Synthesis, Structural Properties and Biological Activities of Novel Hydrazones of 2-, 3-, 4-Iodobenzoic Acid
Abstract
1. Introduction
2. Results
2.1. Chemistry
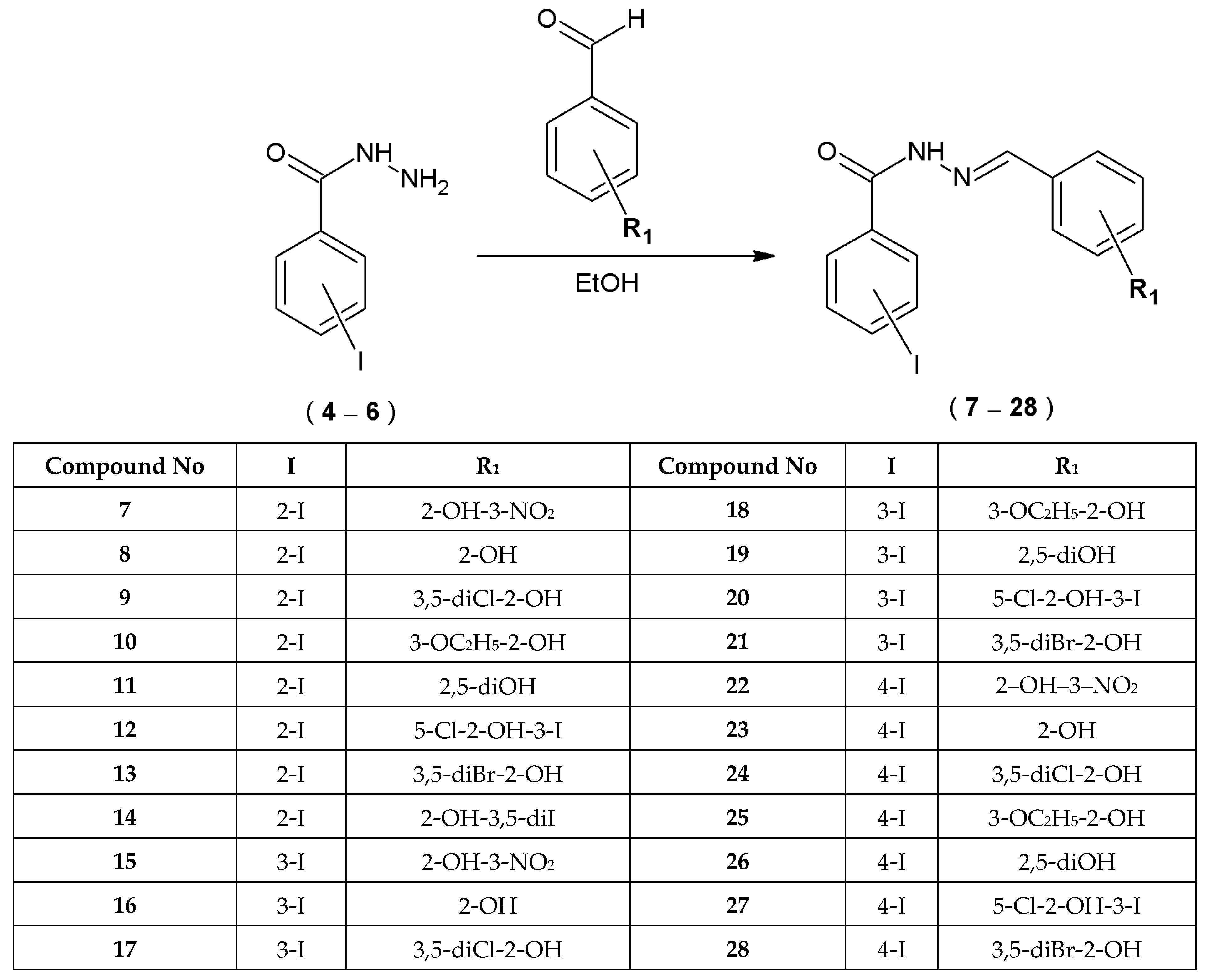
2.1.1. Synthesis and Chemical Characterization
2.1.2. Mechanochemical Preparation of Acylhydrazones
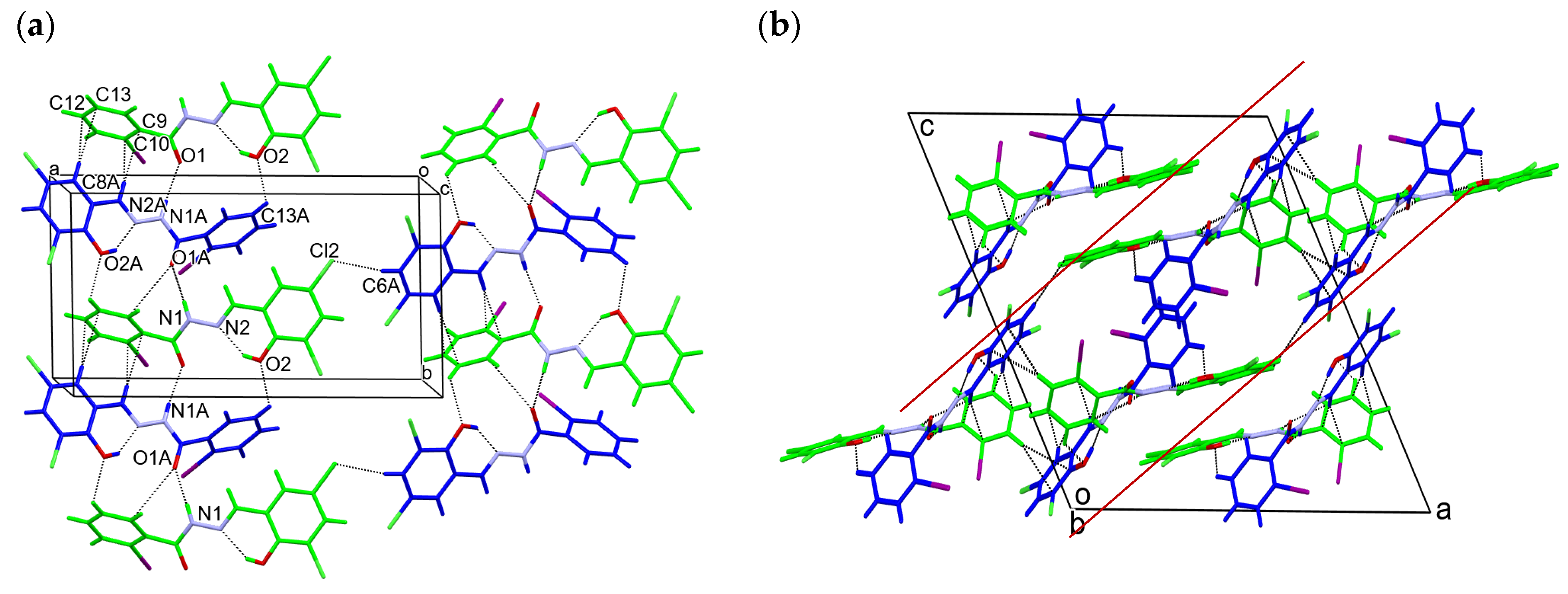
2.1.3. X-ray Diffraction Studies
2.2. Microbiology
2.3. Cytotoxicity Study
3. Discussion
3.1. Chemistry
3.2. Microbiology
3.3. Cytotoxicity Study
4. Materials and Methods
4.1. Chemistry
4.2. Synthesis and Chemical Characterization
4.2.1. Preparation of Hydrazides of 2-, 3-, or 4-Iodobenzoic Acid (4–6)
Physico-Chemical Data of Hydrazides of 2-, 3-, or 4-Iodobenzoic Acid (4–6)
- 2-iodobenzohydrazide (4)
- 3-iodobenzohydrazide (5)
- 4-iodobenzohydrazide (6)
4.2.2. Preparation of Novel 2-, 3-, or 4-Iodobenzoic Acid Hydrazide Derivatives—Acylhydrazones of 2-, 3-, or 4-Iodobenzoic Acid (7–28)
Physico-Chemical Data of Novel Acylhydrazones of 2-, 3-, or 4-Iodobenzoic Acid (7–28)
- N-[(2-hydroxy-3-nitrophenyl)methylidene]-2-iodobenzohydrazide (7)
- N-[(2-hydroxyphenyl)methylidene]-2-iodobenzohydrazide (8)
- N-[(3,5-dichloro-2-hydroxyphenyl)methylidene]-2-iodobenzohydrazide (9)
- N-[(3-ethoxy-2-hydroxyphenyl)methylidene]-2-iodobenzohydrazide (10)
- N-[(2,5-dihydroxyphenyl)methylidene]-2-iodobenzohydrazide (11)
- N-[(5-chloro-2-hydroxy-3-iodophenyl)methylidene]-2-iodobenzohydrazide (12)
- N-[(3,5-dibromo-2-hydroxyphenyl)methylidene]-2-iodobenzohydrazide (13)
- N-[(3,5-diiodo-2-hydroxyphenyl)methylidene]-2-iodobenzohydrazide (14)
- N-[(2-hydroxy-3-nitrophenyl)methylidene]-3-iodobenzohydrazide (15)
- N-[(2-hydroxyphenyl)methylidene]-3-iodobenzohydrazide (16) [38]
- N-[(3,5-dichloro-2-hydroxyphenyl)methylidene]-3-iodobenzohydrazide (17)
- N-[(3-ethoxy-2-hydroxyphenyl)methylidene]-3-iodobenzohydrazide (18)
- N-[(2,5-dihydroxyphenyl)methylidene]-3-iodobenzohydrazide (19)
- N-[(5-chloro-2-hydroxy-3-iodophenyl)methylidene]-3-iodobenzohydrazide (20)
- N-[(3,5-dibromo-2-hydroxyphenyl)methylidene]-3-iodobenzohydrazide (21)
- N-[(2-hydroxy-3-nitrophenyl)methylidene]-4-iodobenzohydrazide (22)
- N-[(2-hydroxyphenyl)methylidene]-4-iodobenzohydrazide (23)
- N-[(3,5-dichloro-2-hydroxyphenyl)methylidene]-4-iodobenzohydrazide (24)
- N-[(3-ethoxy-2-hydroxyphenyl)methylidene]-4-iodobenzohydrazide (25)
- N-[(2,5-dihydroxyphenyl)methylidene]-4-iodobenzohydrazide (26)
- N-[(5-chloro-2-hydroxy-3-iodophenyl)methylidene]-4-iodobenzohydrazide (27)
- N-[(3,5-dibromo-2-hydroxyphenyl)methylidene]-4-iodobenzohydrazide (28)
4.3. Mechanochemistry
4.4. Crystallization Experiments
4.5. X-ray Crystallography
4.6. Powder X-ray Diffraction
4.7. Microbiology
In Vitro Antimicrobial Assay
4.8. Cytotoxicity Study
4.8.1. In Vitro Cytotoxicity Assay
4.8.2. Cell Lines
4.8.3. MTT Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Popiołek, Ł.; Biernasiuk, A. Hydrazide-hydrazones of 3-methoxybenzoic acid and 4-tert-butylbenzoic acid with promising antibacterial activity against Bacillus spp. J. Enzyme Inhib. Med. Chem. 2016, 31 (Suppl. S1), 62–69. [Google Scholar] [CrossRef] [PubMed]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Rollas, S.; Küçükgüzel, S.G. Biological activities of hydrazone derivatives. Molecules 2007, 12, 1910–1939. [Google Scholar] [CrossRef] [PubMed]
