Long Non-Coding RNA AGAP2-AS1: A Comprehensive Overview on Its Biological Functions and Clinical Significances in Human Cancers
Abstract
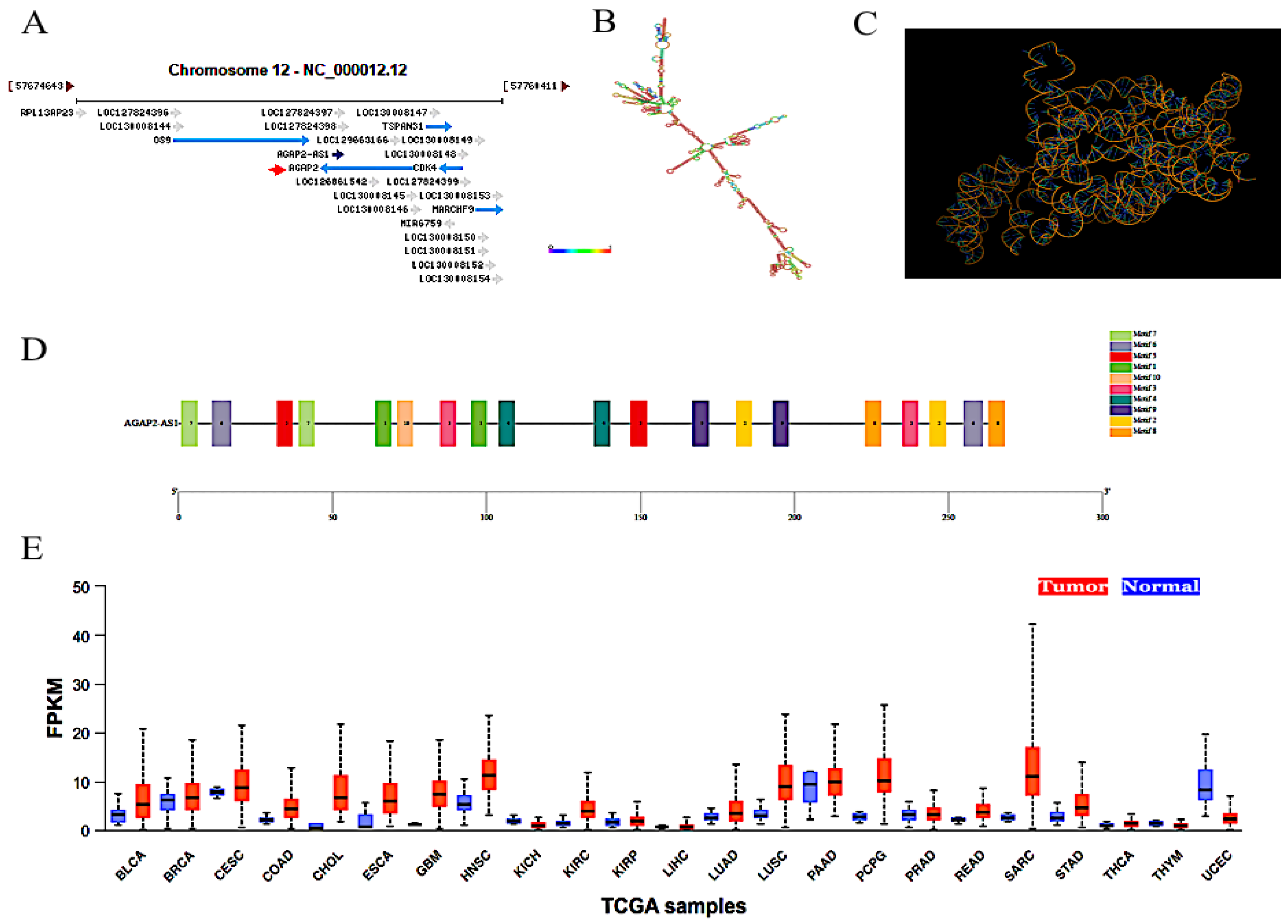
1. Introduction
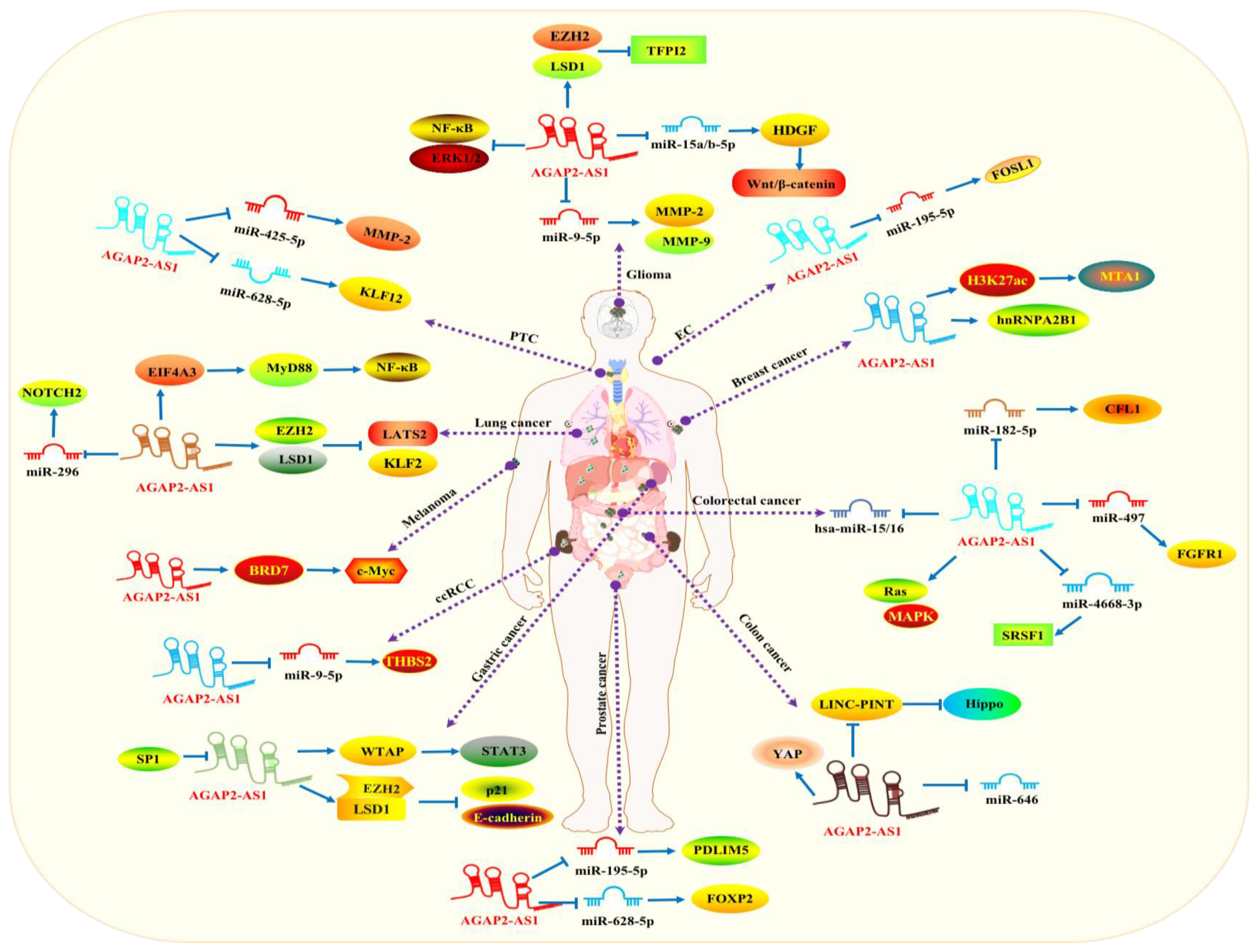
2. Biological Functions
2.1. Glioma
| Cell Lines | Expression Status | Role | Function | Regulatory Mechanism | Ref. |
|---|---|---|---|---|---|
| Glioma | |||||
| U87 and U251 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Metastasis ↑ Invasion ↓ Apoptosis | / | [19] |
| LN229 and U87MG cells | Upregulation | Oncogene | ↑ Proliferation ↑ Metastasis ↑ Invasion ↓ Apoptosis | / | [23] |
| U87, U251 and LN229 cell lines | Upregulation | Oncogene | ↑ Proliferation ↓ Apoptosis | miR-15a/b-5p/HDGF/Wnt/β-catenin axis | [26] |
| U87/MG and U251/MG cells | Upregulation | Oncogene | ↑ Proliferation ↑ Invasion ↓ Apoptosis ↑ Tumor growth | EZH2/LSD1/TFPI2 | [25] |
| / | Upregulation | Oncogene | ↑ Proliferation ↑ Metastasis ↑ Invasion | miR-9-5p/MMP-2/MMP-9 axis | [27] |
| T98G, U251 and LN229 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↑ Invasion | NF-κB/Erk1/2 | [28] |
| LATS2 and KLF2 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Metastasis ↑ Drug resistance ↓ Apoptosis | NOTCH, ERBB, RIG, NOD and JAK/STATA pathways | [29] |
| LC | |||||
| A549, ltp-2, SPCA1, H157 and NIH-H358 cell lines | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↑ Drug resistance | miR-296/NOTCH2 | [30] |
| MRC-5 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Metastasis | EIF4A3/MyD88/NF-κB pathway | [31] |
| H1299 and H1975 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Invasion ↑ Migration ↓ Apoptosis | LATS2/KLF2/EZH2/ LSD1 | [32] |
| CRC | |||||
| LoVo and SW480 cells | Upregulation | Oncogene | ↑ Growth ↑ Migration ↑ Invasion ↑ EMT | E2F4/miR-182-5p/CFL1 axis | [20] |
| DLD-1 and RKO cells | Upregulation | Oncogene | ↑ Proliferation ↓ Apoptosis ↑ Migration ↑ Invasion | miR-497/FGFR1 axis | [33] |
| SW480 and HT29 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration | hsa-miR-15/16 family | [34] |
| DLD-1 and HT29 cells | Upregulation | Oncogene | ↑ Proliferation | Ras/MAPK pathway | [35] |
| SW620 and HT-29 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↑ Invasion ↑ EMT | miR-4668-3p/SRSF1 axis | [36] |
| CLC | |||||
| RKO and HCT116 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Invasion ↑ Migration | LINC-PINT/Hippo signaling | [37] |
| SW480 and HCT-116 cells | Upregulation | Oncogene | ↑ Proliferation ↓ Apoptosis | / | [38] |
| HCT116 cells | Upregulation | Oncogene | ↑ Proliferation ↓ Apoptosis | miR-646 | [39] |
| SW480 and HCT-116 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↓ Apoptosis | YAP pathway | [40] |
| OC | |||||
| OVCAR3 and A2780 cells | Upregulation | Oncogene | ↑ Proliferation | MEG3 | [15] |
| SKOV3-ip, OVCAR3, HO8910, HEY and ES2 cell lines | Downregulation | Anti-oncogene | ↓ Proliferation ↓ Migration ↓ Invasion | KRAS/FGFR4/CTSK/EMT | [16] |
| PCa | |||||
| VCaP, 22Rv1, CRL-1740, CRL-2422 and PC3M cell lines | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↑ Invasion ↑ Tumor growth | miR-195-5p/PDLIM5 axis | [41] |
| PCa cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↑ Invasion ↑ EMT ↓ Apoptosis | miR-628-5p/FOXP2/WNT axis | [42] |
| BC | |||||
| MCF-7, BT-474, SK-BR-3 and MDA-MB-231 cell lines | Upregulation | Oncogene | ↑ Proliferation ↓ Apoptosis | HuR/H3K27ac/MTA1 | [43] |
| SKBR-3 and BT474 cells | Upregulation | Oncogene | ↑ Cell viability ↑ Trastuzumab resistance | hnRNPA2B1 | [12] |
| CHOL | |||||
| RBE and HuCCT1 cells | Upregulation | Oncogene | ↑ Proliferation ↓ Apoptosis | EZH2/CDKN1A | [44] |
| RBE and HuCCT1 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↑ Invasion | / | [45] |
| BLCA | |||||
