Shifting Mycobacterial Serine Hydrolase Activity Visualized Using Multi-Layer In-Gel Activity Assays
Abstract
1. Introduction
2. Results
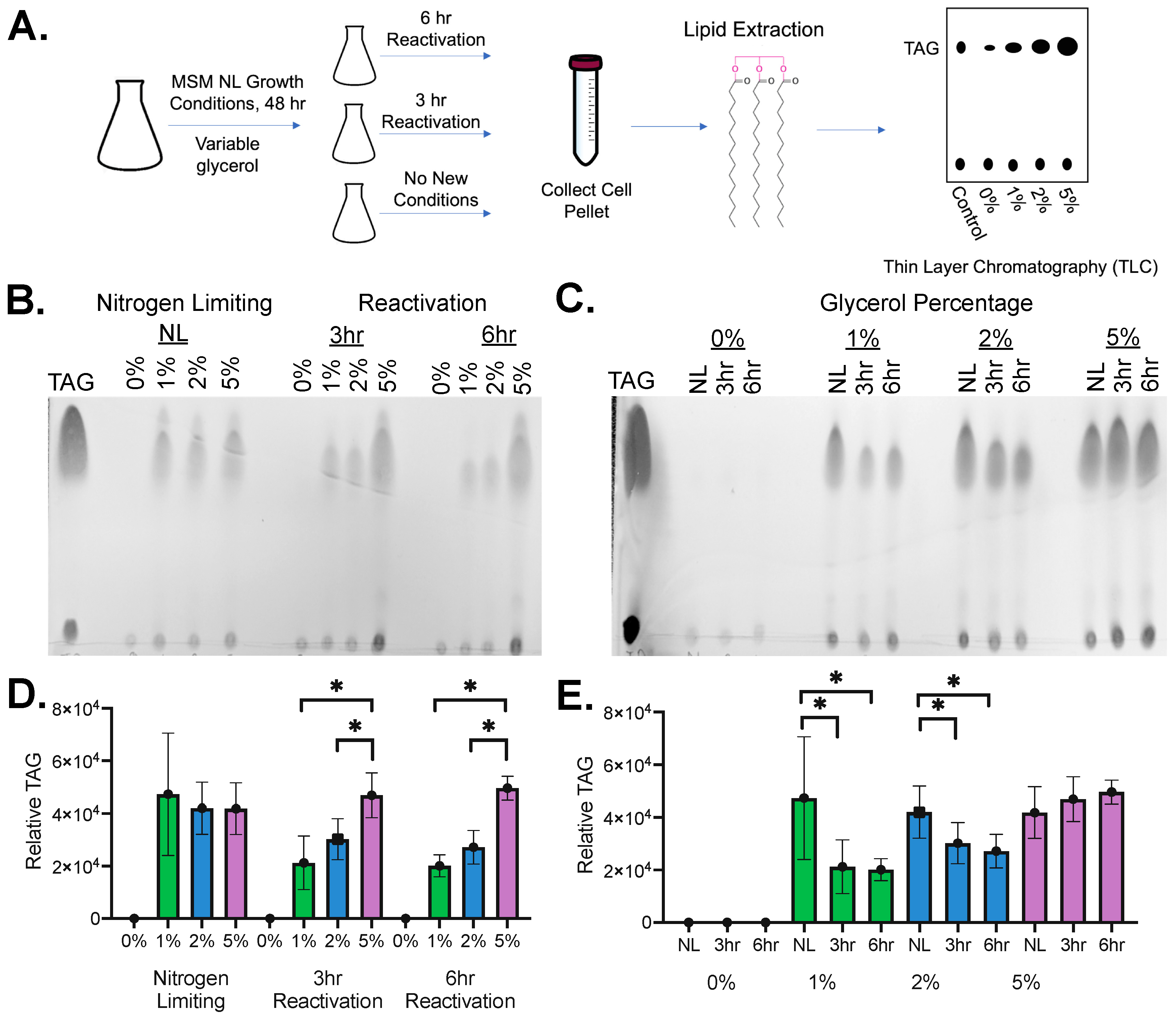
2.1. Dormant and Reactivation Growth System
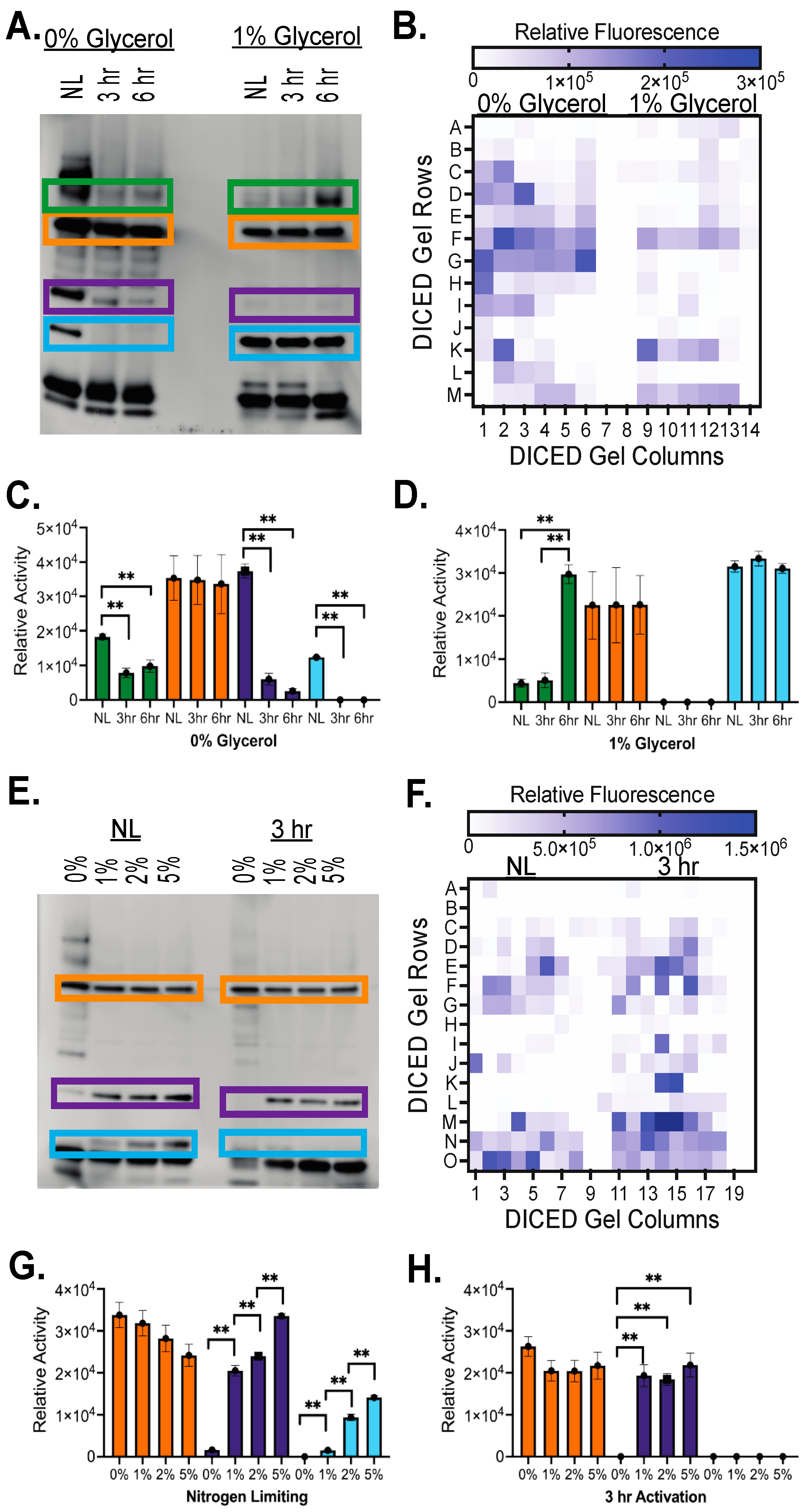
2.2. Shifted Activity of Serine Hydrolases in Dormant versus Active Growth Conditions