- Angelova, V.; Karabeliov, V.; Andreeva-Gateva, P.A.; Tchekalarova, J. Recent Developments of Hydrazide/Hydrazone Derivatives and Their Analogs as Anticonvulsant Agents in Animal Models. Drug Dev. Res. 2016, 77, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Popiołek, Ł. Hydrazide-hydrazones as potential antimicrobial agents: Overview of the literature since 2010. Med. Chem. Res. 2017, 26, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.C.; Sharma, D.; Sharma, A.; Saini, N.; Goyal, R.; Ola, M.; Chawla, R.; Thakur, V.K. Hydrazone comprising compounds as promising anti-infective agents: Chemistry and structure–property relationship. Mater. Today Chem. 2020, 18, 100349. [Google Scholar] [CrossRef]
- Moussa, Z.; Al-Mamary, M.; Al-Juhani, S.; Ahmed, S.A. Preparation and biological assessment of some aromatic hydrazones derived from hydrazides of phenolic acids and aromatic aldehydes. Heliyon 2020, 6, e05019. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, R.d.A.; de Melo, C.V.; Marques, F.J.; Serpa, R.; Evangelista, A.J.; Caetano, E.P.; Mafezoli, J.; de Oliveira, M.C.; da Silva, M.R.; Bandeira, T.d.J.; et al. Synthesis and in vitro antifungal activity of isoniazid-derived hydrazones against Coccidioides posadasii. Microb. Pathog. 2016, 98, 1–5. [Google Scholar] [CrossRef][Green Version]
- Pahonțu, E.; Ilieș, D.C.; Shova, S.; Oprean, C.; Păunescu, V.; Olaru, O.T.; Rădulescu, F.Ș.; Gulea, A.; Roșu, T.; Drăgănescu, D. Synthesis, Characterization, Antimicrobial and Antiproliferative Activity Evaluation of Cu(II), Co(II), Zn(II), Ni(II) and Pt(II) Complexes with Isoniazid-Derived Compound. Molecules 2017, 22, 650. [Google Scholar] [CrossRef]
- Naveen Kumar, H.; Jummat, F.; Asmawi, M.Z. Synthesis and evaluation of isonicotinoyl hydrazone derivatives as antimycobacterial and anticancer agents. Med. Chem. Res. 2014, 23, 269–279. [Google Scholar] [CrossRef]
- Paruch, K.; Popiołek, Ł.; Biernasiuk, A.; Berecka-Rycerz, A.; Malm, A.; Gumieniczek, A.; Wujec, M. Novel Derivatives of 4-Methyl-1,2,3-Thiadiazole-5-Carboxylic Acid Hydrazide: Synthesis, Lipophilicity, and In Vitro Antimicrobial Activity Screening. Appl. Sci. 2021, 11, 1180. [Google Scholar] [CrossRef]
- Bogdanov, A.; Tsivileva, O.; Voloshina, A.; Lyubina, A.; Amerhanova, S.; Burtceva, E.; Bukharov, S.; Samorodov, A.; Pavlov, V. Synthesis and diverse biological activity profile of triethylammonium isatin-3-hydrazones. ADMET DMPK 2022, 10, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, A.V.; Voloshina, A.D.; Khamatgalimov, A.R.; Terekhova, N.V. On the Effect of the Nature of Substituents on the Antimicrobial Activity of Water-Soluble Acylhydrazones on the Isatin Scaffold. Dokl. Chem. 2020, 494, 136–140. [Google Scholar] [CrossRef]
- Popiołek, Ł.; Patrejko, P.; Gawrońska-Grzywacz, M.; Biernasiuk, A.; Berecka-Rycerz, A.; Natorska-Chomicka, D.; Piątkowska-Chmiel, I.; Gumieniczek, A.; Dudka, J.; Wujec, M. Synthesis and in vitro bioactivity study of new hydrazide-hydrazones of 5-bromo-2-iodobenzoic acid. Biomed. Pharmacother. 2020, 130, 110526. [Google Scholar] [CrossRef] [PubMed]