| UM-UC-3 and T24 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration | / | [46] |
| 5637 and T24 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↑ Invasion ↑ Angiogenesis ↑ Tumor growth | IGF2BP2/LRG1 | [47] |
| RCC | |||||
| 786-O and ACHN cells | Upregulation | Oncogene | ↑ M2 polarization ↑ Malignant behavior ↑ Proliferation ↑ Tumor growth | miR-9-5p/THBS2/PI3K-AKT axis | [48] |
| PTC | |||||
| K1 and TPC1 cells | Upregulation | Oncogene | ↑ Migration ↑ Invasion | miR-425-5p/MMP-2 axis | [49] |
| BCPAP, KTC-1, NIM1 and TPC1 cell lines | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↑ Invasion | miR-628-5p/KLF12 axis | [50] |
| GC | |||||
| AGS and HGC-27 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration | WTAP/METTL3/METTL14/STAT3 axis | [51] |
| BGC-823 and AGS cells | Upregulation | Oncogene | ↑ Proliferation ↑ Migration ↑ Invasion ↑ Tumor growth | LSD1/EZH2/CDKN1A/E-cadherin transcription | [52] |
| EC | |||||
| KYSE70, KYSE-510 and EC9706 cell lines | Upregulation | Oncogene | ↑ Proliferation ↓ Apoptosis ↑ Migration ↑ Invasion ↑ Tumorigenesis | miR-195-5p/FOSL1 axis | [53] |
| Melanoma | |||||
| A375 and A875 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Colony formation ↑ Migration | BRD7/c-Myc pathway | [54] |
| LSCC | |||||
| AMC-HN-8 and Tu-177 cells | Upregulation | Oncogene | ↑ Proliferation ↑ Invasion | miR-193a-3p/LOXL4 axis | [55] |
| PC | |||||
| AsPC-1 and BxPC-3 cells | Upregulation | Oncogene | ↑ Invasion ↑ Proliferation ↑ Migration | ANKRD1/ANGPTL4 | [56] |
2.2. Colorectal Carcinoma (CRC)
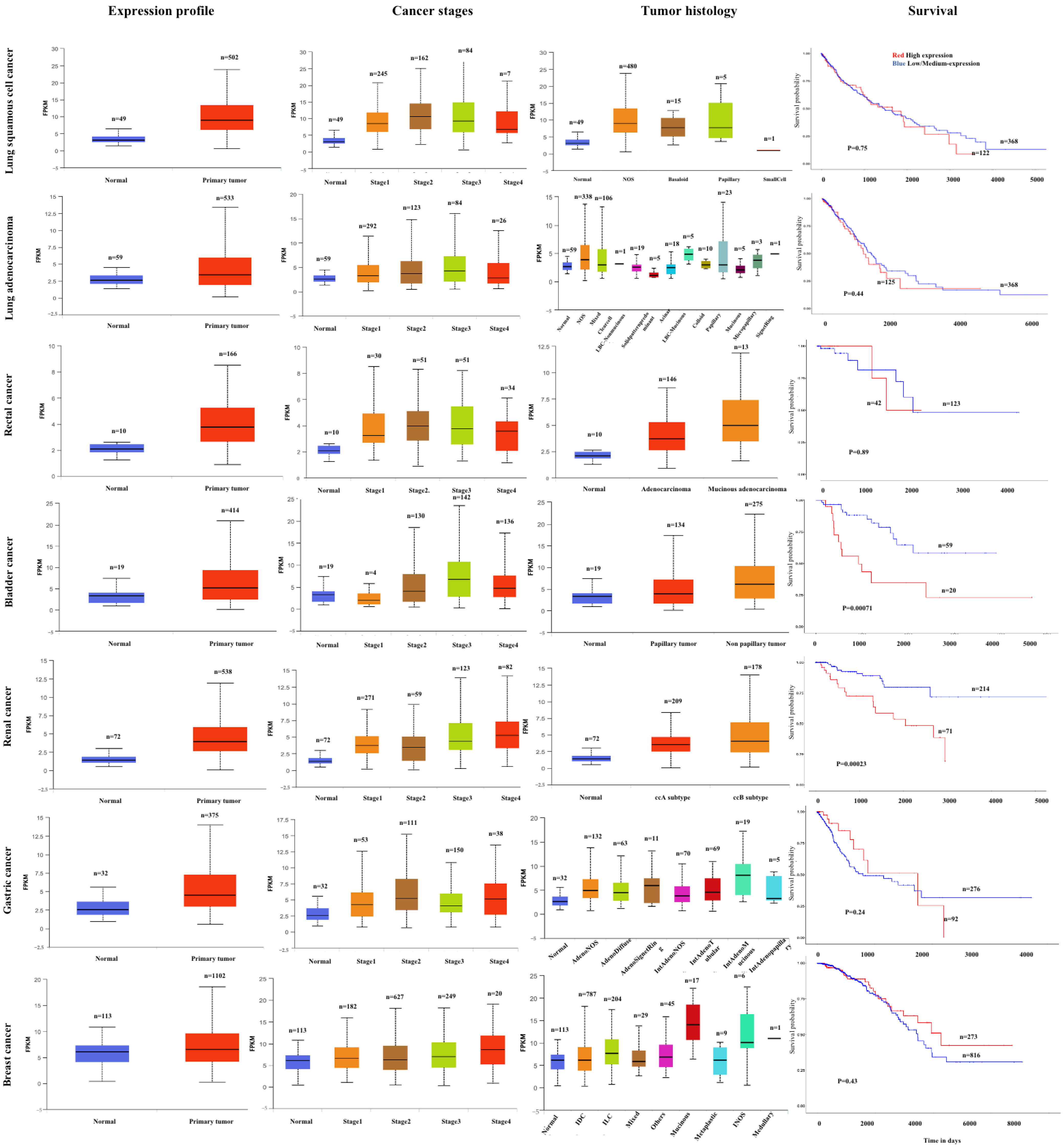
2.3. Lung Carcinoma (LC)
2.4. Ovarian Carcinoma (OC)
2.5. Prostate Cancer (PCa)
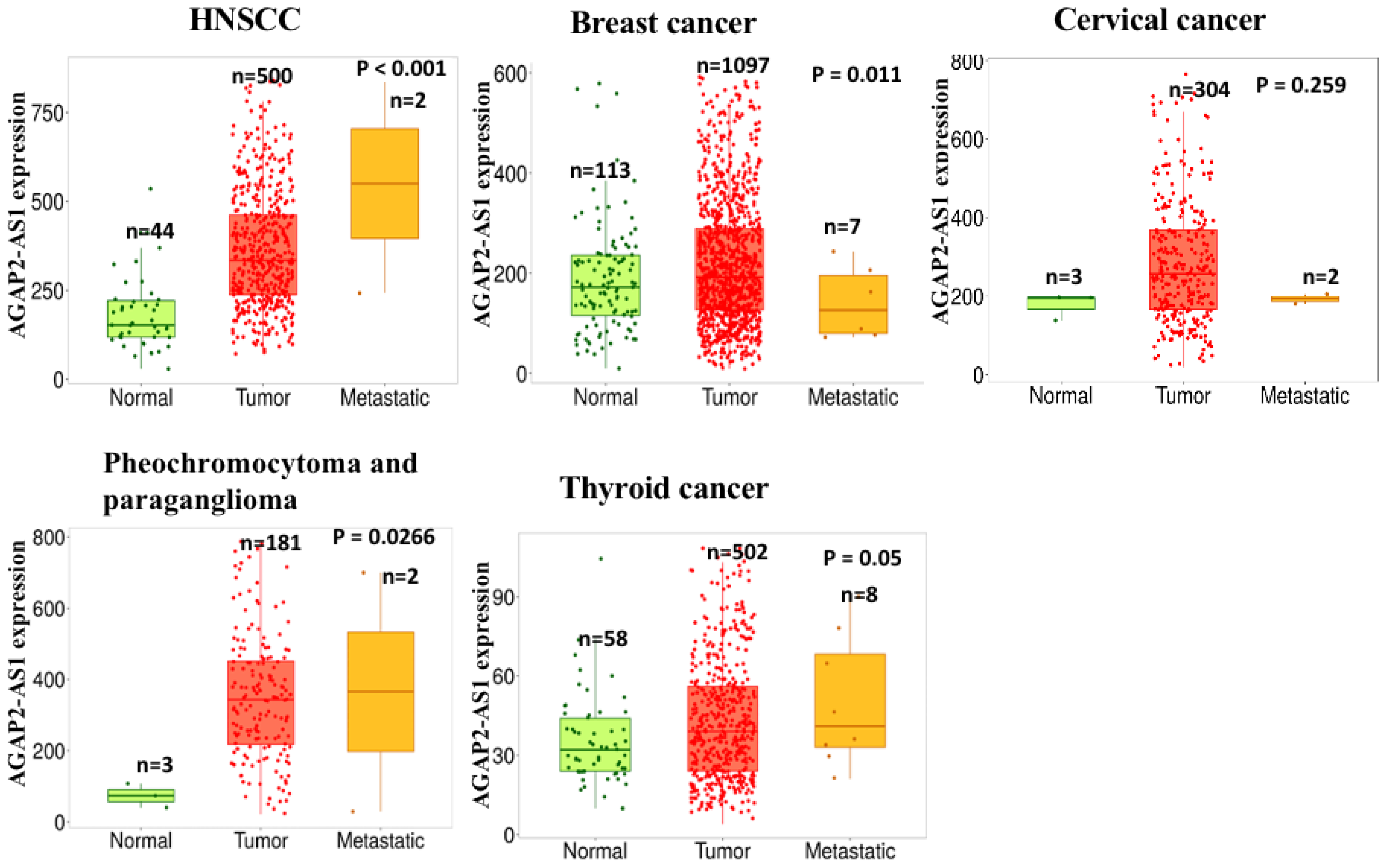
2.6. Breast Carcinoma (BC)
2.7. Cholangiocarcinoma (CHOL)
2.8. Bladder Cancer (BLCA)
2.9. Colon Cancer (CLC)
2.10. Pancreatic Cancer (PC)
2.11. Renal Cell Carcinoma (RCC)
2.12. Laryngeal Squamous Cell Carcinoma (LSCC)
2.13. Melanoma
2.14. Papillary Thyroid Cancer (PTC)
2.15. Esophageal Cancer (EC)
2.16. Gastric Cancer (GC)
3. Clinical Significances
3.1. Gliomas
3.2. Lung Carcinoma
| Cancer | Property | Samples | Clinic-Pathological Features | Ref. |
|---|---|---|---|---|
| Gliomas | Oncogene | GEO: 9 paired tissues TCGA: 169 GBM cases and 5 normal samples | Correlated with occurrence and development of glioblastoma. | [27] |
| Oncogene | GSE16011, CGGA and REMBRANDT datasets | Correlated with TG. | [23] | |
| Oncogene | CGGA dataset: 51 paired tissues | Correlated with TG. | [67] | |
| Oncogene | 136 cancer tissues and 20 normal tissues | Negatively correlated with OS. | [19] | |
| Oncogene | 91 paired tissues | Associated with the advanced TG. | [26] | |
| Oncogene | 58 paired tissues | Correlated with OS. | [25] | |
| LC | Oncogene | 121 LC patients | Related to the TNM stage and LNM. | [30] |
| Oncogene | 198 paired tissues | Correlated with TG and LNM. | [73] | |
| Oncogene | 150 patients with NSCLC, including 86 ADCs and 64 SCCs 150 healthy controls | Correlated with LNM and TNM stage. | [71] | |
| Oncogene | 80 NSCLC patients | Served as a potential independent prognostic value in NSCLC. | [32] | |
| Oncogene | 120 patients with NSCLC 60 healthy controls | Correlated with clinical stage, tumor infiltration and LNM. | [74] | |
| Oncogene | 82 paired tissues | Correlated with occurrence and development of NSCLC. | [72] | |
| Oncogene | 84 NSCLC patients 60 healthy controls | Correlated with LNM, TNM stage and differentiation degree. | [75] | |
| Oncogene | 535 tumor samples and 59 normal samples | Correlated with total patient survival. | [69] | |
| Oncogene | 14 smokers, 17 NSCLC patients and 14 healthy subjects | Correlated with the development of NSCLC. | [70] | |