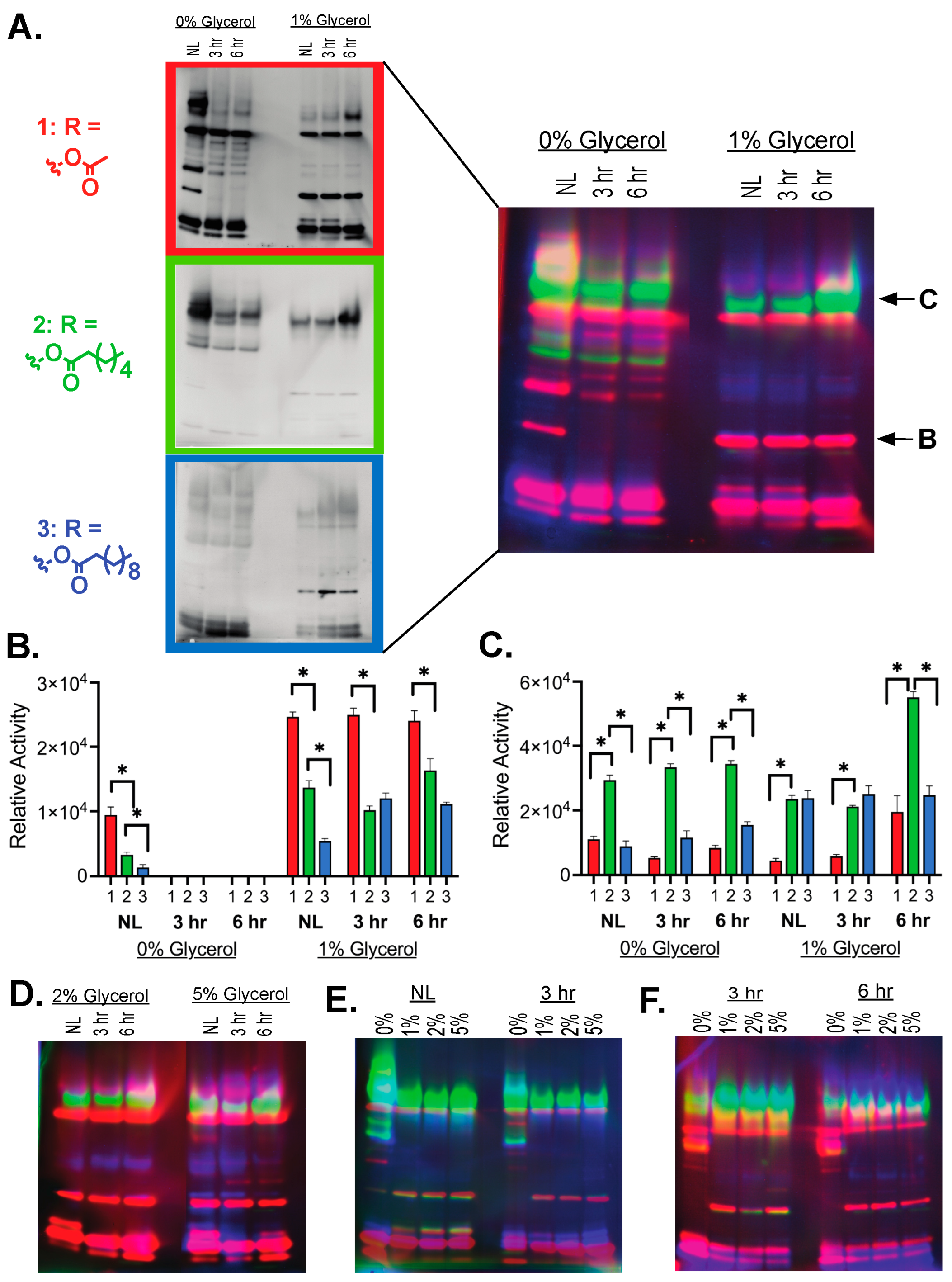
2.3. Patterns of Hydrolase Activation across Ester Substrates and Serine Hydrolase Classes
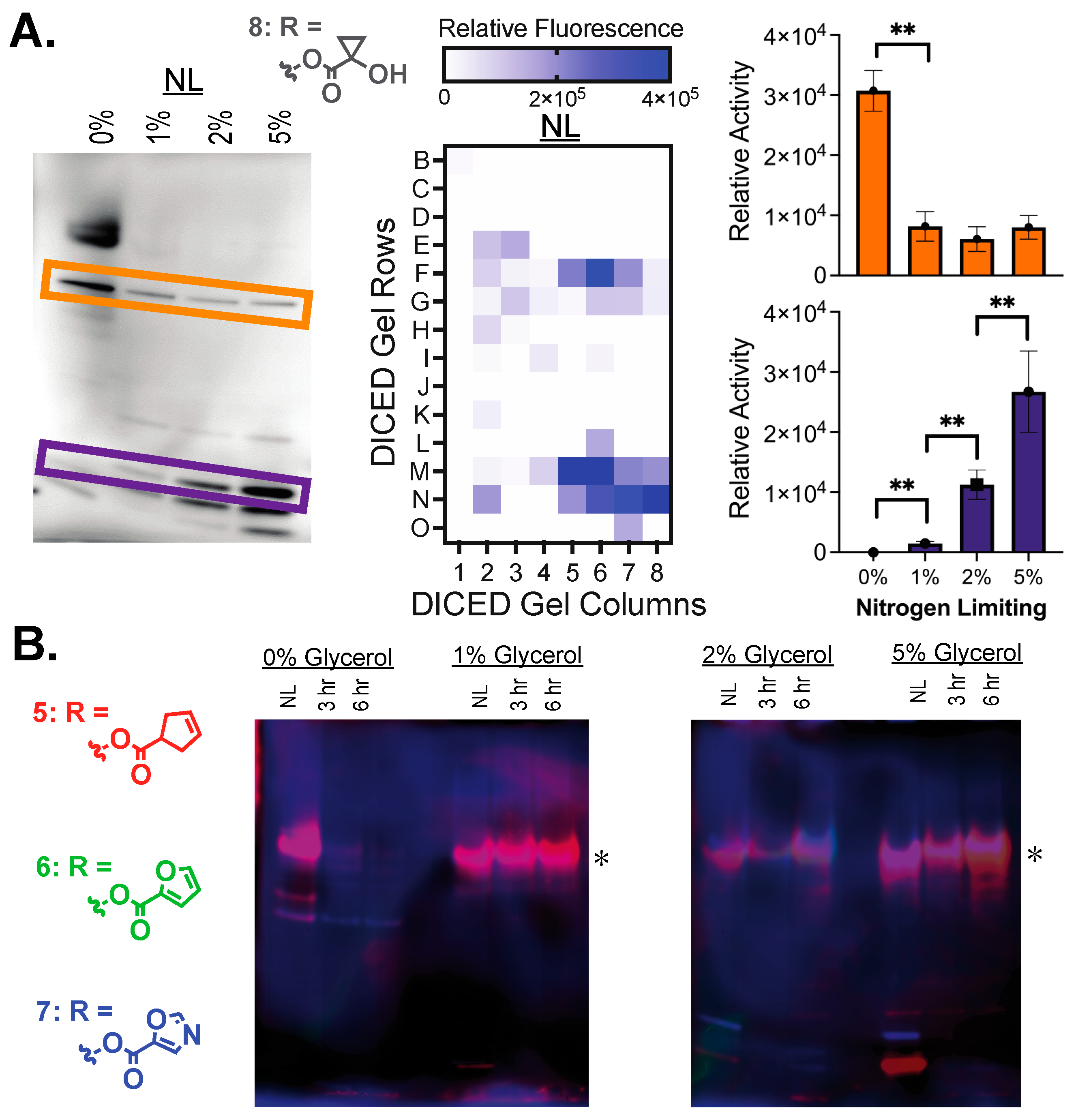
2.4. Identifying Individual Serine Hydrolases
3. Discussion
3.1. Serine Hydrolase Expression Shifts
3.2. Preliminary Assignment of Individual Hydrolase Bands
3.3. Advantages and Limitations of In-Gel Hydrolase Analysis
4. Materials and Methods
4.1. Cell Culturing
4.2. Cell Pellet Collection and Lipid Extraction
4.3. Analysis by Thin Layer Chromatography
4.4. Msmeg Lysis and Measuring Protein Concentration
4.5. Native-PAGE
4.6. Fluorogenic Ester Substrate Exposure and Gel Imaging
4.7. Diced Gel Assay (DEG)
4.8. LipN and pnbA Expression and Purification
4.9. Thermal Stability
4.10. Enzymatic Activity Analysis
4.11. Native-PAGE of Purified Proteins
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2022; World Health Organization: Geneva, Switzerland, 2022.
