Metabolic and Pharmacokinetic Profiling Studies of N, N-Dimethylaniline-Heliamine in Rats by UHPLC-Q-Orbitrap MS/MS
Abstract
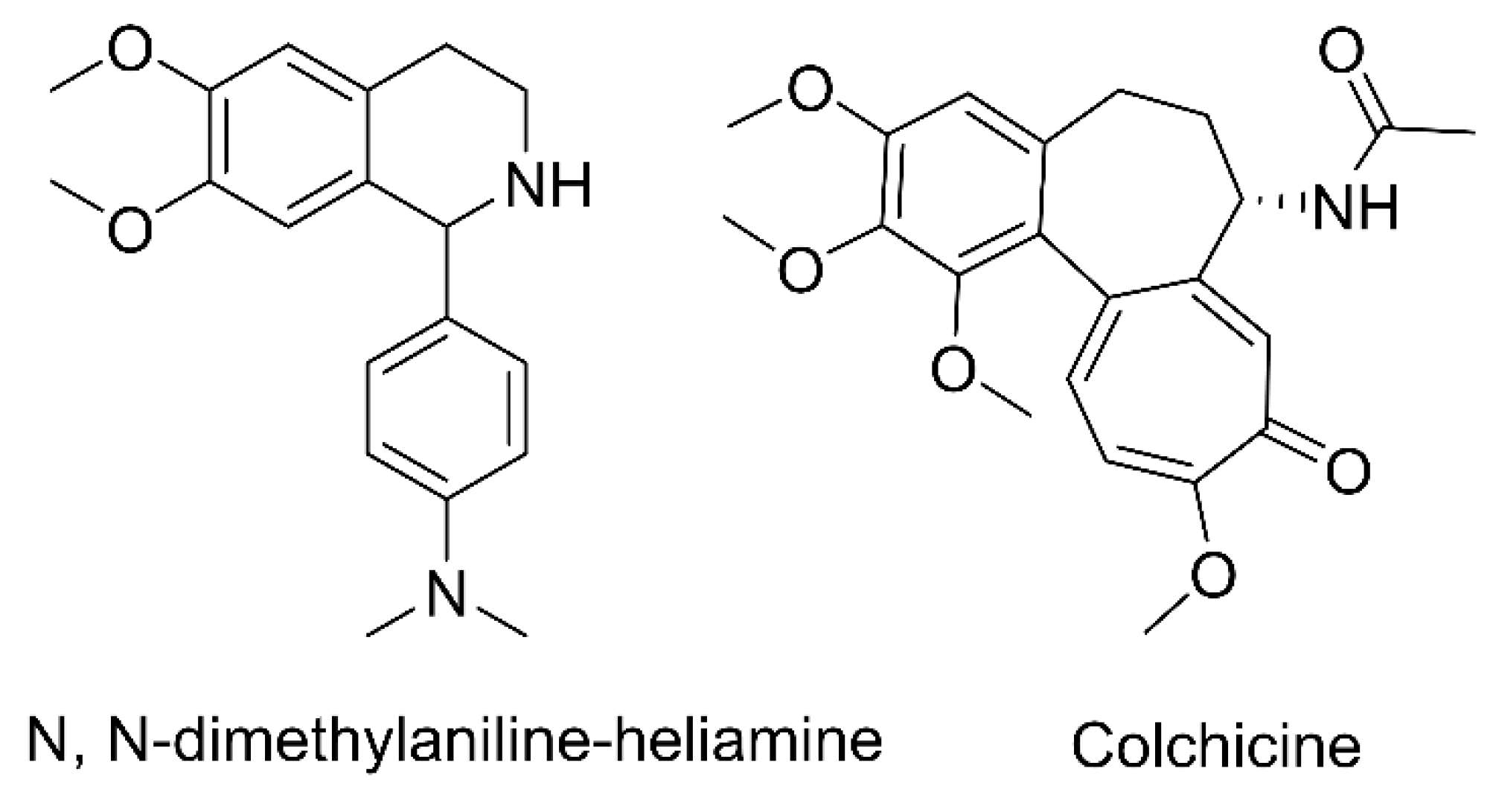
1. Introduction
2. Results and Discussion
2.1. UHPLC-MS/MS Condition
2.2. Metabolites of DH
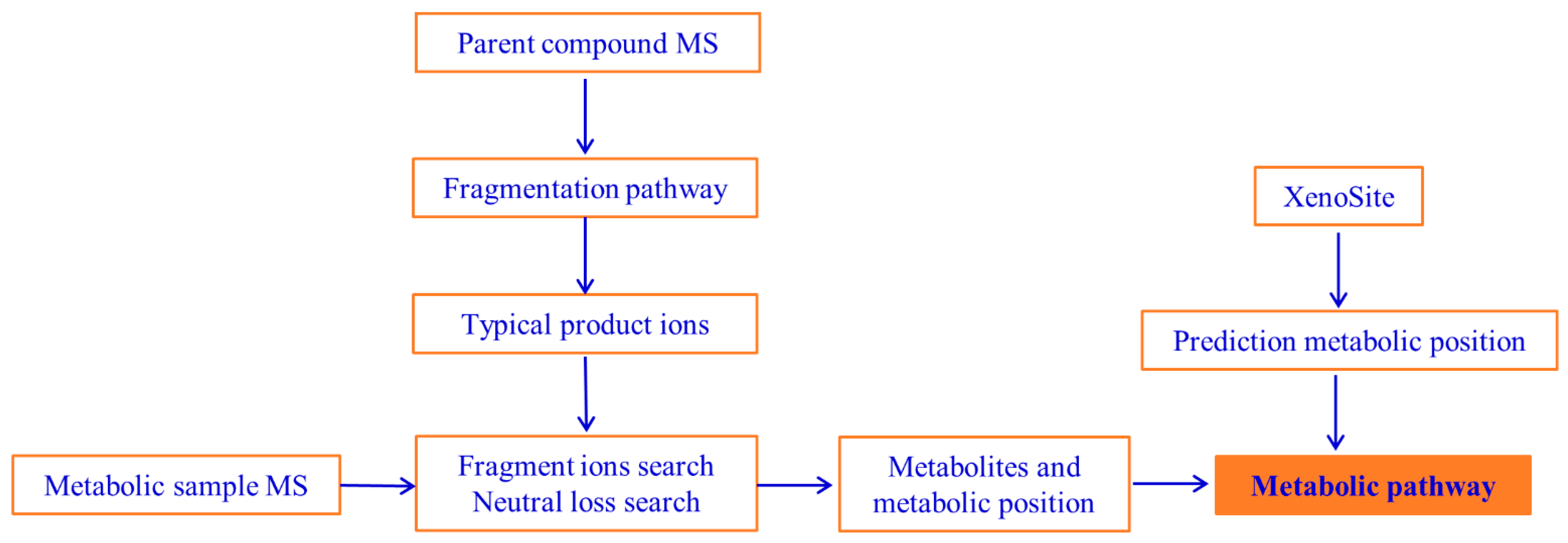
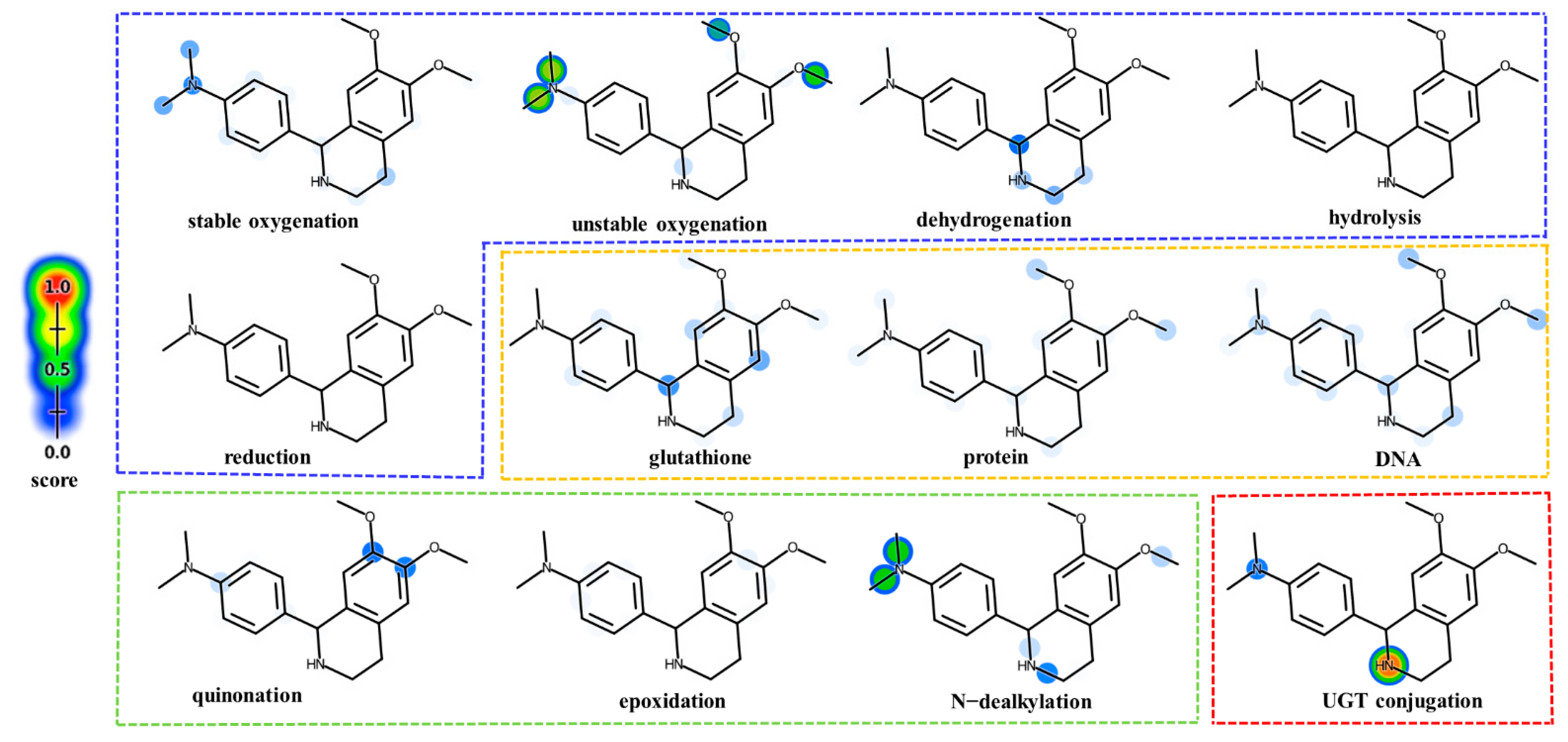
2.2.1. Metabolic Site Prediction of DH
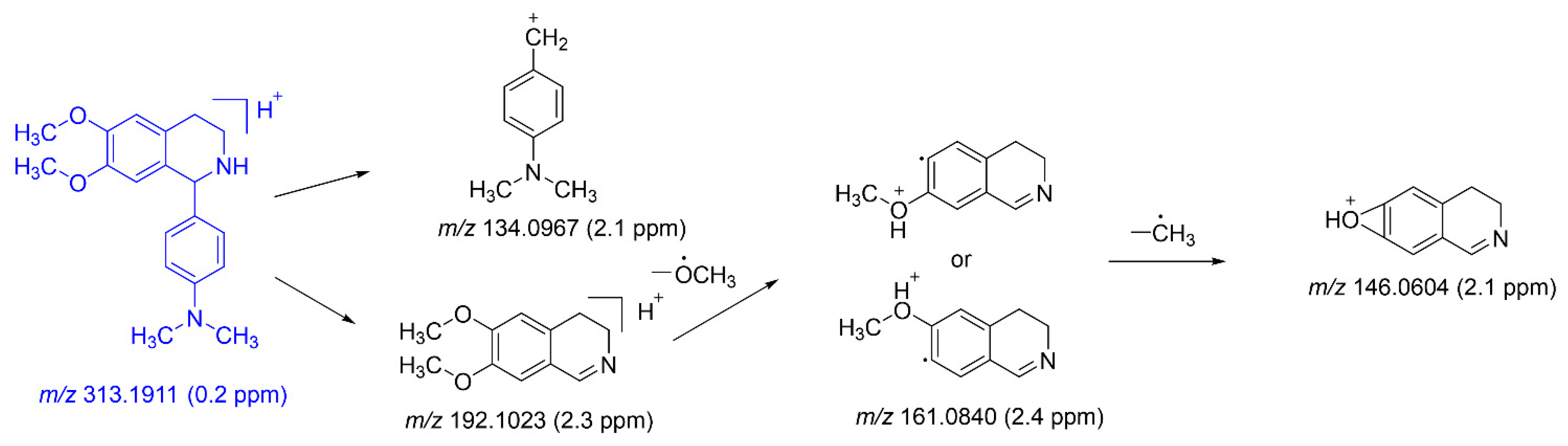
2.2.2. Mass Fragmentation of DH
2.2.3. Metabolites and Pathways of DH
2.3. UHPLC-MS/MS Method Validation
2.3.1. Selectivity
2.3.2. Calibration Curves
2.3.3. Accuracy and Precision
2.3.4. Recovery and Matrix Effect
2.3.5. Stability
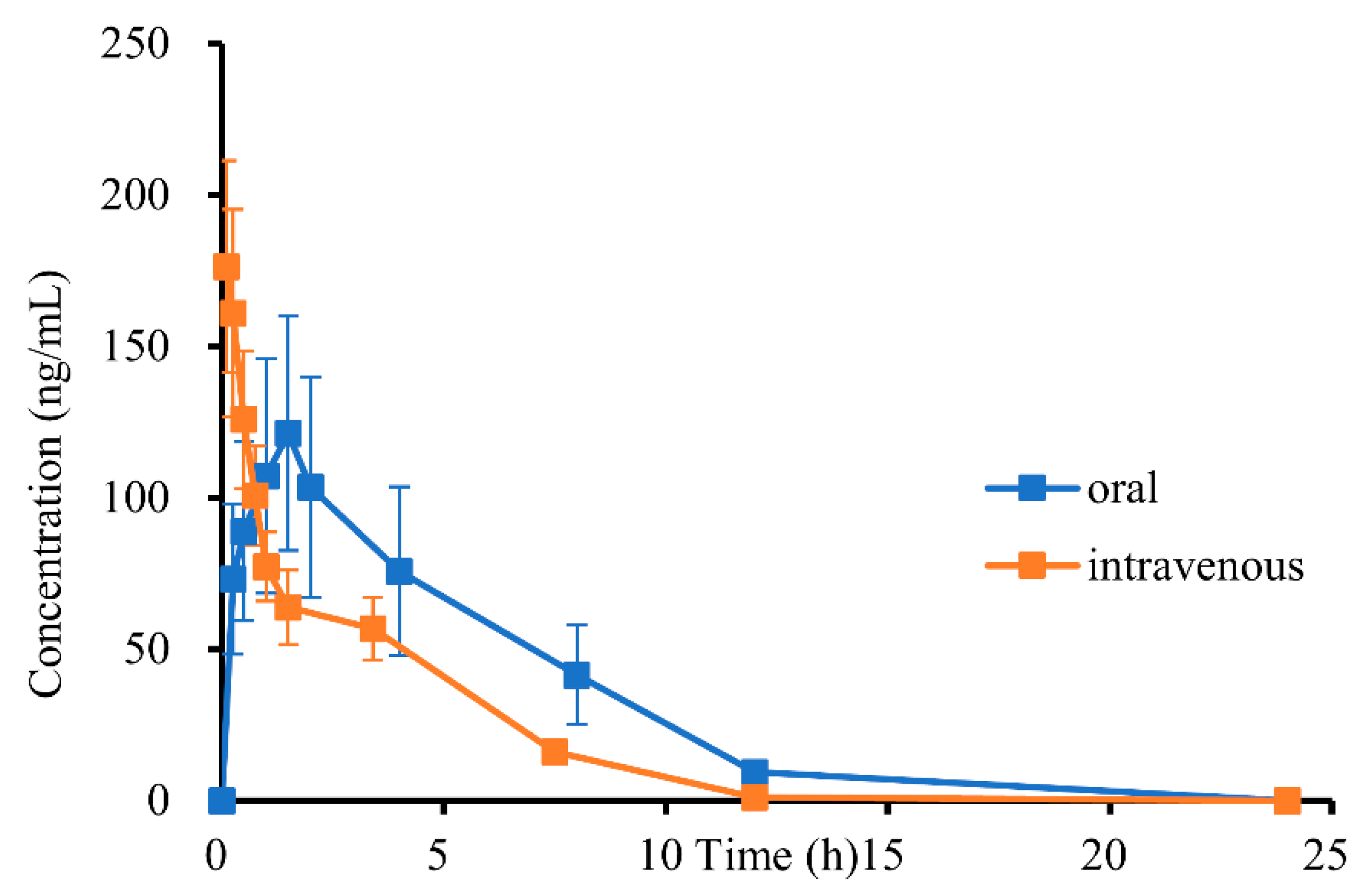
2.4. Pharmacokinetic (PK) Analysis and PK Parameter
3. Materials and Methods
3.1. Reagents and Chemicals
3.2. Animals, Dosing, and Sample Collection
3.3. UHPLC-MS/MS Conditions
3.4. Preparation of Rat Plasma Solutions
3.4.1. Preparation of Calibration Standards and Internal Standard Solutions
3.4.2. Preparation of Quality Control (QC) Solutions
3.4.3. Pharmacokinetic (PK) Sample Preparation
3.5. Metabolic Study
3.5.1. In Silico Metabolism Prediction
3.5.2. In Vitro and In Vivo Experiments
3.6. Method Validation