| OC | Oncogene | 82 paired tissues | Correlated with clinical stage and the expression levels of MEG3. | [15] |
| Anti-oncogene | 80 cancerous tissues and 10 normal tissues | Associated with advanced FIGO stage, high HG, LNM and serous subtype. | [16] | |
| PTC | Oncogene | 110 paired tissues | Associated with invasion and migration. | [49] |
| Oncogene | 31 paired tissues | Not stated. | [50] | |
| CHOL | Oncogene | TCGA and GEO databases (not stated) | Correlated with metabolism-related mechanisms in tumorigenesis. | [76] |
| Oncogene | TCGA: 36 cancer tissues and 9 normal tissues | Correlated with survival time. | [44] | |
| Oncogene | 9 paired tissues | Correlated with survival time. | [45] | |
| BLCA | Oncogene | 33 paired tissues | Associated with higher possibility of recurrence. | [46] |
| Oncogene | 45 paired tissues | Positively correlated with T stage, grade and vascular invasion; negatively correlated with the survival. | [47] | |
| RC | Oncogene | 50 paired tissues | Abnormally overexpressed. | [35] |
| Oncogene | 70 paired tissues | Abnormally overexpressed. | [20] | |
| Oncogene | 116 paired tissues | Highly correlated to TG. | [33] | |
| Oncogene | 100 paired tissues | No association with the clinicopathological characteristics. | [34] | |
| CLC | Oncogene | TCGA: 457 cancer samples and 42 healthy samples 20 paired tissues | Aberrantly overexpressed. | [40] |
| Oncogene | 66 paired tissues | Not significantly correlated with TG. | [37] | |
| ccRCC | Oncogene | 539 ccRCC tissues and 72 adjacent healthy tissues | Associated with poor OS. | [77] |
| Oncogene | 443 paired tissues | Correlated with overall unfavorable survival outcome. | [18] | |
| Oncogene | 50 paired tissues | Associated with poor survival and prognosis. | [48] | |
| Melanoma | Oncogene | 468 cancer tissues and 555 corresponding normal tissues | Associated with shorter OS, disease-specific survival and PFI times. | [54] |
| GC | Oncogene | GSE13911: 69 paired tissues GSE54129: 27 paired tissues | Associated with adverse OS and PFS in advanced-stage (III–IV) GC. | [51] |
| Oncogene | 50 paired tissues | Associated with larger tumors, advanced pathological stage and LNM. | [52] | |
| PC | Oncogene | 16 paired tissues 46 paired tissues | Associated with poor prognosis. | [56] |
| LSCC | Oncogene | 23 paired tissues | Associated with the metastasis of LSCC. | [55] |
| BC | Oncogene | 78 paired tissues | Associated with differentiate malignant states. | [78] |
| Oncogene | 30 paired tissues | Aberrantly overexpressed. | [43] | |
| PCa | Oncogene | 50 cancerous tissues and 20 benign tissues | Aberrantly overexpressed. | [41] |
3.3. OC
3.4. PTC
3.5. CHOL
3.6. BLCA
3.7. Rectal Cancer (RC)
3.8. PC
3.9. CLC
3.10. ccRCC
3.11. Melanoma
3.12. GC
3.13. BC
3.14. Other Cancers
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Wang, N.; Yu, Y.; Xu, B.; Zhang, M.; Li, Q.; Miao, L. Pivotal prognostic and diagnostic role of the long non-coding RNA colon cancer-associated transcript 1 expression in human cancer (Review). Mol. Med. Rep. 2019, 19, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; An, Y.; Liang, Y.; Xie, X.W. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1930–1936. [Google Scholar]
- Chen, B.; Li, J.; Chi, D.; Sahnoune, I.; Calin, S.; Girnita, L.; Calin, G.A. Non-coding RNAs in IGF-1R signaling regulation: The underlying pathophysiological link between diabetes and cancer. Cells 2019, 8, 1638. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, J.; Huang, Z.; Jin, P.; Peng, L.; Luo, M.; Zhang, Z.; Chen, Y.; Xie, N.; Gao, W.; et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/beta-catenin signaling to promote colorectal cancer progression by preventing m(6)A-mediated degradation of STEAP3 mRNA. Mol. Cancer 2022, 21, 168. [Google Scholar] [CrossRef] [PubMed]