- Bacon, J.; Alderwick, L.J.; Allnutt, J.A.; Gabasova, E.; Watson, R.; Hatch, K.A.; Clark, S.O.; Jeeves, R.E.; Marriott, A.; Rayner, E.; et al. Non-replicating Mycobacterium tuberculosis elicits a reduced infectivity profile with corresponding modifications to the cell wall and extracellular matrix. PLoS ONE 2014, 9, e87329. [Google Scholar] [CrossRef]
- Kaufmann, S.H. Tuberculosis vaccines—A new kid on the block. Nat. Med. 2011, 17, 159–160. [Google Scholar] [CrossRef]
- Cotten, K.L.; Davis, K.M. Bacterial heterogeneity and antibiotic persistence: Bacterial mechanisms utilized in the host environment. Microbiol. Mol. Biol. Rev. 2023, 87, e00174-22. [Google Scholar] [CrossRef] [PubMed]
- Santucci, P.; Johansen, M.D.; Point, V.; Poncin, I.; Viljoen, A.; Cavalier, J.-F.; Kremer, L.; Canaan, S. Nitrogen deprivation induces triacylglycerol accumulation, drug tolerance and hypervirulence in mycobacteria. Sci. Rep. 2019, 9, 8667. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.C.; Meena, L.S. Triacylglycerol: Nourishing molecule in endurance of Mycobacterium tuberculosis. J. Biosci. 2018, 43, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Cook, G.M.; Berney, M.; Gebhard, S.; Heinemann, M.; Cox, R.A.; Danilchanka, O.; Niederweis, M. Physiology of mycobacteria. Adv. Microb. Physiol. 2009, 55, 81–319. [Google Scholar] [PubMed]
- Dedieu, L.; Serveau-Avesque, C.; Kremer, L.; Canaan, S. Mycobacterial lipolytic enzymes: A gold mine for tuberculosis research. Biochimie 2013, 95, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Bauer, T.L.; Buchholz, P.C.F.; Pleiss, J. The modular structure of α/β-hydrolases. FEBS J. 2020, 287, 1035–1053. [Google Scholar] [CrossRef] [PubMed]
- Tallman, K.R.; Levine, S.R.; Beatty, K.E. Small-Molecule probes reveal esterases with persistent activity in dormant and reactivating Mycobacterium tuberculosis. ACS Infect. Dis. 2016, 2, 936–944. [Google Scholar] [CrossRef]
- Li, M.; Patel, H.V.; Cognetta, A.B.; Smith, T.C.; Mallick, I.; Cavalier, J.-F.; Previti, M.L.; Canaan, S.; Aldridge, B.B.; Cravatt, B.F. Identification of cell wall synthesis inhibitors active against Mycobacterium tuberculosis by competitive activity-based protein profiling. Cell Chem. Biol. 2022, 29, 883–896.e5. [Google Scholar] [CrossRef]
- Dhouib, R.; Ducret, A.; Hubert, P.; Carriere, F.; Dukan, S.; Canaan, S. Watching intracellular lipolysis in mycobacteria using time lapse fluorescence microscopy. Biochim. Biophys. Acta 2011, 1811, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Ortega, C.; Anderson, L.N.; Frando, A.; Sadler, N.C.; Brown, R.W.; Smith, R.D.; Wright, A.T.; Grundner, C. Systematic survey of serine hydrolase activity in Mycobacterium tuberculosis defines changes associated with persistence. Cell Chem. Biol. 2016, 23, 290–298. [Google Scholar] [CrossRef]
- Ravindran, M.S.; Rao, S.P.; Cheng, X.; Shukla, A.; Cazenave-Gassiot, A.; Yao, S.Q.; Wenk, M.R. Targeting lipid esterases in mycobacteria grown under different physiological conditions using activity-based profiling with tetrahydrolipstatin (THL). Mol. Cell. Proteom. MCP 2014, 13, 435–448. [Google Scholar] [CrossRef]
- Nguyen, P.C.; Delorme, V.; Bénarouche, A.; Martin, B.P.; Paudel, R.; Gnawali, G.R.; Madani, A.; Puppo, R.; Landry, V.; Kremer, L. Cyclipostins and Cyclophostin analogs as promising compounds in the fight against tuberculosis. Sci. Rep. 2017, 7, 11751. [Google Scholar] [CrossRef]
- Lehmann, J.; Vomacka, J.; Esser, K.; Nodwell, M.; Kolbe, K.; Rämer, P.; Protzer, U.; Reiling, N.; Sieber, S. Human lysosomal acid lipase inhibitor lalistat impairs Mycobacterium tuberculosis growth by targeting bacterial hydrolases. MedChemComm 2016, 7, 1797–1801. [Google Scholar] [CrossRef]