3.7. Pharmacokinetic (PK) Study
3.8. Data Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sangster, J.J.; Ruscoe, R.E.; Cosgrove, S.C.; Mangas-Sanchez, J.; Turner, N.J. One-pot chemoenzymatic cascade for the enantioselective C(1)-allylation of tetrahydroisoquinolines. J. Am. Chem. Soc. 2023, 145, 4431–4437. [Google Scholar] [CrossRef] [PubMed]
- Gitto, R.; Barreca, M.L.; De Luca, L.; De Sarro, G.; Ferreri, G.; Quartarone, S.; Russo, E.; Constanti, A.; Chimirri, A. Discovery of a novel and highly potent noncompetitive AMPA receptor antagonist. J. Med. Chem. 2003, 46, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M. Recent studies on chemical constituents of Ophiorrhiza plants. J. Nat. Med. 2022, 76, 748–755. [Google Scholar] [CrossRef]
- Pang, S.; Zhang, Y.; Luo, J.; Tang, M.; Li, Q.; Chen, Y.; Liu, L. Research progress on ion channel mechanism of traditional Chinese medicine monomers inhibiting arrhythmia. Chin. Tradit. Herb. Drugs 2022, 53, 4853–4861. [Google Scholar]
- Al-Khatib, S.M.; Stevenson, W.G.; Ackerman, M.J.; Bryant, W.J.; Callans, D.J.; Curtis, A.B.; Deal, B.J.; Dickfeld, T.; Field, M.E.; Fonarow, G.C.; et al. Correction to: 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: