A Ratiometric Fluorescence Detection Method for Berberine Using Triplex-Containing DNA-Templated Silver Nanoclusters
Abstract
1. Introduction
2. Results
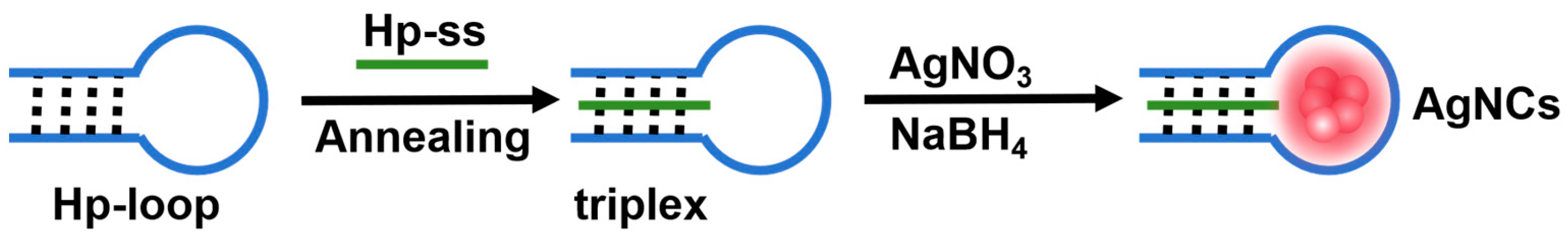
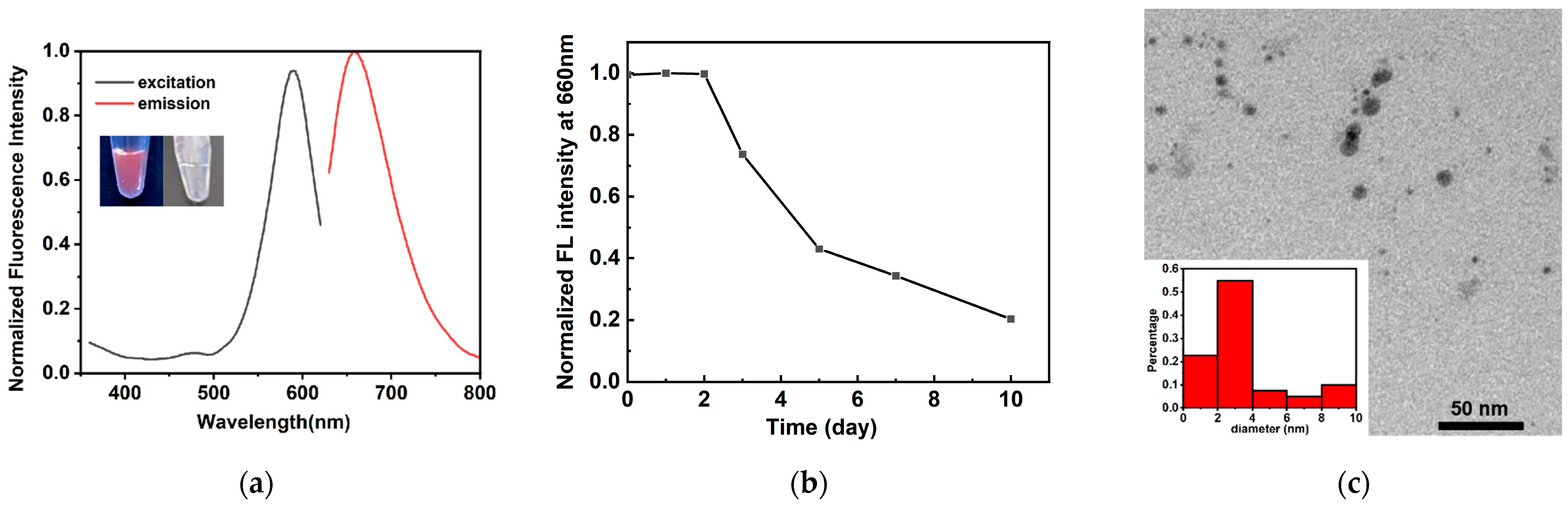
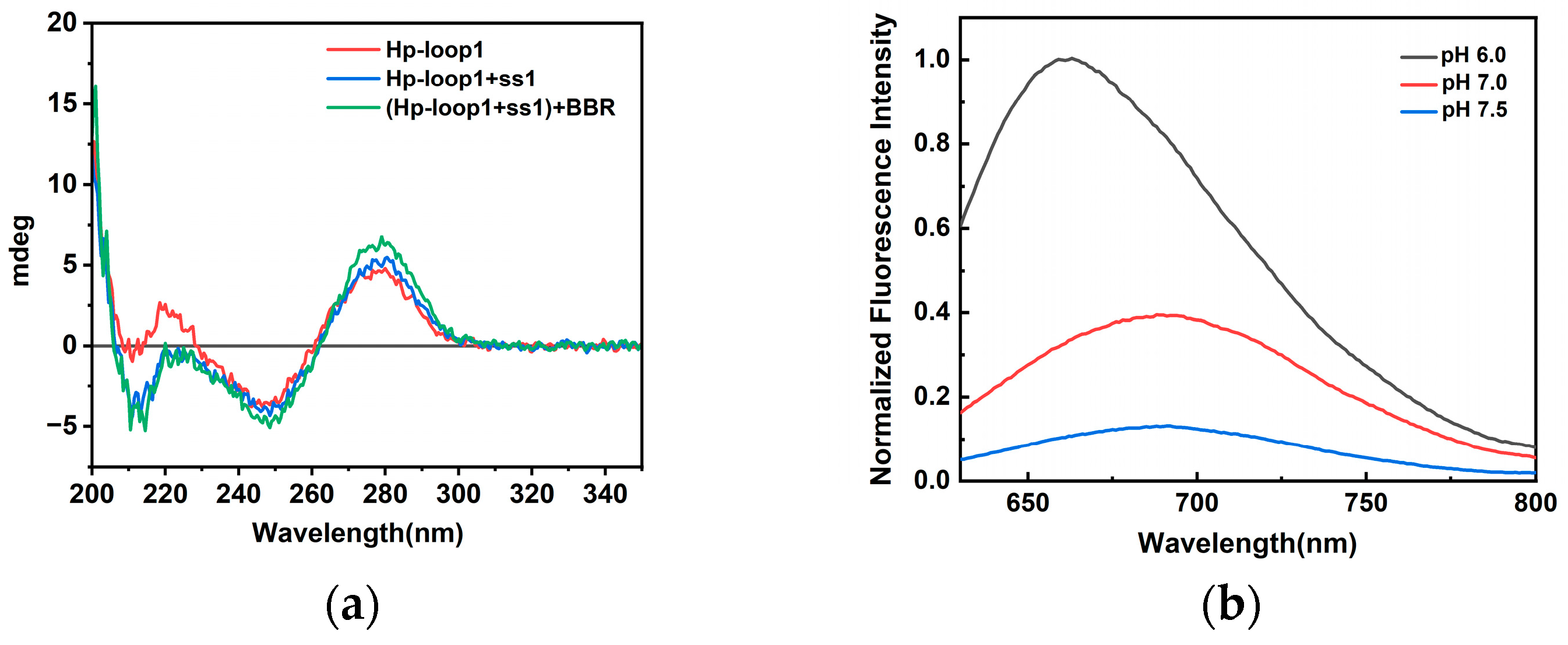
2.1. Characterization of Triplex-Containing DNA-AgNCs
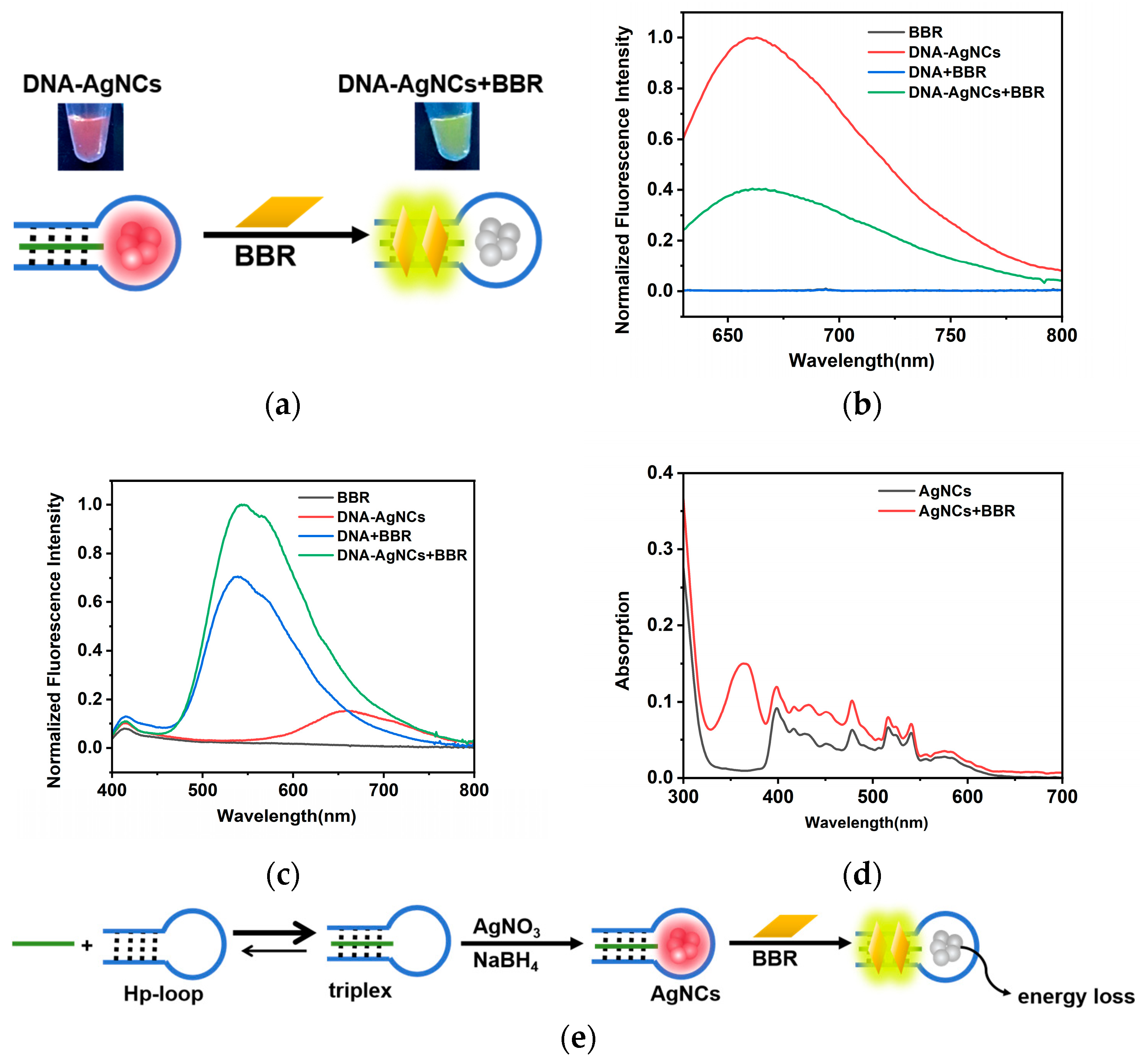
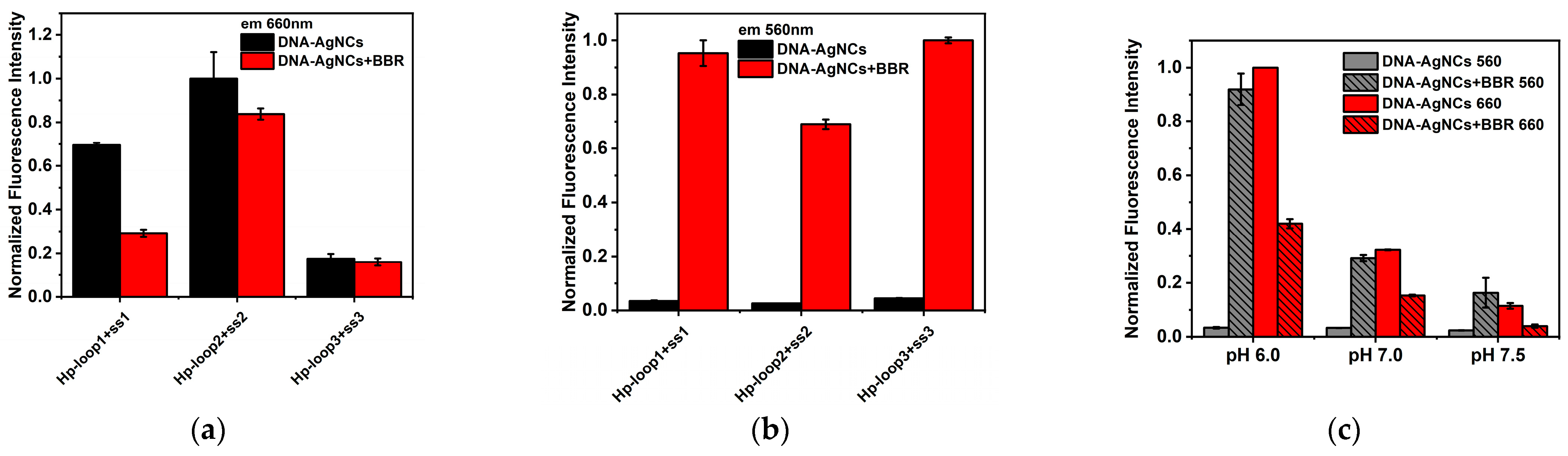
2.2. The Interaction between DNA-AgNCs and BBR
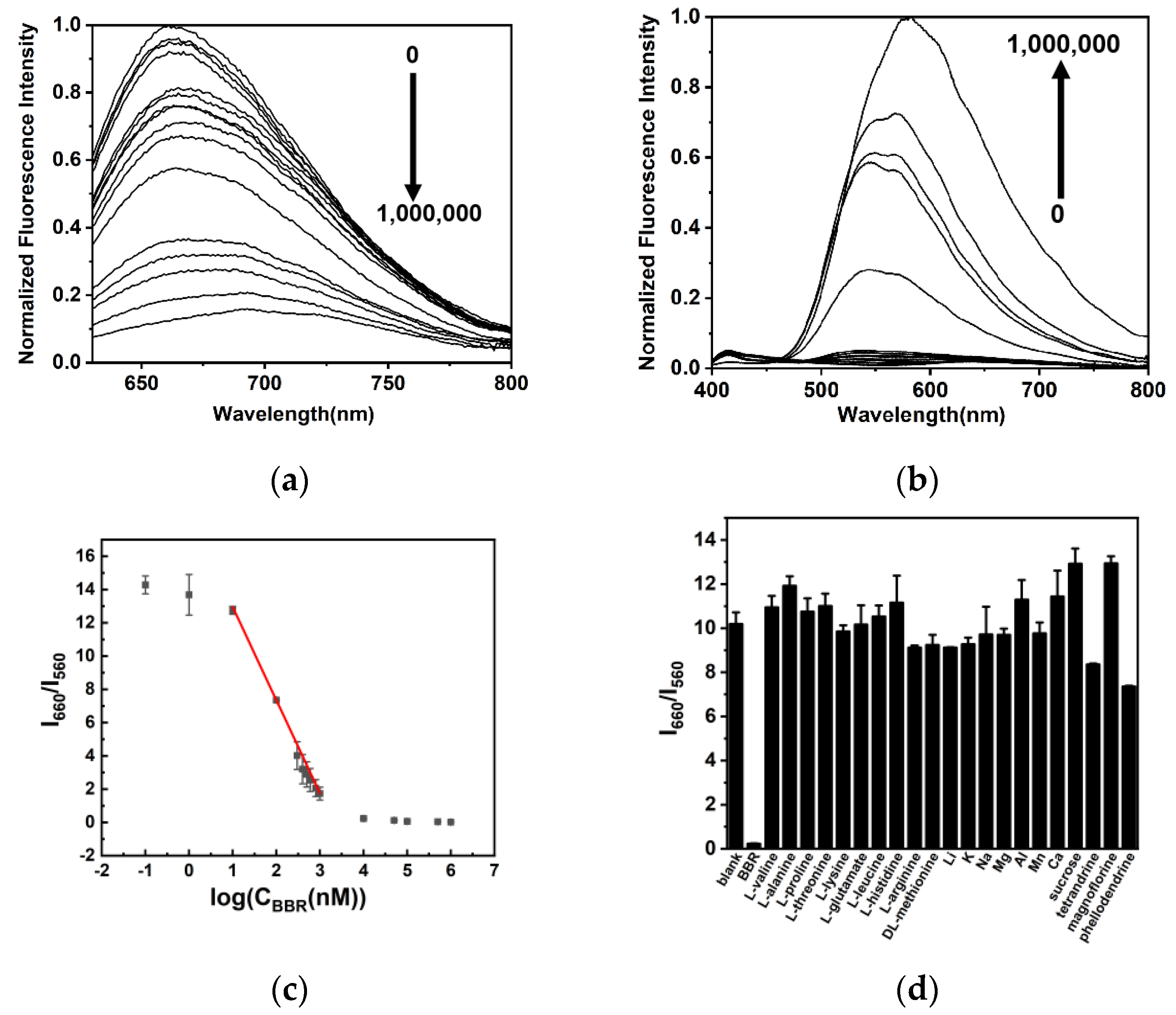
2.3. Detection of BBR
3. Discussion
4. Materials and Methods
4.1. Oligodeoxynucleotides and Materials
4.2. Synthesis of DNA-AgNCs
4.3. Characterization of DNA-AgNCs
4.4. Fluorescence Measurement
4.5. Real Sample Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cicero, A.F.G.; Baggioni, A. Berberine and Its Role in Chronic Disease. Adv. Exp. Med. Biol. 2016, 928, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Khadka, D.B.; Cho, W.-J. Pharmacological effects of berberine and its derivatives: A patent update. Expert Opin. Ther. Pat. 2015, 26, 229–243. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, X.; Zhang, H.; Zhang, S.; Li, Y.; Liu, Y.; Peng, D. Synthesis and anti-inflammatory effects of a series of novel 9-O-substituted berberine derivatives. Med. Chem. Res. 2017, 26, 672–679. [Google Scholar] [CrossRef]
