Synergy HT plate reader (Agilent, Cheshire, UK) using the filters 485 ± 20 nm (excitation) and 528 ± 20 nm (emission). Raw data were analysed in Microsoft Excel and non-linear regression was carried out using GraphPad Prism 6. Statistical analysis (one-way ANOVA with post-hoc Dunnett’s test) was also carried out using GraphPad Prism.
Molecular Docking
Molecular docking was carried out using Autodock Vina version 1.2 and UCSF Chimera [43,44,47]. The human Caf1/CNOT7 protein for docking was prepared from the X-ray structure of Caf1/CNOT7 in complex with CNOT1 and Ccr4/CNOT6L (PDB 7VOI, chain B) [12] by removal of ions and water molecules. Subsequently, the positions of Mg2+ ions were transposed from the active site of the Schizosaccharomyces pombe orthologue Pop2 (PDB 2P51) [9]. The preparation of PDBQT files for the protein was done via UCSF Chimera [47], which was used to assign polar hydrogens and Kollman charges. The grid size was set to 25 × 25 × 25 Å3 centred around the active site Mg2+ ion coordinated by Asp40, Glu42 and Asp230 (coordinates 136, 10, 270). Ligands were built in UCSF Chimera. Following energy minimisation, partial charges were calculated using the Gasteiger method. After docking, poses with a root mean square difference (RMSD) of less than 1.0 Å were clustered and represented by the pose with the most favourable free energy of binding. Ligand poses were analysed using UCSF Chimera and the PoseEdit algorithm accessed through the Proteins Plus portal [48,49]. The receptor-ligand poses were visualised using PyMol [50].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/1420-3049/29/18/4351/s1. References [9,12,45,48,51,52,53] are citied in the Supplementary Materials.
Author Contributions
Conceptualization, G.P.J., P.M.F. and G.S.W.; validation, I.K.; formal analysis, I.K. and G.P.J.; investigation, I.K.; resources, I.K. and G.P.J.; writing—original draft preparation, G.S.W.; writing—review and editing, G.P.J. and G.S.W.; visualization, G.P.J.; supervision, P.M.F. and G.S.W.; funding acquisition, P.M.F. and G.S.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Medical Research Council [grant G1100205].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We gratefully acknowledge the contributions of Ahmad Atieh, who was supported by a Tor Vergata University (Italy)-University of Nottingham (UK) summer exchange programme.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bianchin, C.; Mauxion, F.; Sentis, S.; Seraphin, B.; Corbo, L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA 2005, 11, 487–494. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goldstrohm, A.C.; Wickens, M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008, 9, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Wahle, E.; Winkler, G.S. RNA decay machines: Deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim. Biophys. Acta 2013, 1829, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Raisch, T.; Valkov, E. Regulation of the multisubunit CCR4-NOT deadenylase in the initiation of mRNA degradation. Curr. Opin. Struct. Biol. 2022, 77, 102460. [Google Scholar] [CrossRef]
- Krempl, C.; Lazzaretti, D.; Sprangers, R. A structural biology view on the enzymes involved in eukaryotic mRNA turnover. Biol. Chem. 2023, 404, 1101–1121. [Google Scholar] [CrossRef]
- Yang, W. Nucleases: Diversity of structure, function and mechanism. Q. Rev. Biophys. 2011, 44, 1–93. [Google Scholar] [CrossRef]
- Zuo, Y.; Deutscher, M.P. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001, 29, 1017–1026. [Google Scholar] [CrossRef]
- Horiuchi, M.; Takeuchi, K.; Noda, N.; Muroya, N.; Suzuki, T.; Nakamura, T.; Kawamura-Tsuzuku, J.; Takahasi, K.; Yamamoto, T.; Inagaki, F. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J. Biol. Chem. 2009, 284, 13244–13255. [Google Scholar] [CrossRef]
- Jonstrup, A.T.; Andersen, K.R.; Van, L.B.; Brodersen, D.E. The 1.4-A crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acids Res. 2007, 35, 3153–3164. [Google Scholar] [CrossRef]
- Basquin, J.; Roudko, V.V.; Rode, M.; Basquin, C.; Seraphin, B.; Conti, E. Architecture of the nuclease module of the yeast Ccr4-not complex: The Not1-Caf1-Ccr4 interaction. Mol. Cell 2012, 48, 207–218. [Google Scholar] [CrossRef]