A peport of the american college of cardiology/American heart association task force on clinical practice guidelines and the heart rhythm society. J. Am. Coll. Cardiol. 2018, 138, e415–e418. [Google Scholar]
- Fan, J.; Zhang, K.; Jin, Y.; Li, B.J.N.; Gao, S.H.; Zhu, J.M.; Cui, R.J. Pharmacological effects of berberine on mood disorders. J. Cell. Mol. Med. 2019, 23, 21–28. [Google Scholar] [CrossRef]
- Han, B.J.; Cao, G.Y.; Jia, L.Y.; Zheng, G.; Zhang, L.; Sheng, P.; Xie, J.Z.; Zhang, C.F. Cardioprotective effects of tetrahydropalmatine on acute myocardial infarction in rats. Am. J. Chin. Med. 2022, 50, 1887–1904. [Google Scholar] [CrossRef]
- Jumadilla, R.; Hilola, R.; Aziz, A. Toxicity and anti-arrhythmic activity 1- (4-dimethylaminophenyl) -6, 7-dimethoxy-1,2,3,4 tetrahydroisoquinoline. Am. J. Med. Sci. 2020, 02, 130–133. [Google Scholar] [CrossRef]
- Jumayev, I.; Usmanov, P.; Rustamov, S.; Zhurakulov, S. Comparative inotropic effects of the some isoquinoline alkaloids. Biomed. Biomed. Pharmacol. J. 2020, 13, 325–333. [Google Scholar] [CrossRef]
- Cardiovascular Disease. Available online: https://www.who.int/health-topics/cardiovascular-diseases (accessed on 28 July 2024).