- Malla, R.R. Long noncoding RNAs: Potential mediators of liver cancer metastasis. Crit. Rev. Oncog. 2021, 26, 21–33. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, M.; Weng, J.; Li, S.; Guo, T.; Guo, Y.; Xu, Y. LncRNA GAS6-AS1 contributes to 5-fluorouracil resistance in colorectal cancer by facilitating the binding of PCBP1 with MCM3. Cancer Lett. 2024, 589, 216828. [Google Scholar] [CrossRef]
- Jiang, T.; Qi, J.; Xue, Z.; Liu, B.; Liu, J.; Hu, Q.; Li, Y.; Ren, J.; Song, H.; Xu, Y.; et al. The m(6)A modification mediated-lncRNA POU6F2-AS1 reprograms fatty acid metabolism and facilitates the growth of colorectal cancer via upregulation of FASN. Mol. Cancer 2024, 23, 55. [Google Scholar] [CrossRef]
- Yang, L.; Wang, M.; Wang, Y.; Zhu, Y.; Wang, J.; Wu, M.; Guo, Q.; Han, X.; Pandey, V.; Wu, Z.; et al. LINC00460-FUS-MYC feedback loop drives breast cancer metastasis and doxorubicin resistance. Oncogene 2024, 43, 1249–1262. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Chen, M.; Xing, P.; Yan, X.; Xie, B. Increased expression of exosomal AGAP2-AS1 (AGAP2 antisense RNA 1) in breast cancer cells inhibits trastuzumab-induced cell cytotoxicity. Med. Sci. Monit. 2019, 25, 2211–2220. [Google Scholar] [CrossRef] [PubMed]
- Xian, J.; Shang, M.; Dai, Y.; Wang, Q.; Long, X.; Li, J.; Cai, Y.; Xia, C.; Peng, X. N(6)-methyladenosine-modified long non-coding RNA AGAP2-AS1 promotes psoriasis pathogenesis via miR-424-5p/AKT3 axis. J. Dermatol. Sci. 2022, 105, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xia, X.; Jiang, Y.; Wu, D.; Wang, S.; Fu, S.; Yang, N.; Zhang, Y.; Sun, L. Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. J. Cell Mol. Med. 2020, 24, 4557–4568. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Peng, X.; Dai, Y. The Long non-coding RNA (lncRNA) AGAP2-AS1 is upregulated in ovarian carcinoma and negatively regulates lncRNA MEG3. Med. Sci. Monit. 2019, 25, 4699–4704. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Lin, X.; Tang, X.; Hua, K.; Qiu, J. The antisense long noncoding RNA AGAP2-AS1 regulates cell proliferation and metastasis in epithelial ovarian cancer. J. Cancer 2020, 11, 5318–5328. [Google Scholar]
- Zhong, P.; Hua, H.; Chen, S.; Zhu, Z.; Xie, F. The prognostic value of lncRNA AGAP2-AS1 in cancer patients: A meta-analysis. Medicine 2021, 100, e28425. [Google Scholar] [CrossRef] [PubMed]
- Nakken, S.; Eikrem, O.; Marti, H.P.; Beisland, C.; Bostad, L.; Scherer, A.; Flatberg, A.; Beisvag, V.; Skandalou, E.; Furriol, J.; et al. AGAP2-AS1 as a prognostic biomarker in low-risk clear cell renal cell carcinoma patients with progressing disease. Cancer Cell Int. 2021, 21, 690. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zheng, Y.; Dong, X. AGAP2-AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J. Cell Biochem. 2019, 120, 9056–9062. [Google Scholar] [CrossRef]
- Guo, Z.; Liu, X.; Shao, H. E2F4-induced AGAP2-AS1 up-regulation accelerates the progression of colorectal cancer via miR-182-5p/CFL1 axis. Dig. Liver Dis. 2022, 54, 878–889. [Google Scholar] [CrossRef]
- Liu, Y.; Ali, H.; Khan, F.; Pang, L.; Chen, P. Epigenetic regulation of tumor-immune symbiosis in glioma. Trends Mol. Med. 2024, 30, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Nafari, A.; Ghaffary, E.M.; Shaygannejad, V.; Mirmosayyeb, O. Concurrent glioma and multiple sclerosis: A systematic review of case reports. Mult. Scler. Relat. Disord. 2024, 84, 105455. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, F.; Zhang, L.; Chen, J.; Zhao, Z.; Wang, H.; Wu, F.; Liang, T.; Yan, X.; Li, J.; et al. LncRNA profile study reveals four-lncRNA signature associated with the prognosis of patients with anaplastic gliomas. Oncotarget 2016, 7, 77225–77236. [Google Scholar] [CrossRef]