- Liu, X.; Yu, M.; Liang, J. Research Progress on the Synthesis of Protoberberine Skeleton and Its Anti-inflammatory Activity. Chin. J. Org. Chem. 2023, 43, 1325–1340. [Google Scholar] [CrossRef]
- Zhang, L.C.; Wu, X.Y.; Yang, R.B.; Chen, F.; Liao, Y.; Zhu, Z.F.; Wu, Z.D.; Sun, X.; Wang, L.F. Effects of Berberine on the Gastrointestinal Microbiota. Front. Cell Infect. Microbiol. 2021, 10, 588517. [Google Scholar] [CrossRef]
- Chuang, T.C.; Wu, K.H.; Lin, Y.Y.; Kuo, H.P.; Kao, M.C.; Wang, V.; Hsu, S.C.; Lee, S.L. Dual down-regulation of EGFR and ErbB2 by berberine contributes to suppression of migration and invasion of human ovarian cancer cells. Environ. Toxicol. 2021, 36, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Debnath, D.; Kumar, G.S.; Maiti, M. Circular Dichroism Studies of the Structure of DNA Complex with Berberine. J. Biomol. Struct. Dyn. 1991, 9, 61–79. [Google Scholar] [CrossRef]
- Das, S.; Kumar, G.S.; Ray, A.; Maiti, M. Spectroscopic and thermodynamic studies on the binding of sanguinarine and berberine to triple and double helical DNA and RNA structures. J. Biomol. Struct. Dyn. 2003, 20, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Keppler, M.D.; Neidle, S.; Fox, K.R. Stabilisation of TG- and AG-containing antiparallel DNA triplexes by triplex-binding ligands. Nucleic Acids Res. 2001, 29, 1935–1942. [Google Scholar] [CrossRef]