- Lei, M.; Wu, L.; Terrar, D.A.; Huang, C.L.H. Modernized Classification of Cardiac Antiarrhythmic Drugs. Circulation 2018, 138, 1879–1896. [Google Scholar] [CrossRef]
- Markman, T.M.; Nazarian, S. Arrhythmia and electrophysiological effects of chemotherapy: A review. Oncology 2016, 91, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Hindricks, G.; Potpara, T.; Dagres, N.; Arbelo, E.; Bax, J.J.; Blomstrom-Lundqvist, C.; Boriani, G.; Castella, M.; Dan, G.-A.; Dilaveris, P.E.; et al. 2020 ESC guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS): The task force for the diagnosis and management of atrial fibrillation of the European Society of Cardiology (ESC) Developed with the special contribution of the European Heart Rhythm Association (EHRA) of the ESC. Eur. Heart J. 2021, 42, 373–498. [Google Scholar] [PubMed]
- Wu, S.J.; Fan, Y.F.; Chien, C.Y. Surgical strategies for cardiac perforation after catheter ablation or electrophysiology study a systemic review. Int. Heart J. 2021, 62, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, C.D.; Koneru, J.N.; Gunderson, B.; Kroll, M.W.; Ploux, S.; Ellenbogen, K.A. Impedance in the diagnosis of lead malfunction. Circ. Arrhythmia Electrophysiol. 2020, 13, e008092. [Google Scholar] [CrossRef]
- Li, D.L.; Cox, Z.L.; Richardson, T.D.; Kanagasundram, A.N.; Saavedra, P.J.; Shen, S.T.; Montgomery, J.A.; Murray, K.T.; Roden, D.M.; Stevenson, W.G. Quinidine in the management of recurrent ventricular arrhythmias. JACC-Clin. Electrophysiol. 2021, 7, 1254–1263. [Google Scholar] [CrossRef]
- Ito, N.; Tashiro, T.; Morishige, N.; Nishimi, M.; Hayashida, Y.; Takeuchi, K.; Minematsu, N.; Kuwahara, G.; Sukehiro, Y. Efficacy of propafenone hydrochloride in preventing postoperative atrial aibrillation after coronary artery bypass grafting. Heart Surg. Forum 2010, 13, E223–E227. [Google Scholar] [CrossRef]
- Liu, B.S.; Zhang, R.; Zhang, A.Y.; Wang, G.D.; Xu, J.P.; Zhang, Y.; Liu, Y.P.; Hao, P.P. Effectiveness and safety of four different beta-blockers in patients with chronic heart failure. MedComm 2023, 4, e199. [Google Scholar] [CrossRef]
- Hirose, S.; Makiyama, T.; Melgari, D.; Yamamoto, Y.; Wuriyanghai, Y.; Yokoi, F.; Nishiuchi, S.; Harita, T.; Hayano, M.; Kohjitani, H.; et al. Propranolol attenuates late sodium current in a long QT syndrome type 3-human induced pluripotent stem cell model. Front. Cell. Dev. Biol. 2020, 8, 761. [Google Scholar] [CrossRef]
- Adams, M.J.; Ayers, M.D.; Kean, A.C. The successful use of verapamil in infants with fascicular ventricular tachycardia. Prog. Pediatr. Cardiol. 2022, 67, 101524. [Google Scholar] [CrossRef]
- Nicholson, J.; Czosnowski, Q.; Flack, T.; Pang, P.S.; Billups, K. Hemodynamic comparison of intravenous push diltiazem versus metoprolol for atrial fibrillation rate control. J. Emerg. Med. 2020, 38, 1879–1883. [Google Scholar] [CrossRef]
- Vaughanwilliams, E.M. Classification of antidysrhythmic drugs. Pharmacol. Ther. 1975, 1, 115–138. [Google Scholar]
- Rosen, M.R. The sicilian asmbit-a new approach to the classification of antiarrhythmic drugs based on their actions on arrhythmogenic mechanisms. The task force of the working group on arrhythmias of the European society of cardiology. Eur. Heart J. 1991, 12, 1112–1131. [Google Scholar]
- Huang, C.L.H.; Wu, L.; Jeevaratnam, K.; Lei, M. Update on antiarrhythmic drug pharmacology. J. Cardiovasc. Electrophysiol. 2020, 31, 579–592. [Google Scholar] [CrossRef] [PubMed]
- St-Onge, M. Cardiovascular drug toxicity. Crit. Care Clin. 2021, 37, 563–576. [Google Scholar] [CrossRef]
- Chen, J.C.; Li, P.L.; Zhang, T.Y.; Xu, Z.P.; Huang, X.W.; Wang, R.M.; Du, L.T. Review on strategies and technologies for exosome isolation and purification. Front. Bioeng. Biotechnol. 2022, 9, 35071216. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Zeki, O.C.; Eylem, C.C.; Recber, T.; Kir, S.; Nemutlu, E. Integration of GC-MS and LC-MS for untargeted metabolomics profiling. J. Pharm. Biomed. Anal. 2020, 190, 113509. [Google Scholar] [CrossRef]
- Dang, N.L.; Matlock, M.K.; Hughes, T.B.; Swamidass, S.J. The metabolic rainbow: Deep learning phase I metabolism in five colors. J. Chem. Inf. Model. 2020, 60, 1146–1164. [Google Scholar] [CrossRef]
- Zaretzki, J.; Matlock, M.; Swamidass, S.J. XenoSite: Accurately predicting CYP-mediated sites of metabolism with neural networks. J. Chem. Inf. Model. 2013, 53, 3373–3383. [Google Scholar] [CrossRef]
- Hughes, T.B.; Miller, G.P.; Swamidass, S.J. Modeling epoxidation of drug-like molecules with a deep machine learning network. ACS Cent. Sci. 2015, 1, 168–180. [Google Scholar] [CrossRef]
- Hughes, T.B.; Swamidass, S.J. Deep learning to predict the formation of quinone species in drug metabolism. Chem. Res. Toxicol. 2017, 30, 642–656. [Google Scholar] [CrossRef] [PubMed]
- Dang, N.L.; Hughes, T.B.; Miller, G.P.; Swamidass, S.J. Computationally assessing the bioactivation of drugs by N-dealkylation. Chem. Res. Toxicol. 2018, 31, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.; Morris, T. Pharmaceutical research. Pharm. Res. 1993, 10, 1093–1095. [Google Scholar] [CrossRef] [PubMed]
- Tonoli, D.; Varesio, E.; Hopfgartner, G. Liquid hromatography high-resolution mass spectrometric QUAL/QUAN approaches for drug metabolism and metabolomics. Chimia 2012, 66, 218–222. [Google Scholar] [CrossRef]
- Kaufmann, A. Combining UHPLC and high-resolution MS: A viable approach for the analysis of complex samples? TrAC Trends Anal. Chem. 2014, 63, 113–128. [Google Scholar] [CrossRef]
- Hughes, T.B.; Dang, N.L.; Miller, G.P.; Swamidass, S.J. Modeling reactivity to biological macromolecules with a deep multitask network. ACS Cent. Sci. 2016, 2, 529–537. [Google Scholar] [CrossRef]
- Hughes, T.B.; Miller, G.P.; Swamidass, S.J. Site of reactivity models predict molecular reactivity of diverse chemicals with glutathione. Chem. Res. Toxicol. 2015, 28, 797–809. [Google Scholar] [CrossRef]
- Dang, N.L.; Hughes, T.B.; Krishnamurthy, V.; Swamidass, S.J. A simple model predicts UGT-mediated metabolism. Bioinformatics 2016, 32, 3183–3189. [Google Scholar] [CrossRef]
- Xi, R.; Abdulla, R.; Zhang, M.; Sherzod, Z.; Ivanovna, V.V.; Habasi, M.; Liu, Y. Pharmacokinetic study and metabolite identification of 1-(3′-bromophenyl)-heliamine in rats. Pharmaceuticals 2022, 15, 1483. [Google Scholar] [CrossRef]
- Analytical Procedures and Methods Validation for Drugs and Bologics. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/analytical-procedures-and-methods-validation-drugs-and-biologics (accessed on 28 July 2024).