- Steponaitis, G.; Stakaitis, R.; Valiulyte, I.; Krusnauskas, R.; Dragunaite, R.; Urbanaviciute, R.; Tamasauskas, A.; Skiriute, D. Transcriptome-wide analysis of glioma stem cell specific m6A modifications in long-non-coding RNAs. Sci. Rep. 2022, 12, 5431. [Google Scholar] [CrossRef]
- Luo, W.; Li, X.; Song, Z.; Zhu, X.; Zhao, S. Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in glioblastoma by epigenetically silencing TFPI2 through EZH2 and LSD1. Aging 2019, 11, 3811–3823. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Lu, S.; Xu, Y.; Zheng, J. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/beta-catenin signaling pathway. Int. J. Biol. Macromol. 2019, 128, 521–530. [Google Scholar] [CrossRef]
- Luo, X.; Tu, T.; Zhong, Y.; Xu, S.; Chen, X.; Chen, L.; Yang, F. AGAP2-AS1 may promote the occurrence and development of glioblastoma by sponging miR-9-5p: Evidence from a ceRNA network. Front. Oncol. 2021, 11, 607989. [Google Scholar] [CrossRef]
- Duan, W.W.; Yang, L.T.; Liu, J.; Dai, Z.Y.; Wang, Z.Y.; Zhang, H.; Zhang, X.; Liang, X.S.; Luo, P.; Zhang, J.; et al. A TGF-beta signaling-related lncRNA signature for prediction of glioma prognosis, immune microenvironment, and immunotherapy response. CNS Neurosci. Ther. 2023, 30, e14489. [Google Scholar] [CrossRef]
- Yu, W.; Ma, Y.; Hou, W.; Wang, F.; Cheng, W.; Qiu, F.; Wu, P.; Zhang, G. Identification of immune-related lncRNA prognostic signature and molecular subtypes for glioblastoma. Front. Immunol. 2021, 12, 706936. [Google Scholar] [CrossRef]
- Zhang, F.; Sang, Y.; Chen, D.; Wu, X.; Wang, X.; Yang, W.; Chen, Y. M2 macrophage-derived exosomal long non-coding RNA AGAP2-AS1 enhances radiotherapy immunity in lung cancer by reducing microRNA-296 and elevating NOTCH2. Cell Death Dis. 2021, 12, 467. [Google Scholar] [CrossRef]
- Xu, Q.; Zhao, T.; Han, H.; Fan, J.; Xie, W. EIF4A3 stabilizes the expression of lncRNA AGAP2-AS1 to activate cancer-associated fibroblasts via MyD88/NF-kappaB signaling. Thorac. Cancer 2023, 14, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, M.; Zang, C.; Ma, P.; He, J.; Zhang, M.; Huang, Z.; Ding, Y.; Shu, Y. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016, 7, e2225. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Yan, Z.; Song, Y.; Bi, M.; Li, S. LncRNA AGAP2-AS1 augments cell viability and mobility, and confers gemcitabine resistance by inhibiting miR-497 in colorectal cancer. Aging 2020, 12, 5183–5194. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, T.; Khalaj-Kondori, M.; Hosseinpour Feizi, M.A.; Asadi, P. Long non-coding RNA AGAP2-AS1 is up regulated in colorectal cancer. Nucleos Nucleot Nucl. 2021, 40, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Peng, W.; Li, J.; Gu, Q.; Fu, Z. AGAP2-AS1 promotes colorectal cancer proliferation through Ras/MAPK signaling pathway. Acta Univ. Med. Nanjing 2019, 39, 16–20. [Google Scholar]
- Li, H.; Guo, S.; Zhang, M.; Li, L.; Wang, F.; Song, B. Long non-coding RNA AGAP2-AS1 accelerates cell proliferation, migration, invasion and the EMT process in colorectal cancer via regulating the miR-4,668-3p/SRSF1 axis. J. Gene Med. 2020, 22, e3250. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Chen, S.; Gu, L.; Wang, J.; Zhang, X. LncRNA AGAP2-AS1 promotes cancer cell proliferation, migration and invasion in colon cancer by forming a negative feedback loop with LINC-PINT. Cancer Manag. Res. 2021, 13, 2153–2161. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wu, S. Effects of AGAP2 -AS1 on proliferation and apoptosis of colon cancer cells. China Mod. Doc. 2020, 58, 20–23. [Google Scholar]
- Liu, J.; Yang, H.; Song, C. Effects of proliferation inhibition and apoptosis induction on colon cancer cells by Tribulus terrestris saponins via lncRNA AGAP2-AS1/miR-646 regulation. Chin. Tradit. Pat. Med. 2021, 43, 356–362. [Google Scholar]