- Oliva, R.; Mukherjee, S.; Manisegaran, M.; Campanile, M.; Del Vecchio, P.; Petraccone, L.; Winter, R. Binding Properties of RNA Quadruplex of SARS-CoV-2 to Berberine Compared to Telomeric DNA Quadruplex. Int. J. Mol. Sci. 2022, 23, 5690. [Google Scholar] [CrossRef]
- Xu, L.; Hong, S.; Sun, N.; Wang, K.; Zhou, L.; Ji, L.; Pei, R. Berberine as a novel light-up i-motif fluorescence ligand and its application in designing molecular logic systems. Chem. Commun. 2016, 52, 179–182. [Google Scholar] [CrossRef]
- Hu, Z.; Xie, M.; Yang, D.; Chen, D.; Jian, J.; Li, H.; Yuan, K.; Jiang, Z.; Zhou, H. A simple, fast, and sensitive colorimetric assay for visual detection of berberine in human plasma by NaHSO4-optimized gold nanoparticles. RSC Adv. 2017, 7, 34746–34754. [Google Scholar] [CrossRef]
- Song, Z.; Zhao, T.; Wang, L.; Xiao, Z. Chemiluminescence flow sensor for berberine with immobilized reagents. Bioorg. Med. Chem. 2001, 9, 1701–1705. [Google Scholar] [CrossRef]