- Petit, A.P.; Wohlbold, L.; Bawankar, P.; Huntzinger, E.; Schmidt, S.; Izaurralde, E.; Weichenrieder, O. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex. Nucleic Acids Res. 2012, 40, 11058–11072. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Pavanello, L.; Potapov, A.; Bartlam, M.; Winkler, G.S. Structure of the human Ccr4-Not nuclease module using X-ray crystallography and electron paramagnetic resonance spectroscopy distance measurements. Protein Sci. 2022, 31, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Khazina, E.; Izaurralde, E.; Weichenrieder, O. Crystal structure and functional properties of the human CCR4-CAF1 deadenylase complex. Nucleic Acids Res. 2021, 49, 6489–6510. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.; Valencia-Sanchez, M.A.; Staples, R.R.; Chen, J.; Denis, C.L.; Parker, R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 2001, 104, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.; Staples, R.R.; Valencia-Sanchez, M.A.; Muhlrad, D.; Parker, R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. Embo J. 2002, 21, 1427–1436. [Google Scholar] [CrossRef]
- Chen, J.; Chiang, Y.C.; Denis, C.L. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. Embo J. 2002, 21, 1414–1426. [Google Scholar] [CrossRef]
- Temme, C.; Zaessinger, S.; Meyer, S.; Simonelig, M.; Wahle, E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. Embo J. 2004, 23, 2862–2871. [Google Scholar] [CrossRef]
- Yamashita, A.; Chang, T.C.; Yamashita, Y.; Zhu, W.; Zhong, Z.; Chen, C.Y.; Shyu, A.B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005, 12, 1054–1063. [Google Scholar] [CrossRef]
- Dupressoir, A.; Morel, A.P.; Barbot, W.; Loireau, M.P.; Corbo, L.; Heidmann, T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genom. 2001, 2, 9. [Google Scholar] [CrossRef]
- Airhihen, B.; Pavanello, L.; Jadhav, G.P.; Fischer, P.M.; Winkler, G.S. 1-Hydroxy-xanthine derivatives inhibit the human Caf1 nuclease and Caf1-containing nuclease complexes via Mg2+-dependent binding. FEBS Open Bio 2019, 9, 717–727. [Google Scholar] [CrossRef]
- Maryati, M.; Airhihen, B.; Winkler, G.S. The enzyme activities of Caf1 and Ccr4 are both required for deadenylation by the human Ccr4-Not nuclease module. Biochem. J. 2015, 469, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Pekovic, F.; Rammelt, C.; Kubikova, J.; Metz, J.; Jeske, M.; Wahle, E. RNA binding proteins Smaug and Cup induce CCR4-NOT-dependent deadenylation of the nanos mRNA in a reconstituted system. Nucleic Acids Res. 2023, 51, 3950–3970. [Google Scholar] [CrossRef] [PubMed]
- Niinuma, S.; Fukaya, T.; Tomari, Y. CCR4 and CAF1 deadenylases have an intrinsic activity to remove the post-poly(A) sequence. RNA 2016, 22, 1550–1559. [Google Scholar] [CrossRef] [PubMed]
- Raisch, T.; Chang, C.T.; Levdansky, Y.; Muthukumar, S.; Raunser, S.; Valkov, E. Reconstitution of recombinant human CCR4-NOT reveals molecular insights into regulated deadenylation. Nat. Commun. 2019, 10, 3173. [Google Scholar] [CrossRef]
- Webster, M.W.; Chen, Y.H.; Stowell, J.A.W.; Alhusaini, N.; Sweet, T.; Graveley, B.R.; Coller, J.; Passmore, L.A. mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol. Cell 2018, 70, 1089–1100 e1088. [Google Scholar] [CrossRef]
- Yi, H.; Park, J.; Ha, M.; Lim, J.; Chang, H.; Kim, V.N. PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay. Mol. Cell 2018, 70, 1081–1088 e1085. [Google Scholar] [CrossRef]
- Aslam, A.; Mittal, S.; Koch, F.; Andrau, J.C.; Winkler, G.S. The Ccr4-Not Deadenylase Subunits CNOT7 and CNOT8 Have Overlapping Roles and Modulate Cell Proliferation. Mol. Biol. Cell 2009, 20, 3840–3850. [Google Scholar] [CrossRef]
- Mostafa, D.; Takahashi, A.; Yanagiya, A.; Yamaguchi, T.; Abe, T.; Kureha, T.; Kuba, K.; Kanegae, Y.; Furuta, Y.; Yamamoto, T.; et al. Essential functions of the CNOT7/8 catalytic subunits of the CCR4-NOT complex in mRNA regulation and cell viability. RNA Biol. 2020, 17, 403–416. [Google Scholar] [CrossRef]
- Stoney, P.N.; Yanagiya, A.; Nishijima, S.; Yamamoto, T. CNOT7 Outcompetes Its Paralog CNOT8 for Integration into The CCR4-NOT Complex. J. Mol. Biol. 2022, 434, 167523. [Google Scholar] [CrossRef]
- Shirai, Y.T.; Suzuki, T.; Morita, M.; Takahashi, A.; Yamamoto, T. Multifunctional roles of the mammalian CCR4-NOT complex in physiological phenomena. Front. Genet. 2014, 5, 286. [Google Scholar] [CrossRef]
- Song, X.H.; Liao, X.Y.; Zheng, X.Y.; Liu, J.Q.; Zhang, Z.W.; Zhang, L.N.; Yan, Y.B. Human Ccr4 and Caf1 Deadenylases Regulate Proliferation and Tumorigenicity of Human Gastric Cancer Cells via Modulating Cell Cycle Progression. Cancers 2021, 13, 834. [Google Scholar] [CrossRef] [PubMed]