- Jin, T.; Lei, X.; Tong, X. Long non-coding RNA AGAP2-AS1 affects biological behavior of colon cancer cells through the YAP signaling pathway. Chin. J. Clin. Oncol. 2020, 47, 601–608. [Google Scholar]
- Xie, P.; Liu, M.; Chen, F.; Wu, S.; Shao, T.; Wang, W.; Xu, C.; Zhou, H. Long non-coding RNA AGAP2-AS1 silencing inhibits PDLIM5 expression impeding prostate cancer progression via up-regulation of microRNA-195-5p. Front. Genet. 2020, 11, 1030. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Y.; Luo, C.; Zuo, Y. AGAP2-AS1/miR-628-5p/FOXP2 feedback loop facilitates the growth of prostate cancer via activating WNT pathway. Carcinogenesis 2021, 42, 1270–1280. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wen, L.; Zhou, Y.; Wu, W. Role of lncRNA AGAP2-AS1 in breast cancer cell resistance to apoptosis by the regulation of MTA1 promoter activity. Technol. Cancer Res. Treat. 2022, 21, 15330338221085361. [Google Scholar] [CrossRef]
- Ji, H.; Wang, J.; Lu, B.; Li, J.; Zhou, J.; Wang, L.; Xu, S.; Peng, P.; Hu, X.; Wang, K. SP1 induced long non-coding RNA AGAP2-AS1 promotes cholangiocarcinoma proliferation via silencing of CDKN1A. Mol. Med. 2021, 27, 10. [Google Scholar] [CrossRef]
- Xu, L.; Xiao, T.; Xu, L.; Yao, W. Identification of therapeutic targets and prognostic biomarkers in cholangiocarcinoma via WGCNA. Front. Oncol. 2022, 12, 977992. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, H.; Sun, J.; Mao, W.; Chen, S.; Chen, M. Multi-Omics analysis identifies a lncRNA-related prognostic signature to predict bladder cancer recurrence. Bioengineered 2021, 12, 11108–11125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, J.; Zhang, C.; Xie, G.; Othmane, B.; Kuang, X.; Liu, B. LncRNA AGAP2-AS1 interacts with IGF2BP2 to promote bladder cancer progression via regulating LRG1 mRNA stability. Cell. Signal. 2023, 111, 110839. [Google Scholar] [CrossRef]
- Xu, P.; Feng, D.X.; Wang, J.; Wang, Y.D.; Xie, G.; Zhang, B.; Li, X.H.; Zeng, J.W.; Feng, J.F. LncRNA AGAP2 antisense RNA 1 stabilized by insulin-like growth factor 2 mRNA binding protein 3 promotes macrophage M2 polarization in clear cell renal cell carcinoma through regulation of the microRNA-9-5p/THBS2/PI3K-Akt pathway. Cancer Cell Int. 2023, 23, 330. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Sun, W.; Zhang, H.; Zhang, P.; Wang, Z.; Dong, W.; He, L.; Zhang, T.; Qin, Y. Long non-coding RNA AGAP2-AS1 increases the invasiveness of papillary thyroid cancer. Aging 2020, 12, 18019–18032. [Google Scholar] [CrossRef]
- Xu, C.; Shao, Y.; Liu, J.; Yao, X.; Quan, F.; Zhao, Q.; Zhao, R.; Kou, B.; Li, H.; Han, P.; et al. Long non-coding RNA AGAP2-AS1 promotes proliferation and metastasis in papillary thyroid cancer by miR-628-5p/KLF12 axis. J. Bioenerg. Biomembr. 2021, 53, 235–245. [Google Scholar] [CrossRef]
- Nie, K.; Zheng, Z.; Li, J.; Chang, Y.; Deng, Z.; Huang, W.; Li, X. AGAP2-AS1 promotes the assembly of m6A methyltransferases and activation of the IL6/STAT3 pathway by binding with WTAP in the carcinogenesis of gastric cancer. FASEB J. 2023, 37, e23302. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Liu, X.; Wu, H.; Yu, X.; Wei, C.; Huang, X.; Ji, G.; Nie, F.; Wang, K. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J. Hematol. Oncol. 2017, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Li, K.; Liu, Y.; Liu, X.; Liu, B.; Ba, Y.; Xing, W. Silencing lncRNA AGAP2-AS1 upregulates miR-195-5p to repress migration and invasion of EC cells via the decrease of FOSL1 expression. Mol. Ther. Nucleic Acids 2020, 20, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, S.; Xu, J.; Shen, C.; Qian, Q. AGAP2-AS1/BRD7/c-Myc signaling axis promotes skin cutaneous melanoma progression. Am. J. Transl. Res. 2023, 15, 350–362. [Google Scholar] [PubMed]