- Wen, A.; Peng, X.; Zhang, P.; Long, Y.; Gong, H.; Xie, Q.; Yue, M.; Chen, S. Spectrofluorometric determination of berberine using a novel Au nanocluster with large Stokes shift. Anal. Bioanal. Chem. 2018, 410, 6489–6495. [Google Scholar] [CrossRef]
- Xiong, J.; Yang, L.; Gao, L.X.; Zhu, P.P.; Chen, Q.; Tan, K.J. A highly fluorescent lanthanide metal-organic framework as dual-mode visual sensor for berberine hydrochloride and tetracycline. Anal. Bioanal. Chem. 2019, 411, 5963–5973. [Google Scholar] [CrossRef]
- Wang, J.-S.; Sakthivel, R.; Anbazhagan, R.; Krishnamoorthi, R.; Kubendhiran, S.; Lai, J.-Y.; Tsai, H.-C.; Chen, S.-M. Electroactive polypyrrole-molybdenum disulfide nanocomposite for ultrasensitive detection of berberine in rat plasma. Anal. Chim. Acta 2020, 1125, 210–219. [Google Scholar] [CrossRef]
- Wu, T.-Y.; Chang, F.-R.; Liou, J.-R.; Lo, I.W.; Chung, T.-C.; Lee, L.-Y.; Chi, C.-C.; Du, Y.-C.; Wong, M.-H.; Juo, S.-H.H.; et al. Rapid HPLC Quantification Approach for Detection of Active Constituents in Modern Combinatorial Formula, San-Huang-Xie-Xin-Tang (SHXXT). Front. Pharmacol. 2016, 7, 374. [Google Scholar] [CrossRef]
- Yuan, Z.-W.; Leung, E.L.-H.; Fan, X.-X.; Zhou, H.; Ma, W.-Z.; Liu, L.; Xie, Y. Quantitative evaluation of berberine subcellular distribution and cellular accumulation in non-small cell lung cancer cells by UPLC-MS/MS. Talanta 2015, 144, 20–28. [Google Scholar] [CrossRef]