- Deb, C.; Lee, C.M.; Dubey, V.S.; Daniel, J.; Abomoelak, B.; Sirakova, T.D.; Pawar, S.; Rogers, L.; Kolattukudy, P.E. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS ONE 2009, 4, e6077. [Google Scholar] [CrossRef]
- Tallman, K.R.; Levine, S.R.; Beatty, K.E. Profiling esterases in Mycobacterium tuberculosis using far-red fluorogenic substrates. ACS Chem. Biol. 2016, 11, 1810–1815. [Google Scholar] [CrossRef] [PubMed]
- Bassett, B.; Waibel, B.; White, A.; Hansen, H.; Stephens, D.; Koelper, A.; Larsen, E.M.; Kim, C.; Glanzer, A.; Lavis, L.D.; et al. Measuring the global substrate specificity of mycobacterial serine hydrolases using a library of fluorogenic ester substrates. ACS Infect. Dis. 2018, 4, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Beatty, K.E.; Williams, M.; Carlson, B.L.; Swarts, B.M.; Warren, R.M.; van Helden, P.D.; Bertozzi, C.R. Sulfatase-activated fluorophores for rapid discrimination of mycobacterial species and strains. Proc. Natl. Acad. Sci. USA 2013, 110, 12911–12916. [Google Scholar] [CrossRef]
- Shiloh, M.U.; DiGiuseppe Champion, P.A. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr. Opin. Microbiol. 2010, 13, 86–92. [Google Scholar] [CrossRef]
- Tallman, K.R.; Beatty, K.E. Far-Red fluorogenic probes for esterase and lipase detection. ChemBioChem 2015, 16, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Hedge, M.K.; Gehring, A.M.; Adkins, C.T.; Weston, L.A.; Lavis, L.D.; Johnson, R.J. The structural basis for the narrow substrate specificity of an acetyl esterase from Thermotoga maritima. Biochim. Biophys. Acta 2012, 1824, 1024–1030. [Google Scholar] [CrossRef]
- Lavis, L.D.; Chao, T.-Y.; Raines, R.T. Synthesis and utility of fluorogenic acetoxymethyl ethers. Chem. Sci. 2011, 2, 521–530. [Google Scholar] [CrossRef][Green Version]
- White, A.; Koelper, A.; Russell, A.; Larsen, E.M.; Kim, C.; Lavis, L.D.; Hoops, G.C.; Johnson, R.J. Fluorogenic structure activity library pinpoints molecular variations in substrate specificity of structurally homologous esterases. J. Biol. Chem. 2018, 293, 13851–13862. [Google Scholar] [CrossRef]
- Schemenauer, D.E.; Pool, E.H.; Raynor, S.N.; Ruiz, G.P.; Goehring, L.M.; Koelper, A.J.; Wilson, M.A.; Durand, A.J., Jr.; Kourtoglou, E.C.; Larsen, E.M. Sequence and structural motifs controlling the broad substrate specificity of the mycobacterial hormone-sensitive lipase LipN. ACS Omega 2023, 8, 13252–13264. [Google Scholar] [CrossRef]
- McKary, M.G.; Abendroth, J.; Edwards, T.E.; Johnson, R.J. Structural basis for the strict substrate selectivity of the mycobacterial hydrolase LipW. Biochemistry 2016, 55, 7099–7111. [Google Scholar] [CrossRef] [PubMed]
- Lukowski, J.K.; Savas, C.P.; Gehring, A.M.; McKary, M.G.; Adkins, C.T.; Lavis, L.D.; Hoops, G.C.; Johnson, R.J. Distinct substrate selectivity of a metabolic hydrolase from Mycobacterium tuberculosis. Biochemistry 2014, 53, 7386–7395. [Google Scholar] [CrossRef]
- Bowles, I.E.; Pool, E.H.; Lancaster, B.S.; Lawson, E.K.; Savas, C.P.; Kartje, Z.J.; Severinac, L.; Cho, D.H.; Macbeth, M.R.; Johnson, R.J. Transition metal cation inhibition of Mycobacterium tuberculosis esterase RV0045C. Protein Sci. 2021, 30, 1554–1565. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, T.; Hanaoka, K.; Adibekian, A.; Yoshioka, K.; Terai, T.; Ueno, T.; Kawaguchi, M.; Cravatt, B.F.; Nagano, T. Diced electrophoresis gel assay for screening enzymes with specified activities. J. Am. Chem. Soc. 2013, 135, 6002–6005. [Google Scholar] [CrossRef]