- Popiołek, Ł.; Tuszyńska, K.; Biernasiuk, A. Searching for novel antimicrobial agents among hydrazide-hydrazones of 4-iodosalicylic acid. Biomed. Pharmacother. 2022, 153, 113302. [Google Scholar] [CrossRef] [PubMed]
- Popiołek, Ł.; Gawrońska-Grzywacz, M.; Dziduch, A.; Biernasiuk, A.; Piątkowska-Chmiel, I.; Herbet, M. Design, synthesis, and in vitro and in vivo bioactivity studies of hydrazide-hydrazones of 2,4-dihydroxybenzoic acid. Int. J. Mol. Sci. 2023, 24, 17481. [Google Scholar] [CrossRef]
- Hajipour, A.R.; Mohammadpoor-Baltork, I.; Bigdeli, M. A Convenient and Mild Procedure for the Synthesis of Hydrazones and Semicarbazones from Aldehydes or Ketones under Solvent-Free Conditions. J. Chem. Res. 1999, 9, 570–571. [Google Scholar] [CrossRef]
- Oliveira, P.F.M.; Baron, M.; Chamayou, A.; André-Barrès, C.; Guidetti, B.; Baltas, M. Solvent–Free Mechanochemical Route for Green Synthesis of Pharmaceutically Attractive Phenol-Hydrazones. RSC Adv. 2014, 4, 56736–56742. [Google Scholar] [CrossRef]
- Pisk, J.; Đilović, I.; Hrenar, T.; Cvijanović, D.; Pavlović, G.; Vrdoljak, V. Effective methods for the synthesis of hydrazones, quinazolines, and Schiff bases: Reaction monitoring using a chemometric approach. RSC Adv. 2020, 10, 38566. [Google Scholar] [CrossRef]
- Crawford, D.E.; Porcheddu, A.; McCalmont, A.S.; Delogu, F.; James, S.L.; Colacino, E. Solvent–Free, Continuous Synthesis of Hydrazone-Based Active Pharmaceutical Ingredients by Twin-Screw Extrusion. ACS Sustain. Chem. Eng. 2020, 8, 12230–12238. [Google Scholar] [CrossRef]
- Tan, D.; Loots, L.; Friščić, T. Towards medicinal mechanochemistry: Evolution of milling from pharmaceutical solid form screening to the synthesis of active pharmaceutical ingredients (APIs). Chem. Commun. 2016, 52, 7760. [Google Scholar] [CrossRef] [PubMed]
- Kaupp, G.; Schmeyers, J.; Boy, J. Iminium Salts in Solid-State Syntheses Giving 100% Yield. J. Prakt. Chem. 2000, 342, 269–280. [Google Scholar] [CrossRef]
- Nun, P.; Martin, C.; Martinez, J.; Lamaty, F. Solvent-Free Synthesis of Hydrazones and Their Subsequent N-Alkylation in a Ball-Mill. Tetrahedron 2011, 67, 8187–8194. [Google Scholar] [CrossRef]
- Oliveira, P.F.M.; Guidetti, B.; Chamayou, A.; André-Barrès, C.; Madacki, J.; Korduláková, J.; Mori, G.; Orena, B.S.; Chiarelli, L.R.; Pasca, M.R.; et al. Mechanochemical Synthesis and Biological Evaluation of Novel Isoniazid Derivatives with Potent Antitubercular Activity. Molecules 2017, 22, 1457. [Google Scholar] [CrossRef] [PubMed]
- Pisk, J.; Hrenar, T.; Rubčić, M.; Pavlović, G.; Damjanović, V.; Lovrić, J.; Cindrić, M.; Vrdoljak, V. Comparative studies on conventional and solvent-free synthesis toward hydrazones: Application of PXRD and chemometric data analysis in mechanochemical reaction monitoring. CrystEngComm 2018, 20, 1804–1817. [Google Scholar] [CrossRef]