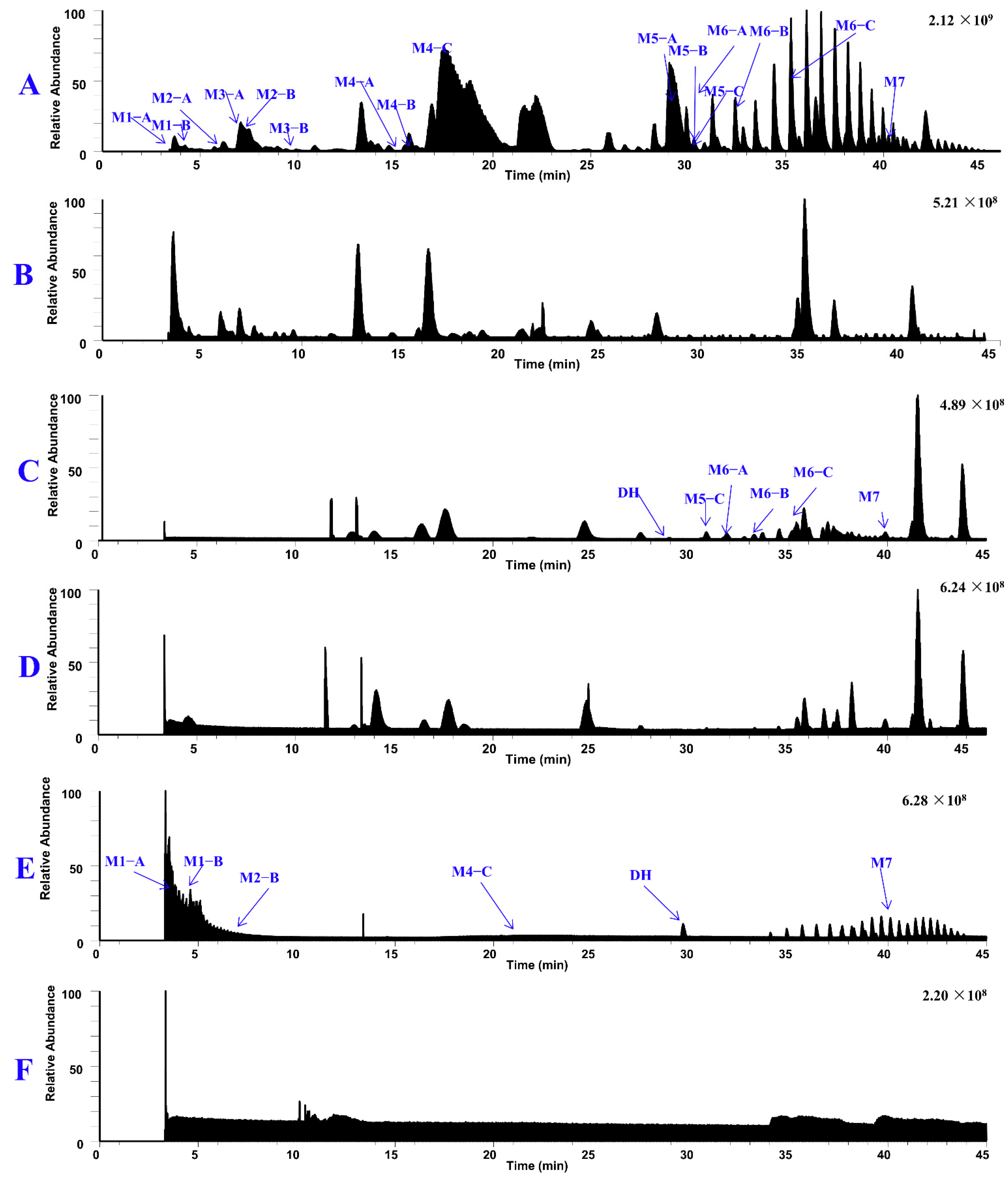
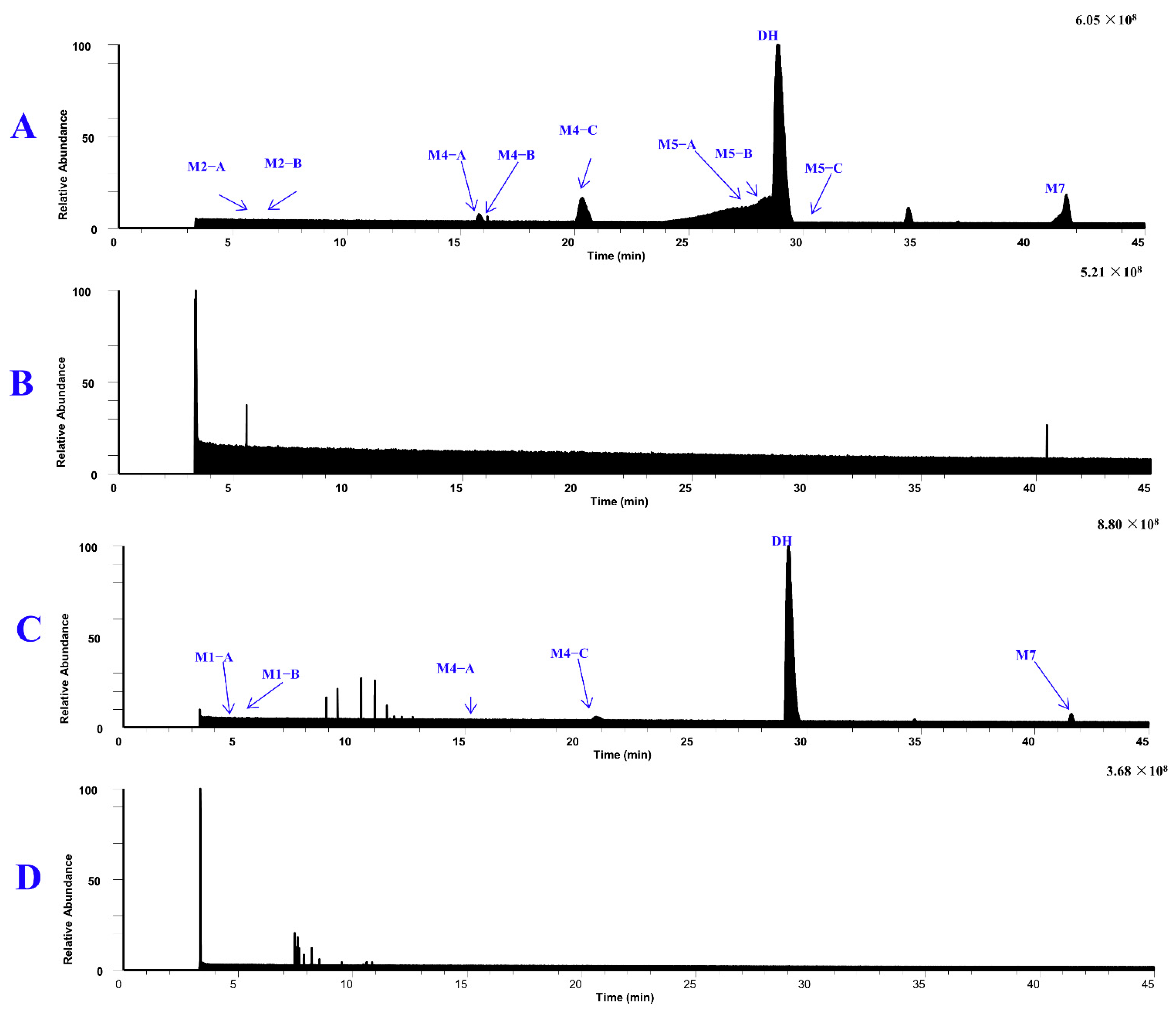
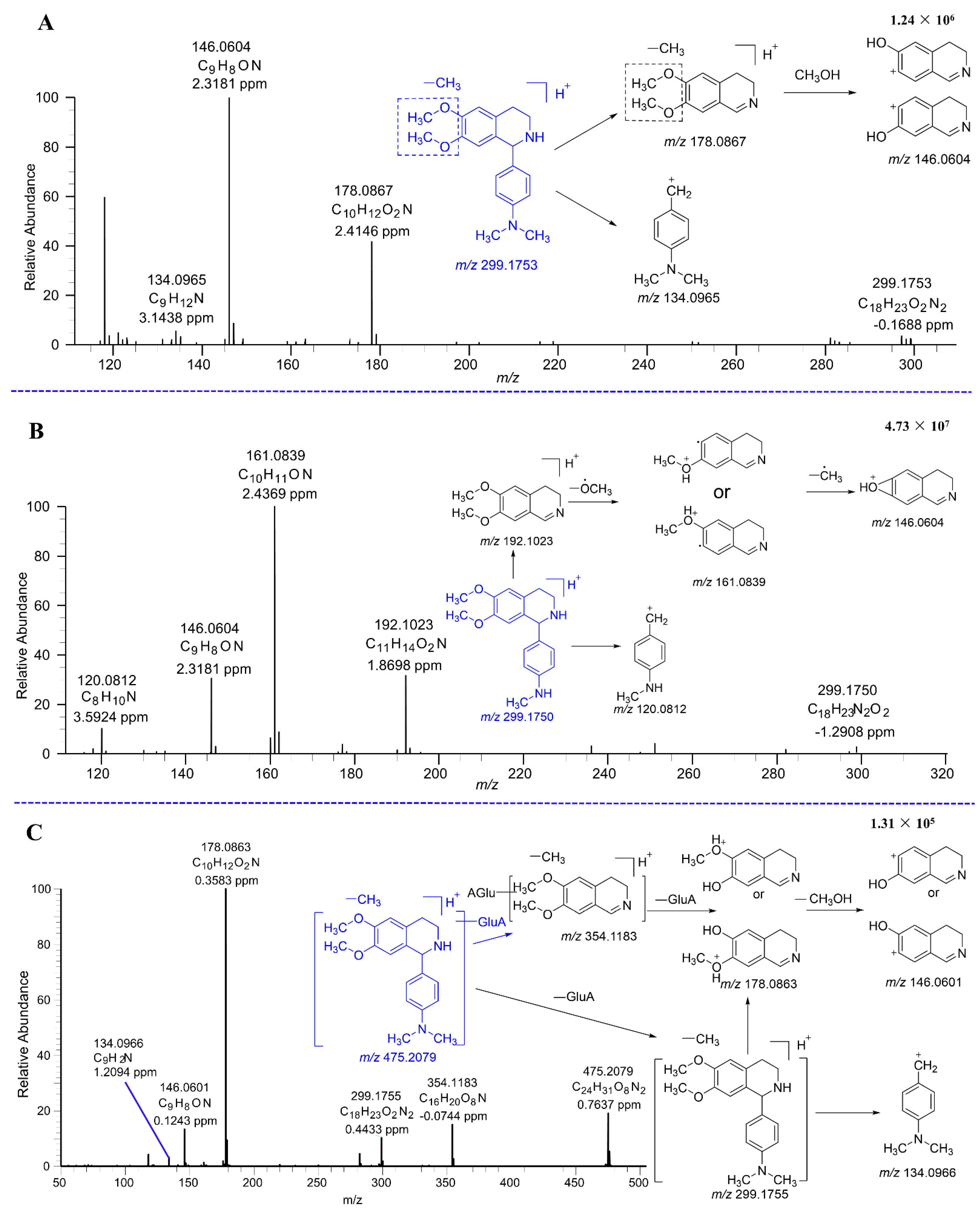
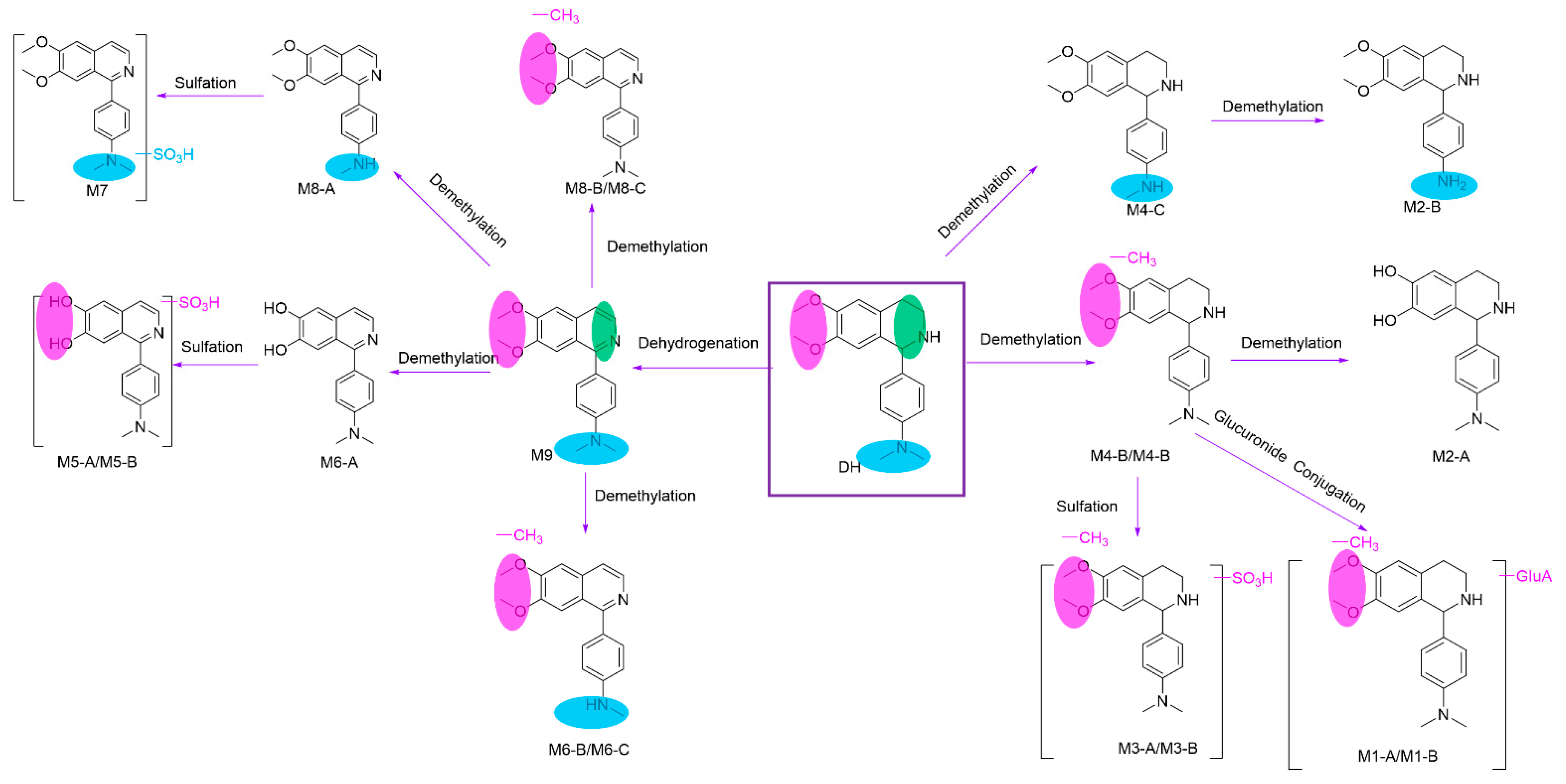
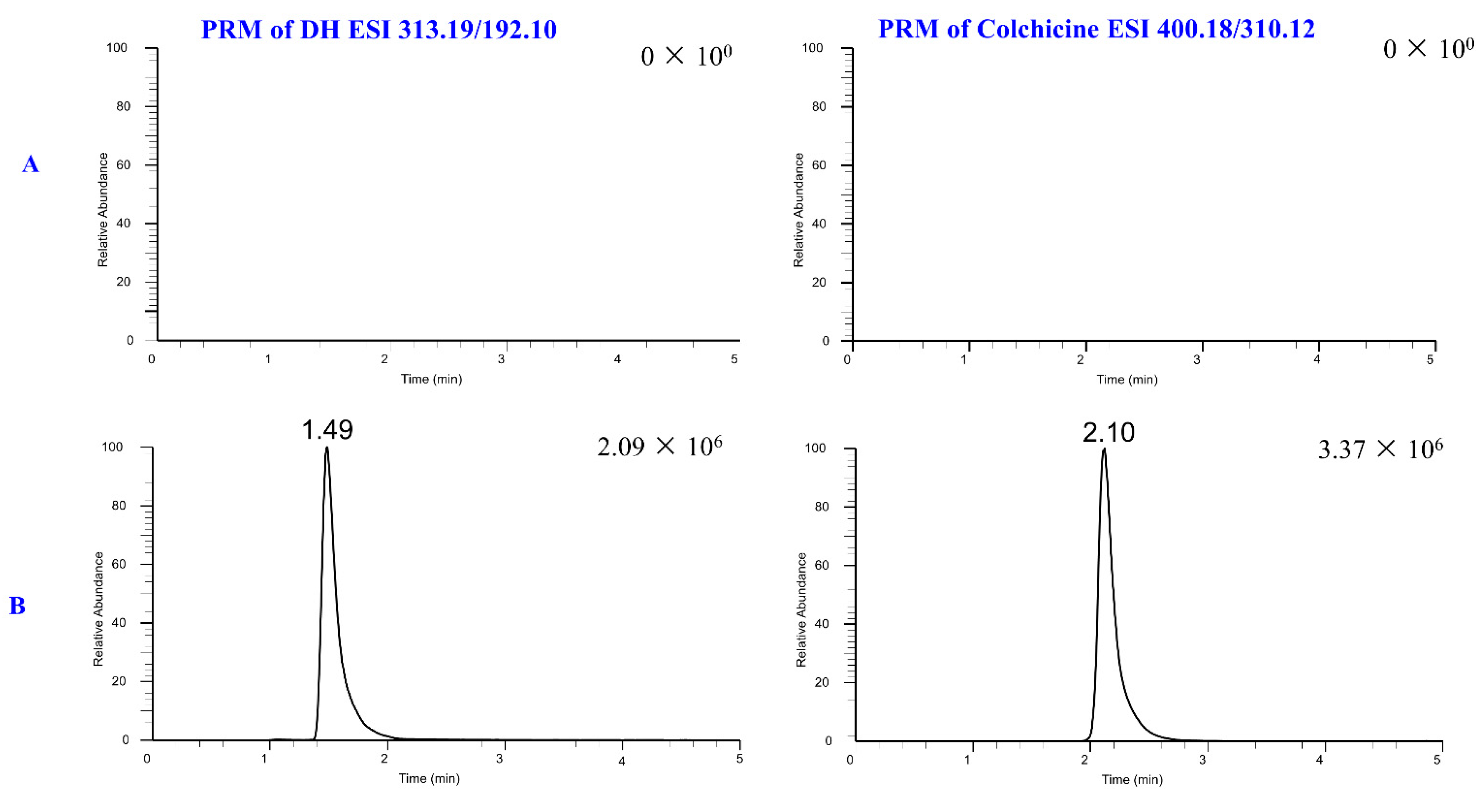

| Metabolites | tR (min) | Formula (M+H+) | Observed Mass (m/z) | Calculated Mass | Error (ppm) | Fragmention | Transformations | Rat and LM* |
|---|---|---|---|---|---|---|---|---|
| DH | 28.88 | C19H25N2O2 | 313.1911 | 313.1911 | 0.2 | 192.1023 (71), 161.0840 (100), 146.0604 (30), 134.0967 (8) | Parent | P*, U*, F*, LM* |
| M1–A | 3.65 | C24H31N2O8 | 475.2081 | 475.2075 | 1.2 | 354.1183 (44), 178.0864 (100), 146.0600 (48), 134.0966 (5) | Demethylation, Glucuronide Conjugation | P*, U*, LM* |
| M1–B | 4.83 | C24H31N2O8 | 475.2081 | 475.2075 | 1.2 | 354.1182 (36), 178.0865 (100), 146.0603 (55), 134.0960 (5) | Demethylation, Glucuronic Conjugation | P*, U*, LM* |
| M2–A | 5.61 | C17H21N2O2 | 285.1598 | 285.1598 | 0.2 | 283.1449 (5), 164.0710 (97), 146.0604 (100), 134.0964 (6), 122.0969 (54) | Demethylation | U*, LM* |
| M2–B | 6.93 | C17H21N2O2 | 285.1598 | 285.1598 | 0.3 | 192.1026 (67), 178.0866 (28), 161.0836 (100), 146.0635 (62), | Demethylation, | P*, U*, LM* |
| M3–A | 6.51 | C18H23N2O5S | 379.1316 | 379.1322 | −1.6 | 258.0431 (32), 178.0863 (96), 146.0601 (100), 122.0967 (15) | Demethylation, Sulfation | U* |
| M3–B | 9.84 | C18H23N2O5S | 379.1323 | 379.1323 | 0.2 | 258.0431 (24), 178.0864 (100), 146.0602 (98), 122.0967 (14) | Demethylation, Sulfation | U* |