- Komatsu, T.; Shimoda, M.; Kawamura, Y.; Urano, Y.; Nagano, T. Development and validation of an improved diced electrophoresis gel assay cutter-plate system for enzymomics studies. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2019, 1867, 82–87. [Google Scholar] [CrossRef]
- Ino, Y.; Kagawa, H.; Akiyama, T.; Nakai, Y.; Ito, S.; Shimoda, M.; Kawamura, M.; Hirano, H.; Kimura, Y. Protein fractionation for proteomics using the SAINOME-plate. J. Electrophor. 2018, 62, 11–15. [Google Scholar] [CrossRef]
- Long, J.Z.; Cravatt, B.F. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 2011, 111, 6022–6063. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Yang, Y.; Wysocki, L.M.; Arnold, A.C.; Hu, A.; Ravichandran, B.; Sternson, S.M.; Looger, L.L.; Lavis, L.D. Selective esterase-ester pair for targeting small molecules with cellular specificity. Proc. Natl. Acad. Sci. USA 2012, 109, 4756–4761. [Google Scholar] [CrossRef]
- Jones, K.A.; Kentala, K.; Beck, M.W.; An, W.; Lippert, A.R.; Lewis, J.C.; Dickinson, B.C. Development of a split esterase for protein–protein interaction-dependent small-molecule activation. ACS Cent. Sci. 2019, 5, 1768–1776. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.A.; Snodgrass, H.M.; Belsare, K.; Dickinson, B.C.; Lewis, J.C. Phage-assisted continuous evolution and selection of enzymes for chemical synthesis. ACS Cent. Sci. 2021, 7, 1581–1590. [Google Scholar] [CrossRef] [PubMed]
- Hetrick, K.J.; Raines, R.T. Assessing and utilizing esterase specificity in antimicrobial prodrug development. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2022; Volume 664, pp. 199–220. [Google Scholar]
- Hetrick, K.J.; Aguilar Ramos, M.A.; Raines, R.T. Endogenous enzymes enable antimicrobial activity. ACS Chem. Biol. 2021, 16, 800–805. [Google Scholar] [CrossRef]
- Rock, J.M.; Hopkins, F.F.; Chavez, A.; Diallo, M.; Chase, M.R.; Gerrick, E.R.; Pritchard, J.R.; Church, G.M.; Rubin, E.J.; Sassetti, C.M.; et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2017, 2, 16274. [Google Scholar] [CrossRef]
- Dong, B.; He, Z.; Li, Y.; Xu, X.; Wang, C.; Zeng, J. Improved conventional and new approaches in the diagnosis of tuberculosis. Front. Microbiol. 2022, 13, 924410. [Google Scholar] [CrossRef]
- Campo-Pérez, V.; Guallar-Garrido, S.; Luquin, M.; Sánchez-Chardi, A.; Julián, E. The high plasticity of nonpathogenic Mycobacterium brumae induces rapid changes in its lipid profile during pellicle maturation: The potential of this bacterium as a versatile cell factory for lipid compounds of therapeutic interest. Int. J. Mol. Sci. 2022, 23, 13609. [Google Scholar] [CrossRef]
- Moorey, A.R.; Besra, G.S. The role of triacylglycerols and repurposing DGAT1 inhibitors for the treatment of Mycobacterium tuberculosis. Cell Surf. 2022, 8, 100083. [Google Scholar] [CrossRef] [PubMed]
- Deb, C.; Daniel, J.; Sirakova, T.D.; Abomoelak, B.; Dubey, V.S.; Kolattukudy, P.E. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 3866–3875. [Google Scholar] [CrossRef] [PubMed]
- Long, N.P.; Anh, N.K.; Yen, N.T.H.; Phat, N.K.; Park, S.; Thu, V.T.A.; Cho, Y.-S.; Shin, J.-G.; Oh, J.Y.; Kim, D.H. Comprehensive lipid and lipid-related gene investigations of host immune responses to characterize metabolism-centric biomarkers for pulmonary tuberculosis. Sci. Rep. 2022, 12, 13395. [Google Scholar] [CrossRef] [PubMed]
- Kolbe, K.; Veleti, S.K.; Johnson, E.E.; Cho, Y.W.; Oh, S.; Barry, C.E., 3rd. Role of chemical biology in tuberculosis drug discovery and diagnosis. ACS Infect. Dis. 2018, 4, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Larsen, E.M.; Stephens, D.C.; Clarke, N.H.; Johnson, R.J. Ester-prodrugs of ethambutol control its antibacterial activity and provide rapid screening for mycobacterial hydrolase activity. Bioorg Med. Chem. Lett. 2017, 27, 4544–4547. [Google Scholar] [CrossRef] [PubMed]