- Kapusterynska, A.; Bijani, C.; Paliwoda, D.; Vendier, L.; Bourdon, V.; Imbert, N.; Cojean, S.; Loiseau, P.M.; Recchia, D.; Scoffone, V.C.; et al. Mechanochemical Studies on Coupling of Hydrazines and Hydrazine Amides with Phenolic and Furanyl Aldehydes-Hydrazones with Antileishmanial and Antibacterial Activities. Molecules 2023, 28, 5284. [Google Scholar] [CrossRef] [PubMed]
- Wardell, S.M.S.V.; de Souza, M.V.N.; Wardell, J.L.; Low, J.N.; Glidewell, C. Supramolecular structures in N-isonicotinoyl arylaldehydehydrazones: Multiple hydrogen-bonding modes in series of geometric isomers. Acta Crystallogr. Sect. B 2007, 63, 879–895. [Google Scholar] [CrossRef] [PubMed]
- Mazur, L.; Jarzembska, K.N.; Kamiński, R.; Woźniak, K.; Pindelska, E.; Zielinska-Pisklak, M. Substituent and Solvent Effects on Intermolecular Interactions in Crystals of N-Acylhydrazone Derivatives: Single-crystal X-ray, Solid-State NMR and Computational Studies. Cryst. Growth Des. 2014, 14, 2263–2281. [Google Scholar] [CrossRef]
- Mazur, L.; Jarzembska, K.N.; Kamiński, R.; Hoser, A.A.; Madsen, A.Ø.; Pindelska, E.; Zielinska-Pisklak, M. Crystal structures and thermodynamic properties of polymorphs and hydrates of selected 2-pyridinecarboxaldehyde hydrazones. Cryst. Growth Des. 2016, 16, 3101–3112. [Google Scholar] [CrossRef]
- Cruz-Cabeza, A.J.; Bernstein, J. Conformational polymorphism. Chem. Rev. 2014, 114, 2170–2191. [Google Scholar] [CrossRef] [PubMed]
- Siddique, M.; Saeed, A.B.; Dogar, N.A.; Ahmad, S. Biological Potential of Synthetic Hydrazide Based Schiff Bases. J. Sci. Innov. Res. 2013, 2, 651–657. [Google Scholar]
- Manikandan, V.; Balaji, S.; Senbagam, R.; Vijayakumar, R.; Rajarajan, M.; Vanangamudi, G.; Arulkumaran, R.; Sundararajan, R.; Thirunarayanan, G. Synthesis and antimicrobial activities of some (E)-N′-1-(substituted benzylidene)benzohydrazides. Inter. J. Adv. Chem. 2017, 5, 17–24. [Google Scholar] [CrossRef]
- Backes, G.L.; Neumann, D.M.; Jursic, B.S. Synthesis and antifungal activity of substituted salicylaldehyde hydrazones, hydrazides and sulfohydrazides. Bioorg. Med. Chem. 2014, 22, 4629–4636. [Google Scholar] [CrossRef] [PubMed]
- Puskullu, M.O.; Celik, I.; Erol, M.; Fatullayev, H.; Uzunhisarcikli, E.; Kuyucuklu, G. Antimicrobial and antiproliferative activity studies of some new quinoline-3-carbaldehyde hydrazone derivatives. Bioorg. Chem. 2020, 101, 104014. [Google Scholar] [CrossRef] [PubMed]
- Al Rasheed, H.H.; Malebari, A.M.; Dahlous, K.A.; El–Faham, A. Synthesis and Characterization of New Series of 1,3,5-Triazine Hydrazone Derivatives with Promising Antiproliferative Activity. Molecules 2020, 25, 2708. [Google Scholar] [CrossRef] [PubMed]
- Castrillón-López, W.; Herrera-Ramírez, A.; Moreno-Quintero, G.; Coa, J.C.; Naranjo, T.W.; Cardona–Galeano, W. Resveratrol/Hydrazone Hybrids: Synthesis and Chemopreventive Activity against Colorectal Cancer Cells. Pharmaceutics 2022, 14, 2278. [Google Scholar] [CrossRef]
- Al Rasheed, H.H.; Dahlous, K.; Sharma, A.; Sholkamy, E.; El-Faham, A.; de la Torre, B.G.; Albericio, F. Barbiturate- and Thiobarbituarte-Based s-Triazine Hydrazone Derivatives with Promising Antiproliferative Activities. ACS Omega 2020, 5, 15805–15811. [Google Scholar] [CrossRef] [PubMed]