- Washio-Oikawa, K.; Nakamura, T.; Usui, M.; Yoneda, M.; Ezura, Y.; Ishikawa, I.; Nakashima, K.; Noda, T.; Yamamoto, T.; Noda, M. Cnot7-null mice exhibit high bone mass phenotype and modulation of BMP actions. J. Bone Miner. Res. 2007, 22, 1217–1223. [Google Scholar] [CrossRef]
- Washio-Oikawa, K.; Nakamura, T.; Usui, M.; Yoneda, M.; Ezura, Y.; Ishikawa, I.; Nakashima, K.; Yamamoto, T.; Noda, M. Expression analysis of LacZ gene placed in the locus of Cnot7 exhibits its activity in osteoblasts in vivo and in mineralized nodules in vitro. J. Cell Biochem. 2006, 99, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Levy, R.; Mott, R.F.; Iraqi, F.A.; Gabet, Y. Collaborative cross mice in a genetic association study reveal new candidate genes for bone microarchitecture. BMC Genom. 2015, 16, 1013. [Google Scholar] [CrossRef] [PubMed]
- Faraji, F.; Hu, Y.; Wu, G.; Goldberger, N.E.; Walker, R.C.; Zhang, J.; Hunter, K.W. An integrated systems genetics screen reveals the transcriptional structure of inherited predisposition to metastatic disease. Genome Res. 2014, 24, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Faraji, F.; Hu, Y.; Yang, H.H.; Lee, M.P.; Winkler, G.S.; Hafner, M.; Hunter, K.W. Post-transcriptional Control of Tumor Cell Autonomous Metastatic Potential by CCR4-NOT Deadenylase CNOT7. PLoS Genet. 2016, 12, e1005820. [Google Scholar] [CrossRef]
- Maryati, M.; Kaur, I.; Gopal, J.; Olotu-Umoren, L.; Oveh, B.; Hashmi, L.; Fischer, P.M.; Winkler, G.S. A fluorescence-based assay suitable for quantitative analysis of deadenylase enzyme activity. Nucleic Acids Res. 2014, 42, e30. [Google Scholar] [CrossRef]
- Jadhav, G.P.; Kaur, I.; Maryati, M.; Airhihen, B.; Fischer, P.M.; Winkler, G.S. Discovery, synthesis and biochemical profiling of purine-2,6-dione derivatives as inhibitors of the human poly(A)-selective ribonuclease Caf1. Bioorganic Med. Chem. Lett. 2015, 25, 4219–4224. [Google Scholar] [CrossRef]
- Exell, J.C.; Thompson, M.J.; Finger, L.D.; Shaw, S.J.; Debreczeni, J.; Ward, T.A.; McWhirter, C.; Sioberg, C.L.; Martinez Molina, D.; Abbott, W.M.; et al. Cellularly active N-hydroxyurea FEN1 inhibitors block substrate entry to the active site. Nat. Chem. Biol. 2016, 12, 815–821. [Google Scholar] [CrossRef]
- Klumpp, K.; Hang, J.Q.; Rajendran, S.; Yang, Y.; Derosier, A.; Wong Kai In, P.; Overton, H.; Parkes, K.E.; Cammack, N.; Martin, J.A. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 2003, 31, 6852–6859. [Google Scholar] [CrossRef]
- Parkes, K.E.; Ermert, P.; Fassler, J.; Ives, J.; Martin, J.A.; Merrett, J.H.; Obrecht, D.; Williams, G.; Klumpp, K. Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J. Med. Chem. 2003, 46, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Tumey, L.N.; Bom, D.; Huck, B.; Gleason, E.; Wang, J.; Silver, D.; Brunden, K.; Boozer, S.; Rundlett, S.; Sherf, B.; et al. The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorganic Med. Chem. Lett. 2005, 15, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.T.L.; Stowell, J.A.W.; Hill, C.H.; Passmore, L.A. The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases. Nat. Struct. Mol. Biol. 2019, 26, 433–442. [Google Scholar] [CrossRef]
- Airhihen, B.; Pavanello, L.; Maryati, M.; Winkler, G.S. Quantitative Biochemical Analysis of Deadenylase Enzymes Using Fluorescence and Chemiluminescence-Based Assays. Methods Mol. Biol. 2024, 2723, 55–68. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Diedrich, K.; Krause, B.; Berg, O.; Rarey, M. PoseEdit: Enhanced ligand binding mode communication by interactive 2D diagrams. J. Comput. Aided Mol. Des. 2023, 37, 491–503. [Google Scholar] [CrossRef]
- Schoning-Stierand, K.; Diedrich, K.; Fahrrolfes, R.; Flachsenberg, F.; Meyder, A.; Nittinger, E.; Steinegger, R.; Rarey, M. ProteinsPlus: Interactive analysis of protein-ligand binding interfaces. Nucleic Acids Res. 2020, 48, W48–W53. [Google Scholar] [CrossRef]
- Schrödinger, L.L.C. The PyMol Molecular Graphics System; version 2.5.0 (open source); De-Lano Scientific: San Carlos, CA, USA, 2021. [Google Scholar]
- SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [CrossRef]
- iLOGP: A simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [CrossRef]
- A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).