- Liu, S.P.; Yang, Z.; Liu, Z.F.; Liu, J.T.; Shi, Y. Resonance Rayleigh scattering study on the interaction of gold nanoparticles with berberine hydrochloride and its analytical application. Anal. Chim. Acta 2006, 572, 283–289. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, W. Sub-nanometre sized metal clusters: From synthetic challenges to the unique property discoveries. Chem. Soc. Rev. 2012, 41, 3594–3623. [Google Scholar] [CrossRef]
- Shang, L.; Dong, S.; Nienhaus, G.U. Ultra-small fluorescent metal nanoclusters: Synthesis and biological applications. Nano Today 2011, 6, 401–418. [Google Scholar] [CrossRef]
- Zhang, L.B.; Wang, E.K. Metal nanoclusters: New fluorescent probes for sensors and bioimaging. Nano Today 2014, 9, 132–157. [Google Scholar] [CrossRef]
- Tao, Y.; Li, M.Q.; Ren, J.S.; Qu, X.G. Metal nanoclusters: Novel probes for diagnostic and therapeutic applications. Chem. Soc. Rev. 2015, 44, 8636–8663. [Google Scholar] [CrossRef]
- Diez, I.; Ras, R.H.A. Fluorescent silver nanoclusters. Nanoscale 2011, 3, 1963–1970. [Google Scholar] [CrossRef]
- Richards, C.I.; Choi, S.; Hsiang, J.C.; Antoku, Y.; Vosch, T.; Bongiorno, A.; Tzeng, Y.L.; Dickson, R.M. Oligonucleotide-stabilized Ag nanocluster fluorophores. J. Am. Chem. Soc. 2008, 130, 5038–5039. [Google Scholar] [CrossRef]