- Czyżewska, I.; Mazur, L.; Popiołek, Ł. Synteza i badania strukturalne nowej pochodnej z grupy hydrazonów. In Nauka i Przemysł: Lubelskie Spotkania Studenckie; UMCS: Lublin, Poland, 2022; pp. 234–237. [Google Scholar]
- CrysAlisPRO Software System; Rigaku: Oxford, UK, 2016.
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Faruggia, L.J. WinGX suite for small–molecule single–crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Cryst. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- PANalytical. HighScore Plus, Software Ver. 3.0.5; PANalytical: Almelo, The Netherlands, 2012.
- European Committee for Antimicrobial Susceptibility Testing (EUCAST). Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. EUCAST discussion document E. Dis 5.1. Clin. Microbiol. Infect. 2003, 9, 1–7. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. M27–S4; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Popiołek, Ł; Biernasiuk, A.; Malm, A. Synthesis and antimicrobial activity of new 1,3-thiazolidin-4-one derivatives obtained from carboxylic acid hydrazides. Phosphorus Sulfur Silicon Relat. Elem. 2015, 190, 251–260. [Google Scholar]
- O’Donnell, F.; Smyth, T.J.; Ramachandran, V.N.; Smyth, W.F. A study of the antimicrobial activity of selected synthetic and naturally occurring quinolines. Int. J. Antimicrob. Agents 2010, 35, 30–38. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar]

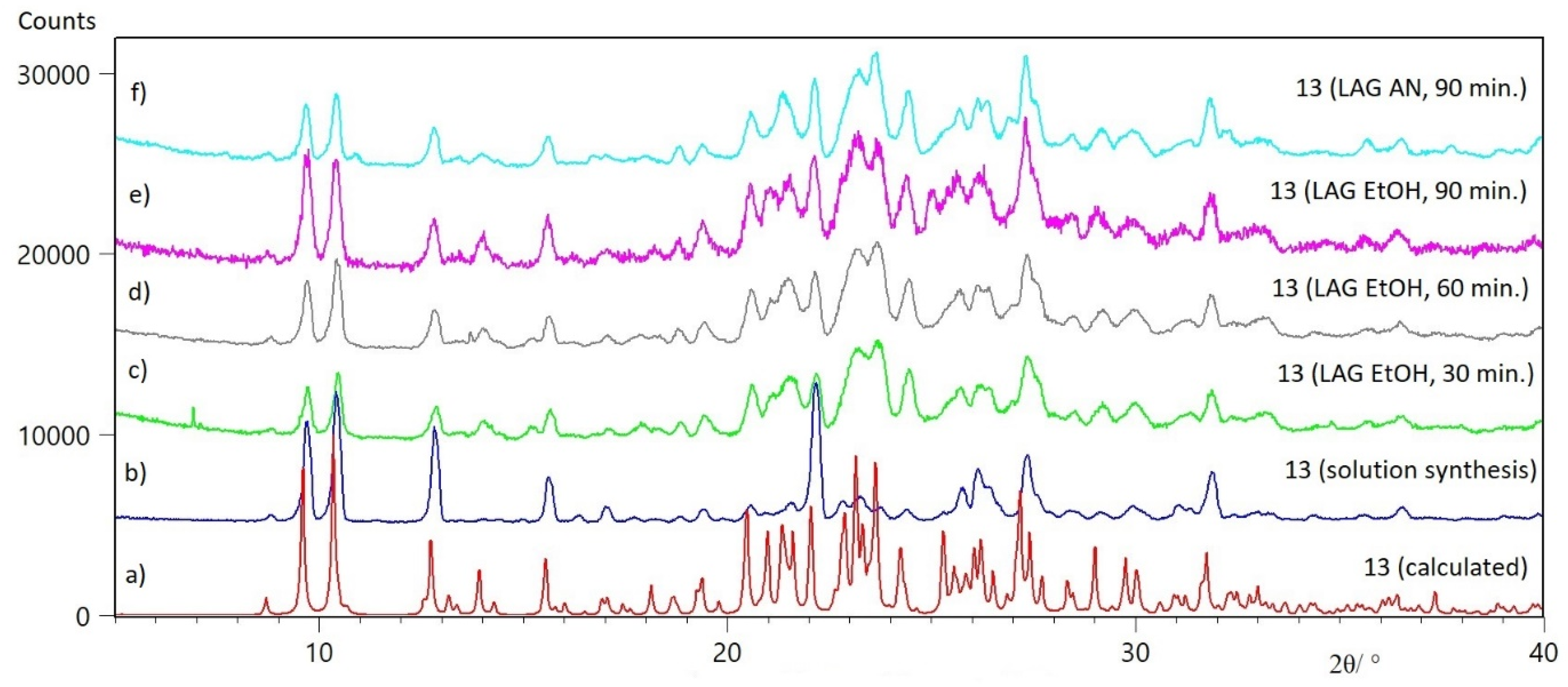
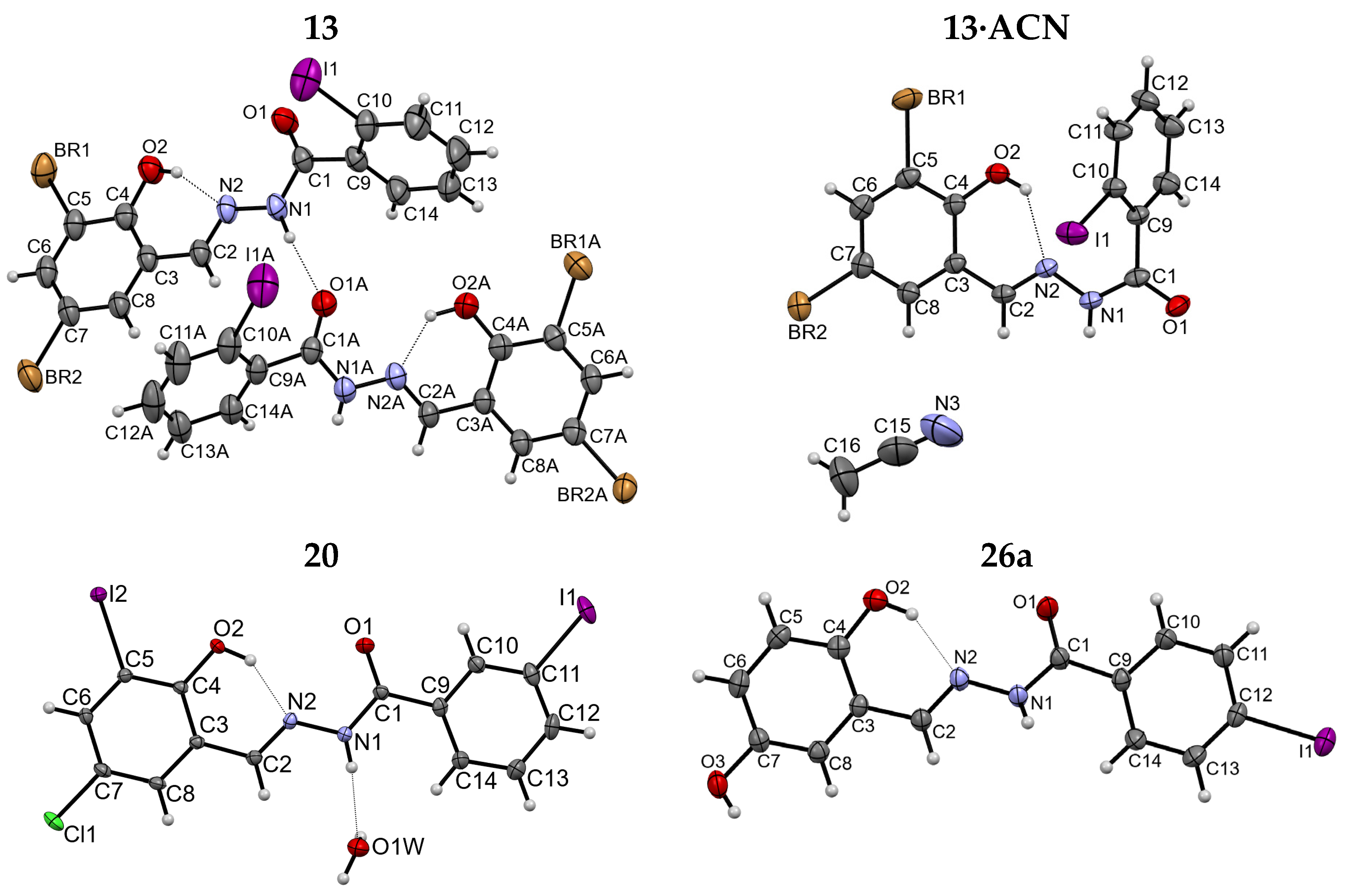

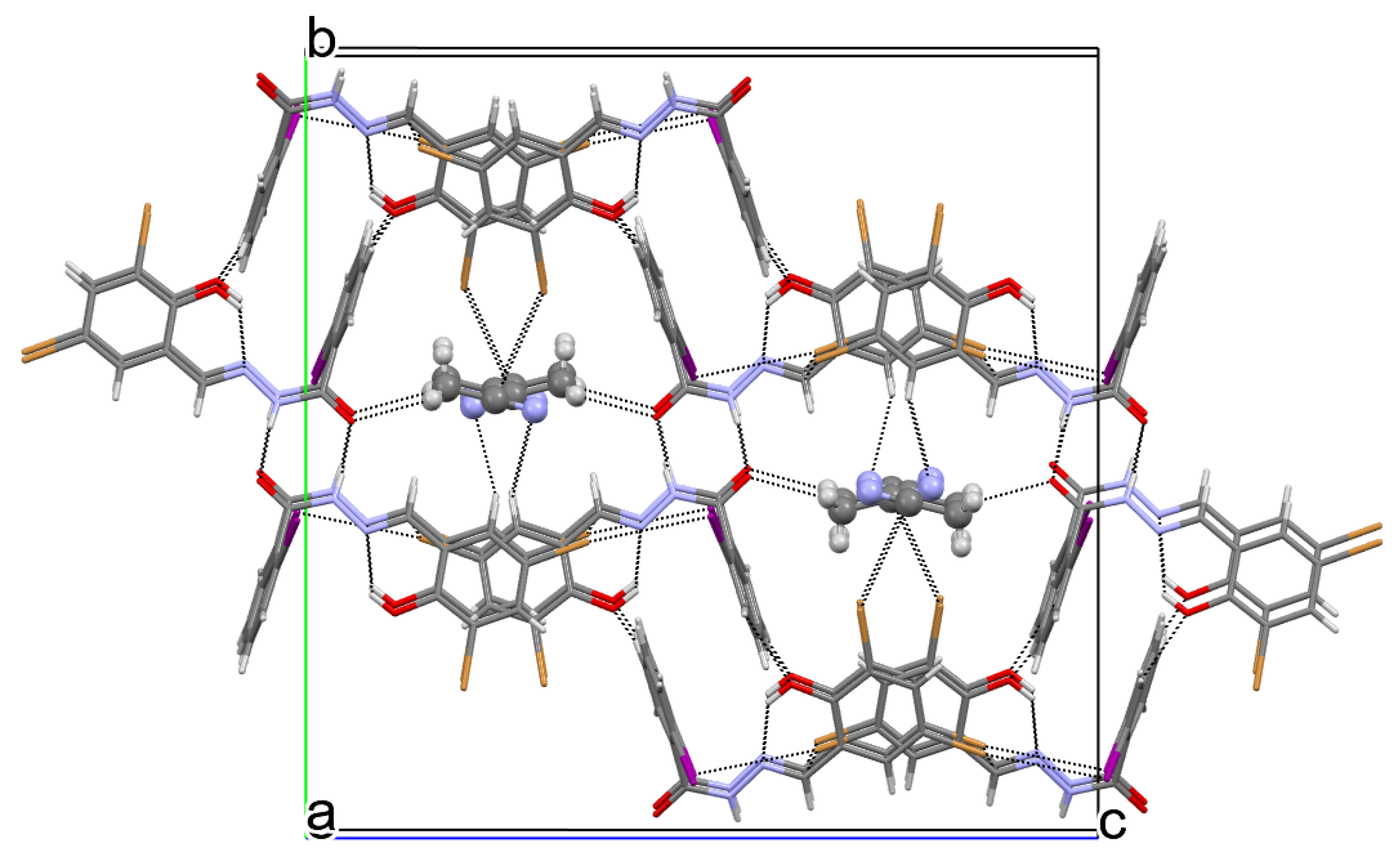

| Crystal Structure | 9 | 12 | 13 | 13∙ACN | 14 |
|---|---|---|---|---|---|
| Chemical formula | C14H9N2O2ICl2 | C14H9N2O2I2Cl | C14H9N2O2IBr2 | C14H9N2O2IBr2 ∙CH3CN | C14H9N2O2I3 |
| Formula weight | 435.03 | 526.48 | 523.95 | 565.01 | 617.93 |
| Crystal system | monoclinic | monoclinic | monoclinic | orthorhombic | monoclinic |
| Space group | P21/c | P21/n | P21/c | Pbca | P21/c |
| a/Å | 18.7006(4) | 18.9192(4) | 18.9820(7) | 7.2352(1) | 19.4686(3) |
| b/Å | 9.5210(2) | 9.5250(1) | 9.5183(3) | 22.9495(2) | 9.6874(1) |
| c/Å | 20.2936(4) | 20.7505(4) | 20.4590(6) | 23.2354(3) | 20.4776(3) |
| β/° | 115.229(2) | 116.442(3) | 115.753(4) | 90 | 115.951(2) |
| V/Å3 | 3268.6(1) | 3348.2(1) | 3329.3(2) | 3858.10(8) | 3472.7(1) |
| Z/Z’ | 8/2 | 8/2 | 8/2 | 8/1 | 8/2 |
| dcalc/g·cm−3 | 1.768 | 2.089 | 2.091 | 1.945 | 2.364 |
| R1 [F2 > 2σ(F2)] | 0.060 | 0.060 | 0.053 | 0.048 | 0.055 |
| wR2 (all data) | 0.170 | 0.171 | 0.138 | 0.123 | 0.157 |
| CCDC number | 2357466 | 2357467 | 2357468 | 2357469 | 2357470 |
| Crystal Structure | 20 | 24 | 26a | 26b | 26c |
| Chemical formula | C14H9N2O2I2Cl H2O | C14H9N2O2ICl2 | C14H11N2O3I | C14H11N2O3I | C14H11N2O3I |
| Formula weight | 544.50 | 435.03 | 382.15 | 382.15 | 382.15 |
| Crystal system | monoclinic | monoclinic | orthorhombic | monoclinic | monoclinic |
| Space group | P21/c | C2/c | Pca21 | P21 | P21 |
| a/Å | 15.3068(3) | 21.3618(3) | 33.8455(4) | 4.5303(1) | 4.8826(1) |
| b/Å | 7.6017(1) | 9.9728(2) | 4.7071(1) | 10.5368(2) | 5.0674(1) |
| c/Å | 15.2558(3) | 29.6378(4) | 8.6261(1) | 14.3691(3) | 27.2710(4) |
| β/° | 110.106(2) | 101.705(1) | 90 | 90.737(2) | 90.047(1) |