- Bachovchin, D.A.; Cravatt, B.F. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat. Rev. Drug Discov. 2012, 11, 52–68. [Google Scholar] [CrossRef]
- Pala, L.; Sirec, T.; Spitz, U. Modified enzyme substrates for the detection of bacteria: A review. Molecules 2020, 25, 3690. [Google Scholar] [CrossRef] [PubMed]
- Chyan, W.; Raines, R.T. Enzyme-activated fluorogenic probes for live-cell and in vivo imaging. ACS Chem. Biol. 2018, 13, 1810–1823. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, T.; Urano, Y. Chemical toolbox for ‘live’ biochemistry to understand enzymatic functions in living systems. J. Biochem. 2020, 167, 139–149. [Google Scholar] [CrossRef]
- Johnson, R.J.; Hoops, G.C.; Savas, C.J.; Kartje, Z.; Lavis, L.D. A sensitive and robust enzyme kinetic experiment using microplates and fluorogenic ester substrates. J. Chem. Educ. 2014, 92, 385–388. [Google Scholar] [CrossRef]
- Komatsu, T.; Urano, Y. Evaluation of enzymatic activities in living systems with small-molecular fluorescent substrate probes. Anal. Sci. 2015, 31, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Larsen, E.M.; Johnson, R.J. Microbial esterases and ester prodrugs: An unlikely marriage for combating antibiotic resistance. Drug Dev. Res. 2019, 80, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Kamiya, M.; Yoshioka, T.; Ogasawara, A.; Hino, R.; Kojima, R.; Ueo, H.; Urano, Y. Rapid and accurate visualization of breast tumors with a fluorescent probe targeting α-mannosidase 2C1. ACS Cent. Sci. 2020, 6, 2217–2227. [Google Scholar] [CrossRef] [PubMed]
- Ichihashi, Y.; Komatsu, T.; Kyo, E.; Matsuzaki, H.; Hata, K.; Watanabe, T.; Ueno, T.; Hanaoka, K.; Urano, Y. Separation-based enzymomics assay for the discovery of altered peptide-metabolizing enzymatic activities in biosamples. Anal. Chem. 2019, 91, 11497–11501. [Google Scholar] [CrossRef] [PubMed]
- Kuriki, Y.; Yoshioka, T.; Kamiya, M.; Komatsu, T.; Takamaru, H.; Fujita, K.; Iwaki, H.; Nanjo, A.; Akagi, Y.; Takeshita, K. Development of a fluorescent probe library enabling efficient screening of tumour-imaging probes based on discovery of biomarker enzymatic activities. Chem. Sci. 2022, 13, 4474–4481. [Google Scholar] [CrossRef]
- Johnson, R.J.; Savas, C.J.; Kartje, Z.; Hoops, G.C. Rapid and adaptable measurement of protein thermal stability by differential scanning fluorimetry: Updating a common biochemical laboratory experiment. J. Chem. Educ. 2014, 91, 1077–1080. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goss, A.L.; Shudick, R.E.; Johnson, R.J. Shifting Mycobacterial Serine Hydrolase Activity Visualized Using Multi-Layer In-Gel Activity Assays. Molecules 2024, 29, 3386. https://doi.org/10.3390/molecules29143386
Goss AL, Shudick RE, Johnson RJ. Shifting Mycobacterial Serine Hydrolase Activity Visualized Using Multi-Layer In-Gel Activity Assays. Molecules. 2024; 29(14):3386. https://doi.org/10.3390/molecules29143386
Chicago/Turabian StyleGoss, Allison L., Renee E. Shudick, and R. Jeremy Johnson. 2024. "Shifting Mycobacterial Serine Hydrolase Activity Visualized Using Multi-Layer In-Gel Activity Assays" Molecules 29, no. 14: 3386. https://doi.org/10.3390/molecules29143386
APA StyleGoss, A. L., Shudick, R. E., & Johnson, R. J. (2024). Shifting Mycobacterial Serine Hydrolase Activity Visualized Using Multi-Layer In-Gel Activity Assays. Molecules, 29(14), 3386. https://doi.org/10.3390/molecules29143386