- Ren, P.; Niu, X.; Zhao, R.; Liu, J.; Ren, W.; Dai, H.; Chen, J.; Yan, J.; Li, B.; Shao, Y.; et al. Long non-coding RNA AGAP2-AS1 promotes cell proliferation and invasion through regulating miR-193a-3p/LOXL4 axis in laryngeal squamous cell carcinoma. Cell Cycle 2022, 21, 697–707. [Google Scholar] [CrossRef]
- Hui, B.; Ji, H.; Xu, Y.; Wang, J.; Ma, Z.; Zhang, C.; Wang, K.; Zhou, Y. RREB1-induced upregulation of the lncRNA AGAP2-AS1 regulates the proliferation and migration of pancreatic cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death Dis. 2019, 10, 207. [Google Scholar] [CrossRef]
- Rastin, F.; Javid, H.; Oryani, M.A.; Rezagholinejad, N.; Afshari, A.R.; Karimi-Shahri, M. Immunotherapy for colorectal cancer: Rational strategies and novel therapeutic progress. Int. Immunopharmacol. 2024, 126, 111055. [Google Scholar] [CrossRef] [PubMed]
- Latimer, K.M.; Mott, T.F. Lung cancer: Diagnosis, treatment principles, and screening. Am. Fam. Physician 2015, 91, 250–256. [Google Scholar] [PubMed]
- Davidson, B.; Trope, C.G.; Reich, R. The clinical and diagnostic role of microRNAs in ovarian carcinoma. Gynecol. Oncol. 2014, 133, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Skotheim, R.I.; Bogaard, M.; Carm, K.T.; Axcrona, U.; Axcrona, K. Prostate cancer: Molecular aspects, consequences, and opportunities of the multifocal nature. Biochim. Biophys. Acta Rev. Cancer 2024, 1879, 189080. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Canadian Cancer Statistics Advisory Committee. Canadian Cancer Statistics: A 2022 Special Report on Cancer Prevalence; Canadian Cancer Society: Toronto, ON, Canada, 2022. [Google Scholar]
- Latenstein, A.E.J.; van der Geest, L.G.M.; Bonsing, B.A.; Groot Koerkamp, B.; Haj Mohammad, N.; de Hingh, I.; de Meijer, V.E.; Molenaar, I.Q.; van Santvoort, H.C.; van Tienhoven, G.; et al. Nationwide trends in incidence, treatment and survival of pancreatic ductal adenocarcinoma. Eur. J. Cancer 2020, 125, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Bartha, A.; Gyorffy, B. TNMplot.com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, Z.; Yang, F.; Wang, H.; Wu, F.; Liang, T.; Yan, X.; Li, J.; Lan, Q.; Wang, J.; et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J. Neurooncol. 2018, 136, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pan, Y.; Gao, H.; Yi, Y.; Qin, S.; Ma, F.; Zhou, X.; Guan, M. Mechanistic insights into super-enhancer-driven genes as prognostic signatures in patients with glioblastoma. J. Cancer Res. Clin. Oncol. 2023, 149, 12315–12332. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Chen, Z.; Chen, Z.; Chen, J.; Liu, M.; Xu, X.; Liu, Y.; Yang, S.; Hu, Z.; He, F. Construction of an immune-related lncRNA signature pair for predicting oncologic outcomes and the sensitivity of immunosuppressor in treatment of lung adenocarcinoma. Respir. Res. 2022, 23, 123. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yin, Z.; Yang, L.; Fan, J.; Xu, J.; Jin, Y.; Yu, J.; Zhang, D.; Yang, G. Smoking induced extracellular vesicles release and their distinct properties in non-small cell lung cancer. J. Cancer 2019, 10, 3435–3443. [Google Scholar] [CrossRef]
- Tao, Y.; Tang, Y.; Yang, Z.; Wu, F.; Wang, L.; Yang, L.; Lei, L.; Jing, Y.; Jiang, X.; Jin, H.; et al. Exploration of serum exosomal lncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of non-small cell lung cancer. Int. J. Biol. Sci. 2020, 16, 471–482. [Google Scholar] [CrossRef]
- Ma, X.; Wu, J.; Zhu, H.; Zhang, Z.; Li, X.; Liu, Z. Relationship between KLF9, PTPRJ, AGAP2-AS1 expression levels in non-small cell lung cancer tissues and clinicopathological features and prognosis. J. Qiqihar Med. Univ. 2021, 42, 1387–1391. [Google Scholar]