| V/Å3 | 1666.95(5) | 6182.7(2) | 1374.26(4) | 685.85(2) | 674.74(2) |
| Z/Z’ | 4/1 | 16/2 | 4/1 | 2/1 | 2/1 |
| dcalc/g·cm−3 | 2.170 | 1.869 | 1.847 | 1.850 | 1.881 |
| R1 [F2 > 2σ(F2)] | 0.043 | 0.036 | 0.029 | 0.031 | 0.038 |
| wR2 (all data) | 0.118 | 0.095 | 0.073 | 0.082 | 0.101 |
| CCDC number | 2357471 | 2357472 | 2357473 | 2357474 | 2357475 |
| Species | MIC (MBC or MFC) [µg/mL] and {MBC/MIC or MFC/MIC} Values of the Studied Compounds and Positive Controls | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 7 | 10 | 11 | 12 | 14 | CIP/NY* | NIT | CFX | APE | ||
| Gram-positive bacteria | Staphylococcus aureus ATCC 25923 | 500 (>1000) {>2} | 500 (>1000) {>2} | – | – | 125 (>1000) {>8} | 125 (250) {2} | 0.48 (0.48) {1} | 15.62 (15.62) | 0.49 | nd |
| Staphylococcus aureus ATCC 29213 | – | – | – | – | 1000 (>1000) {>1} | 125 (500) {4} | 0.48 (0.48) {1} | nd | nd | nd | |
| Staphylococcus aureus ATCC 43300 | – | – | – | – | 1000 (>1000) {>1} | 250 (1000) {4} | 0.24 (0.24) {1} | 7.81 | nd | nd | |
| Staphylococcus epidermidis ATCC 12228 | 1000 (>1000) {>1} | 125 (1000) {8} | – | – | 250 (500) {2} | 125 (250) {2} | 0.12 (0.12) {1} | 3.91 (7.81) | 0.24 | nd | |
| Micrococcus luteus ATCC 10240 | 1000 (>1000) {>1} | 1000 (>1000) {>1} | – | – | 31.25 (250) {8} | 62.5 (125) {2} | 0.98 (1.95) {2} | 62.5 (62.5) | 0.98 | nd | |
| Bacillus subtilis ATCC 6633 | – | 1000 (>1000) {>1} | – | – | 1000 (>1000) {>1} | 500 (500) {1} | 0.03 (0.03) {1} | 3.91 (3.91) | 15.62 | 62.5 | |
| Bacillus cereus ATCC 10876 | – | – | – | – | 500 (>1000) {>2} | 125 (250) {2} | 0.06 (0.12) {2} | 7.81 (15,62) | 31.25 | nd | |
| Fungi | Candida albicans ATCC 10231 | 1000 (>1000) {>1} | 1000 (>1000) {>1} | – | 1000 (>1000) {>1} | 500 (>1000) {>2} | 500 (1000) {2} | 0.48 * (0.48) {1} | na | na | na |
| Candida albicans ATCC 2091 | – | – | – | – | 500 (>1000) {>2} | 500 (>1000) {>2} | 0.24 * (0.24) {1} | na | na | na | |
| Candida parapsilosis ATCC 22019 | – | – | – | – | 1000 (>1000) {>1} | 1000 (>1000) {>1} | 0.24 * (0.48) {2} | na | na | na | |
| Candida glabrata ATCC 90030 | – | – | 1000 (>1000) {>1} | 1000 (>1000) {>1} | 250 (>1000) {>4} | 250 (1000) {4} | 0.24 * (0.48) {2} | na | na | na | |
| Candida krusei ATCC 14243 | 1000 (>1000) {>1} | – | 1000 (>1000) {>1} | – | 500 (>1000) {>2} | 500 (>1000) {>2} | 0.24 * (0.24) {1} | na | na | na | |
| Species | MIC (MBC or MFC) [µg/mL] and {MBC/MIC or MFC/MIC} Values of the Studied Compounds and Positive Controls | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 15 | 17 | 18 | 19 | 20 | 21 | CIP/NY* | NIT | CFX | APC | ||
| Gram-positive bacteria | Staphylococcus aureus ATCC 25923 | – | 250 (>1000) {>4} | 31.25 (31.25) {1} | – | – | 1.95 (3.91) {2} | 7.81 (31.25) {4} | 0.48 (0.48) {1} | 15.62 (15.62) | 0.49 | nd |
| Staphylococcus aureus ATCC 29213 | – | 250 (>1000) {>4} | 15.62 (15.62) {1} | – | – | 3.91 (3.91) {1} | 15.62 (15.62) {1} | 0.48 (0.48) {1} | nd | nd | nd | |
| Staphylococcus aureus ATCC 43300 | – | 1000 (>1000) {>1} | 15.62 (15.62) {1} | – | – | 3.91 (7.81) {2} | 15.62 (62.5) {4} | 0.24 (0.24) {1} | 7.81 | nd | nd | |