- Zhou, B.; Khan, I.M.; Ding, X.; Niazi, S.; Zhang, Y.; Wang, Z. Fluorescent DNA-Silver nanoclusters in food safety detection: From synthesis to application. Talanta 2024, 273, 125834. [Google Scholar] [CrossRef]
- Pandya, A.; Lad, A.N.; Singh, S.P.; Shanker, R. DNA assembled metal nanoclusters: Synthesis to novel applications. RSC Adv. 2016, 6, 113095–113114. [Google Scholar] [CrossRef]
- Guo, W.; Yuan, J.; Dong, Q.; Wang, E. Highly Sequence-Dependent Formation of Fluorescent Silver Nanoclusters in Hybridized DNA Duplexes for Single Nucleotide Mutation Identification. J. Am. Chem. Soc. 2010, 132, 932–934. [Google Scholar] [CrossRef]
- Xu, L.J.; Guo, Y.H.; Wang, J.N.; Zhou, L.; Zhang, Y.Y.; Hong, S.N.; Wang, Z.L.; Zhang, J.Y.; Pei, R.J. A H+/Ag+ Dual-Target Responsive Label-Free Light-Up Probe Based on a DNA Triplex. Chem. Asian J. 2015, 10, 1126–1129. [Google Scholar] [CrossRef]
| Name | Sequences(5′-3′) |
|---|---|
| Hp-loop1 | CTTTCTTCCTTCCCCCCAAGGAAGAAAG |
| Hp-ss1 | TTCCTTCTTTC |
| Hp-loop2 | CTTCCTTCCCTCCCCCCAGGGAAGGAAG |
| Hp-ss2 | TCCCTTCCTTC |
| Hp-loop3 | CTTTTTTCTTTCCCCCCAAAGAAAAAAG |
| Hp-ss3 | TTTCTTTTTTC |
| Method | BBR Content (mg) | Average Content (mg) | RSD (%) |