| M4–A | 15.05 | C18H23N2O2 | 299.1753 | 299.1754 | −0.3 | 178.0867 (100), 146.0604 (98), 134.0965 (5) | Demethylation | U*, LM* |
| M4–B | 15.71 | C18H23N2O2 | 299.1754 | 299.1754 | −0.2 | 178.0866 (100), 146.0604 (90), 134.0969 (7) | Demethylation | U*, LM* |
| M4–C | 19.47 | C18H23N2O2 | 299.1750 | 299.1754 | −1.3 | 192.1023 (72), 161.0839 (100), 146.0604 (28), 120.0812 (13) | Demethylation | P*, U*, LM* |
| M5–A | 26.68 | C17H17N2O2 | 281.1283 | 281.1285 | −0.45 | 266.1046 (22), 238.1101 (24) | Dehydrogenation, Demethylation | U*, LM* |
| M5–B | 29.29 | C17H17N2O2 | 281.1288 | 281.1285 | 1.0 | 266.1054 (27), 238.1105 (23) | Dehydrogenation, Demethylation | U*, LM* |
| M5–C | 30.44 | C17H17N2O2 | 281.1289 | 281.1285 | 1.3 | 266.1054 (40), 238.1105 (39) | Dehydrogenation, Demethylation | U*, F*, LM* |
| M6–A | 30.99 | C18H19N2O2 | 295.1446 | 295.1441 | 1.5 | 280.1209 (22), 252.1283 (20) | Dehydrogenation, Demethylation | F* |
| M6–B | 34.13 | C18H19N2O2 | 295.1447 | 295.1441 | 1.2 | 280.1211 (20), 252.1263 (17) | Dehydrogenation, Demethylation | U*, F* |
| M6–C | 35.58 | C18H19N2O2 | 295.1440 | 295.1441 | −0.31 | 280.1202 (30), 252.1256 (25) | Dehydrogenation, Demethylation | U*, F* |
| M7 | 40.10 | C19H21N2O2 | 309.1602 | 309.1598 | 1.4 | 293.1288 (9), 265.1340 (16) | Dehydrogenation | P*, U*, F*, LM* |