| Staphylococcus epidermidis ATCC 12228 | 1000 (>1000) {>1} | 31.25 (125) {4} | 15.62 (62.5) {4} | 500 (>1000) {>2} | – | 1.95 (7.81) {4} | 7.81 (31.25) {4} | 0.12 (0.12) {1} | 3.91 (7.81) | 0.24 | nd | |
| Micrococcus luteus ATCC 10240 | 1000 (>1000) {>1} | 31.25 (125) {4} | 15.62 (15.62) {1} | 31.25 (250) {8} | – | 1.95 (7.81) {4} | 3.91 (15.62) {4} | 0.98 (1.95) {2} | 62.5 (62.5) | 0.98 | nd | |
| Bacillus subtilis ATCC 6633 | 1000 (>1000) {>1} | 125 (500) {4} | 7.81 (15.62) {1} | 62.5 (>1000) {>16} | – | 7.81 (31.25) {4} | 7.81 (31.25) {4} | 0.03 (0.03) {1} | 3.91 (3.91) | 15.62 | 62.5 | |
| Bacillus cereus ATCC 10876 | – | 125 (250) {2} | 15.62 (15.62) {1} | – | – | 7.81 (31.25) {4} | 15.62 (62.5) {4} | 0.06 (0.12) {2} | 7.81 (15.62) | 31.25 | nd | |
| Gram-negative bacteria | Bordetella bronchiseptica ATCC 4617 | – | – | – | – | – | 500 (>1000) {>2} | – | 0.98 (0.98) {1} | 125 | nd | nd |
| Klebsiella pneumoniae ATCC 13883 | – | – | – | – | – | 500 (>1000) {>2} | – | 0.12 (0.24) {2} | 15.62 | nd | nd | |
| Proteus mirabilis ATCC 12453 | – | – | – | – | – | 500 (1000) {2} | – | 0.03 (0.03) {1) | 62.5 | nd | nd | |
| Escherichia coli ATCC 25922 | – | – | – | – | – | 1000 (>1000) {>1} | – | 0.06 (0.06) {1} | 7.81 | nd | nd | |
| Salmonella typhimurium ATCC 14028 | – | – | – | – | – | 1000 (>1000) {>1} | – | 0.004 (0.008) {2} | 31.25 | nd | nd | |
| Pseudomonas aeruginosa ATCC 9027 | – | – | – | – | – | 500 (>1000) {>2} | – | 0.48 (0.98) {2} | nd | nd | nd | |
| Fungi | Candida albicans ATCC 10231 | – | – | – | – | 250 (>1000) {>4} | 250 (>1000) {>4} | 250 (>1000) {>4} | 0.48 * (0.48) {1} | na | na | na |
| Candida albicans ATCC 2091 | – | – | – | – | 500 (>1000) {>2} | 500 (>1000) {>2} | 500 (>1000) {>2} | 0.24 * (0.24) {1} | na | na | na | |
| Candida parapsilosis ATCC 22019 | – | – | – | – | 125 (>1000) {>8} | 125 (>1000) {>8} | 250 (>1000) {>4} | 0.24 * (0.48) {2} | na | na | na | |
| Candida glabrata ATCC 90030 | – | – | – | – | 500 (>1000) {>2} | 125 (>1000) {>8} | 125 (>1000) {>8} | 0.24 * (0.48) {2} | na | na | na | |
| Candida krusei ATCC 14243 | – | – | – | – | 250 (>1000) {>4} | 500 (>1000) {>2} | 500 (>1000) {>2} | 0.24 * (0.24) {1} | na | na | na | |
| Species | MIC (MBC or MFC) [µg/mL] and {MBC/MIC or MFC/MIC} Values of the Studied Compounds and Positive Controls | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 23 | 24 | 25 | 26 | 27 | 28 | CIP/ NY* | NIT | CFX | APC | ||
| Gram-positive bacteria | Staphylococcus aureus ATCC 25923 | – | 1000 (>1000) {>1} | – | – | – | 62.5 (250) {4} | – | 0.48 (0.48) {1} | 15.62 (15.62) | 0.49 | nd |
| Staphylococcus aureus ATCC 29213 | – | 500 (>1000) {>2} | 1000 (>2000) | – | – | 62.5 (125) {2} | – | 0.48 (0.48) {1} | nd | nd | nd | |
| Staphylococcus aureus ATCC 43300 | – | 500 (>1000) {>2} | – | – | 1000 (>1000) {>1} | 62.5 (250) {4} | – | 0.24 (0.24) {1} | 7.81 | nd | nd | |
| Staphylococcus epidermidis ATCC 12228 | 1000 (>1000) {>1} | 1000 (>1000) {>1} | – | 1000 (>1000) {>1} | 1000 (>1000) {>1} | 31.25 (31.25) {1} | – | 0.12 (0.12) {1} | 3.91 (7.81) | 0.24 | nd | |