- Fan, K.J.; Liu, Y.; Yang, B.; Tian, X.D.; Li, C.R.; Wang, B. Prognostic and diagnostic significance of long non-coding RNA AGAP2-AS1 levels in patients with non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2392–2396. [Google Scholar]
- Zhang, X.; Wei, H.; Wang, W.; Shi, J.; Zhang, Y. Changes of serum exosomes miR-152-5p and AGAP2-AS1 levels and their evaluation value for prognosis in non-small cell lung cancer. Med. J. West. China 2021, 33, 1844–1853. [Google Scholar]
- Wu, C.; Fu, F.; Lin, W.; Guo, N.; Ma, X. Expression of serum lipocalin-2, periodin and agap2-as1 in non-small cell lung cancer and their correlation with pathological features. Prog. Mod. Biomed. 2022, 22, 3577–3581. [Google Scholar]
- Zou, W.; Wang, Z.; Wang, F.; Li, L.; Liu, R.; Hu, M. A metabolism-related 4-lncRNA prognostic signature and corresponding mechanisms in intrahepatic cholangiocarcinoma. BMC Cancer 2021, 21, 608. [Google Scholar] [CrossRef]
- Gao, L.; Zhao, A.; Wang, X. Upregulation of lncRNA AGAP2-AS1 is an independent predictor of poor survival in patients with clear cell renal carcinoma. Oncol. Lett. 2020, 19, 3993–4001. [Google Scholar] [CrossRef] [PubMed]
- Mohebi, M.; Ghafouri-Fard, S.; Modarressi, M.H.; Dashti, S.; Zekri, A.; Kholghi-Oskooei, V.; Taheri, M. Expression analysis of vimentin and the related lncRNA network in breast cancer. Exp. Mol. Pathol. 2020, 115, 104439. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Cai, B.; Chen, J.; Zhao, K.; Li, M.; Lang, J.; Wang, K.; Pan, S.; Zhu, K. A Notch signaling-related lncRNA signature for predicting prognosis and therapeutic response in clear cell renal cell carcinoma. Sci. Rep. 2023, 13, 21141. [Google Scholar] [CrossRef] [PubMed]
- Hisey, C.L.; Tomek, P.; Nursalim, Y.N.S.; Chamley, L.W.; Leung, E. Towards establishing extracellular vesicle-associated RNAs as biomarkers for HER2+ breast cancer. F1000Research 2020, 9, 1362. [Google Scholar] [CrossRef]
- Liu, F.; Chu, H.X.; Han, J.S.; Sun, X.; Chen, J.; Qiu, X.L.; Zheng, X.H.; Jia, B.; Zhao, J.J. Inhibitory effect of the Notch pathway-inhibitor DAPT on invasion and metastasis of tongue cancer via lncRNA-KAT14 regulation. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 189–199. [Google Scholar]
- Dai, W.; Mu, L.; Cui, Y.; Li, Y.; Chen, P.; Xie, H.; Wang, X. Berberine promotes apoptosis of colorectal cancer via regulation of the long non-coding RNA (lncRNA) cancer susceptibility candidate 2 (CASC2)/AU-binding factor 1 (AUF1)/B-cell CLL/lymphoma 2 (Bcl-2) axis. Med. Sci. Monit. 2019, 25, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Huang, Z.; Qian, C.; Wang, M.; Zheng, Y.; Jiang, R.; Yu, C. LncRNA WEE2-AS1 promotes proliferation and inhibits apoptosis in triple negative breast cancer cells via regulating miR-32-5p/TOB1 axis. Biochem. Biophys. Res. Commun. 2020, 526, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, F.; Zhang, B.; Wang, Y.; Lou, C. Long Non-Coding RNA AGAP2-AS1: A Comprehensive Overview on Its Biological Functions and Clinical Significances in Human Cancers. Molecules 2024, 29, 3461. https://doi.org/10.3390/molecules29153461
Ma F, Zhang B, Wang Y, Lou C. Long Non-Coding RNA AGAP2-AS1: A Comprehensive Overview on Its Biological Functions and Clinical Significances in Human Cancers. Molecules. 2024; 29(15):3461. https://doi.org/10.3390/molecules29153461
Chicago/Turabian StyleMa, Feng, Bingbing Zhang, Yiqi Wang, and Chenghua Lou. 2024. "Long Non-Coding RNA AGAP2-AS1: A Comprehensive Overview on Its Biological Functions and Clinical Significances in Human Cancers" Molecules 29, no. 15: 3461. https://doi.org/10.3390/molecules29153461
APA StyleMa, F., Zhang, B., Wang, Y., & Lou, C. (2024). Long Non-Coding RNA AGAP2-AS1: A Comprehensive Overview on Its Biological Functions and Clinical Significances in Human Cancers. Molecules, 29(15), 3461. https://doi.org/10.3390/molecules29153461