| Analytes | Nominal Concentration (ng/mL) | Intra-Day Mean | RSD (%) | RE (%) | Inter-Day Mean | RSD (%) | RE (%) |
|---|---|---|---|---|---|---|---|
| DH | 1 | 1.08 | 7.41 | 8.00 | 1.02 | 8.16 | 2.00 |
| 10 | 9.51 | 6.03 | −4.90 | 9.93 | 7.84 | −0.70 | |
| 100 | 98.38 | 5.64 | −1.62 | 104.5 | 5.44 | 4.50 | |
| 500 | 497.00 | 3.91 | −0.60 | 540.82 | 2.49 | 8.00 |
| Analytes | Nominal Concentration (ng/mL) | Recovery | Matrix Effect | ||
|---|---|---|---|---|---|
| Mean (%) | RSD (%) | Mean (%) | RSD (%) | ||
| DH | 10 | 89.22 | 12.67 | 87.55 | 6.30 |
| 100 | 87.26 | 4.51 | 88.16 | 2.97 | |
| 500 | 85.43 | 0.97 | 84.42 | 1.85 | |
| IS | 300 | 86.95 | 1.32 | 85.47 | 4.75 |
| Analytes | Nominal Concentration (ng/mL) | Accuracy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DH | 4 h | 8 h | 12 h | 1 day | 7 days | 15 days | Stock solution | Room temper | Thaw | |
| 10 | 110.30 | 108.80 | 110.57 | 109.12 | 108.43 | 106.68 | 104.96 | 109.44 | 100.00 | |
| 100 | 106.65 | 106.69 | 103.81 | 103.58 | 106.23 | 101.13 | 99.49 | 101.36 | 104.56 | |
| 500 | 105.03 | 103.54 | 103.34 | 99.17 | 105.59 | 94.99 | 96.24 | 97.46 | 97.37 | |
| Parameters | Unit | P.O. (19.2 mg/kg) | I.V. (1.9 mg/kg) |
|---|---|---|---|
| AUC0–t * | h* (ng/mL) | 707.28 ± 264.72 | 439.08 ± 30.11 |
| AUC0–∞ * | h* (ng/mL) | 748.44 ± 278.98 | 494.64 ± 36.40 |
| MRT0–t * | h | 4.07 ± 0.34 | 2.49 ± 0.33 |
| MRT0–∞ * | h | 4.76 ± 0.40 | 3.56 ± 0.26 |