| Micrococcus luteus ATCC 10240 | 500 (>1000) {>2} | 500 (>1000) {>2} | 500 (>1000) {>2} | 31.25 (62.5) {2} | 500 (>1000) {>2} | 125 (250) {2} | – | 0.98 (1.95) {2} | 62.5 (62.5) | 0.98 | nd | |
| Bacillus subtilis ATCC 6633 | 1000 (>1000) {>1} | 1000 (>1000) {>1} | 500 (>1000) {>2} | – | 500 (>1000) {>2} | 125 (500) {4} | – | 0.03 (0.03) {1} | 3.91 (3.91) | 15.62 | 62.5 | |
| Bacillus cereus ATCC 10876 | – | 1000 (>1000) {>1} | – | – | – | 62.5 (250) {4} | – | 0.06 (0.12) {2} | 7.81 (15,62) | 31.25 | nd | |
| Gram-negative bacteria | Bordetella bronchiseptica ATCC 4617 | – | – | – | – | – | 500 (1000) {2} | – | 0.98 (0.98) {1} | 125 | nd | nd |
| Klebsiella Pneumoniae ATCC 13883 | – | – | – | – | – | 500 (>1000) {>2} | – | 0.12 (0.24) {2} | 15.62 | nd | nd | |
| Proteus mirabilis ATCC 12453 | – | – | – | – | – | 500 (1000) {2} | – | 0.03 (0.03) {1) | 62.5 | nd | nd | |
| Escherichia coli ATCC 25922 | – | – | – | – | – | 500 (500) {1} | – | 0.06 (0.06) {1} | 7.81 | nd | nd | |
| Salmonella typhimurium ATCC 14028 | – | – | – | – | – | 500 (1000) {2} | – | 0.004 (0.008) {2} | 31.25 | nd | nd | |
| Pseudomonas aeruginosa ATCC 9027 | – | – | – | – | – | 1000 (>1000) {>1} | – | 0.48 (0.98) {2} | nd | nd | nd | |
| Fungi | Candida albicans ATCC 10231 | – | – | 500 (>1000) {>2} | – | 500 (>1000) {>2} | 15.62 (31.25) {2} | 500 (>1000) {>2} | 0.48 * (0.48) {1} | na | na | na |
| Candida albicans ATCC 2091 | – | – | 1000 (>1000) {>1} | – | 1000 (>1000) {>1} | 62.5 (62.5) {1} | 1000 (>1000) {>1} | 0.24 * (0.24) {1} | na | na | na | |
| Candida parapsilosis ATCC 22019 | 250 (>1000) {>8} | 1000 (>1000) {>1} | 1000 (>1000) {>1} | 250 (>1000) {>8} | 250 (>1000) {>4} | 31.25 (62.5) {2} | 500 (>1000) {>2} | 0.24 * (0.48) {2} | na | na | na | |
| Candida glabrata ATCC 90030 | – | – | 250 (>1000) {>4} | – | 1000 (>1000) {>1} | 31.25 (62.5) {2} | 1000 (>1000) {>1} | 0.24 * (0.48) {2} | na | na | na | |
| Candida krusei ATCC 14243 | – | – | – | – | 500 (>1000) {>2} | 62.5 (62.5) {1} | 1000 (>1000) {>1} | 0.24 * (0.24) {1} | na | na | na | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czyżewska, I.; Mazur, L.; Biernasiuk, A.; Hordyjewska, A.; Popiołek, Ł. Synthesis, Structural Properties and Biological Activities of Novel Hydrazones of 2-, 3-, 4-Iodobenzoic Acid. Molecules 2024, 29, 3814. https://doi.org/10.3390/molecules29163814
Czyżewska I, Mazur L, Biernasiuk A, Hordyjewska A, Popiołek Ł. Synthesis, Structural Properties and Biological Activities of Novel Hydrazones of 2-, 3-, 4-Iodobenzoic Acid. Molecules. 2024; 29(16):3814. https://doi.org/10.3390/molecules29163814
Chicago/Turabian StyleCzyżewska, Izabela, Liliana Mazur, Anna Biernasiuk, Anna Hordyjewska, and Łukasz Popiołek. 2024. "Synthesis, Structural Properties and Biological Activities of Novel Hydrazones of 2-, 3-, 4-Iodobenzoic Acid" Molecules 29, no. 16: 3814. https://doi.org/10.3390/molecules29163814
APA StyleCzyżewska, I., Mazur, L., Biernasiuk, A., Hordyjewska, A., & Popiołek, Ł. (2024). Synthesis, Structural Properties and Biological Activities of Novel Hydrazones of 2-, 3-, 4-Iodobenzoic Acid. Molecules, 29(16), 3814. https://doi.org/10.3390/molecules29163814