|---|---|---|---|
| Ratiometric fluorescence with DNA-AgNCs | 29.90 | 29.80 | 2.71 |
| 30.64 | |||
| 29.02 | |||
| HPLC | 29.23 | 29.23 | 0.07 |
| 29.26 | |||
| 29.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, M.; Sun, M.; Liu, J.; Chen, C.; Yang, Y.; Teng, Y. A Ratiometric Fluorescence Detection Method for Berberine Using Triplex-Containing DNA-Templated Silver Nanoclusters. Molecules 2024, 29, 3459. https://doi.org/10.3390/molecules29153459
Zhu M, Sun M, Liu J, Chen C, Yang Y, Teng Y. A Ratiometric Fluorescence Detection Method for Berberine Using Triplex-Containing DNA-Templated Silver Nanoclusters. Molecules. 2024; 29(15):3459. https://doi.org/10.3390/molecules29153459
Chicago/Turabian StyleZhu, Ming, Mingyang Sun, Juntong Liu, Changbao Chen, Yonggang Yang, and Ye Teng. 2024. "A Ratiometric Fluorescence Detection Method for Berberine Using Triplex-Containing DNA-Templated Silver Nanoclusters" Molecules 29, no. 15: 3459. https://doi.org/10.3390/molecules29153459
APA StyleZhu, M., Sun, M., Liu, J., Chen, C., Yang, Y., & Teng, Y. (2024). A Ratiometric Fluorescence Detection Method for Berberine Using Triplex-Containing DNA-Templated Silver Nanoclusters. Molecules, 29(15), 3459. https://doi.org/10.3390/molecules29153459