| Tmax * | h | 1.50 ± 0.00 | — |
| T1/2 * | h | 2.96 ± 0.32 | 2.79 ± 0.34 |
| Cmax * | ng/mL | 121.35 ± 42.41 | 183.37 ± 33.56 |
| CL * | mL/h/kg | 30,148.65 ± 15,354.27 | 3859.65 ± 301.53 |
| Vd * | mL/kg | 126,728.09 ± 56,867.09 | 15,514.58 ± 2159.74 |
| Fabs (%) * | 16.11 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, R.; Abdulla, R.; Sherzod, J.; Ivanovna, V.V.; Habasi, M.; Liu, Y. Metabolic and Pharmacokinetic Profiling Studies of N, N-Dimethylaniline-Heliamine in Rats by UHPLC-Q-Orbitrap MS/MS. Molecules 2024, 29, 4324. https://doi.org/10.3390/molecules29184324
Xi R, Abdulla R, Sherzod J, Ivanovna VV, Habasi M, Liu Y. Metabolic and Pharmacokinetic Profiling Studies of N, N-Dimethylaniline-Heliamine in Rats by UHPLC-Q-Orbitrap MS/MS. Molecules. 2024; 29(18):4324. https://doi.org/10.3390/molecules29184324
Chicago/Turabian StyleXi, Ruqi, Rahima Abdulla, Jurakulov Sherzod, Vinogradova Valentina Ivanovna, Maidina Habasi, and Yongqiang Liu. 2024. "Metabolic and Pharmacokinetic Profiling Studies of N, N-Dimethylaniline-Heliamine in Rats by UHPLC-Q-Orbitrap MS/MS" Molecules 29, no. 18: 4324. https://doi.org/10.3390/molecules29184324
APA StyleXi, R., Abdulla, R., Sherzod, J., Ivanovna, V. V., Habasi, M., & Liu, Y. (2024). Metabolic and Pharmacokinetic Profiling Studies of N, N-Dimethylaniline-Heliamine in Rats by UHPLC-Q-Orbitrap MS/MS. Molecules, 29(18), 4324. https://doi.org/10.3390/molecules29184324







