Pinus koraiensis Essential Oil Attenuates the Pathogenicity of Superbacteria by Suppressing Virulence Gene Expression
Abstract
1. Introduction
2. Results
2.1. Chemical Composition of PKEO
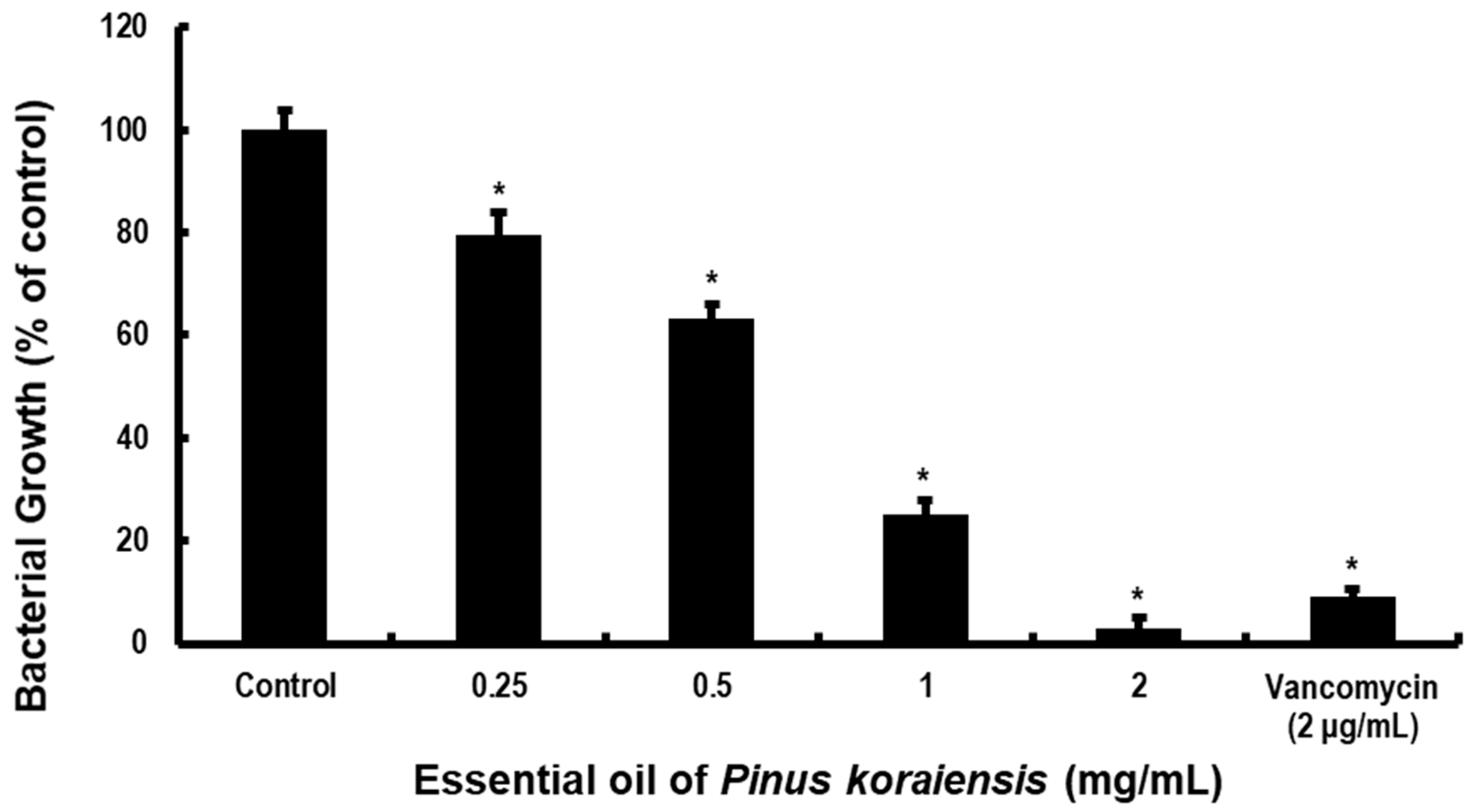
2.2. PKEO Suppresses MRSA Growth and Acidogenicity
2.3. Destructive Effects of PKEO on MRSA Biofilm Growth
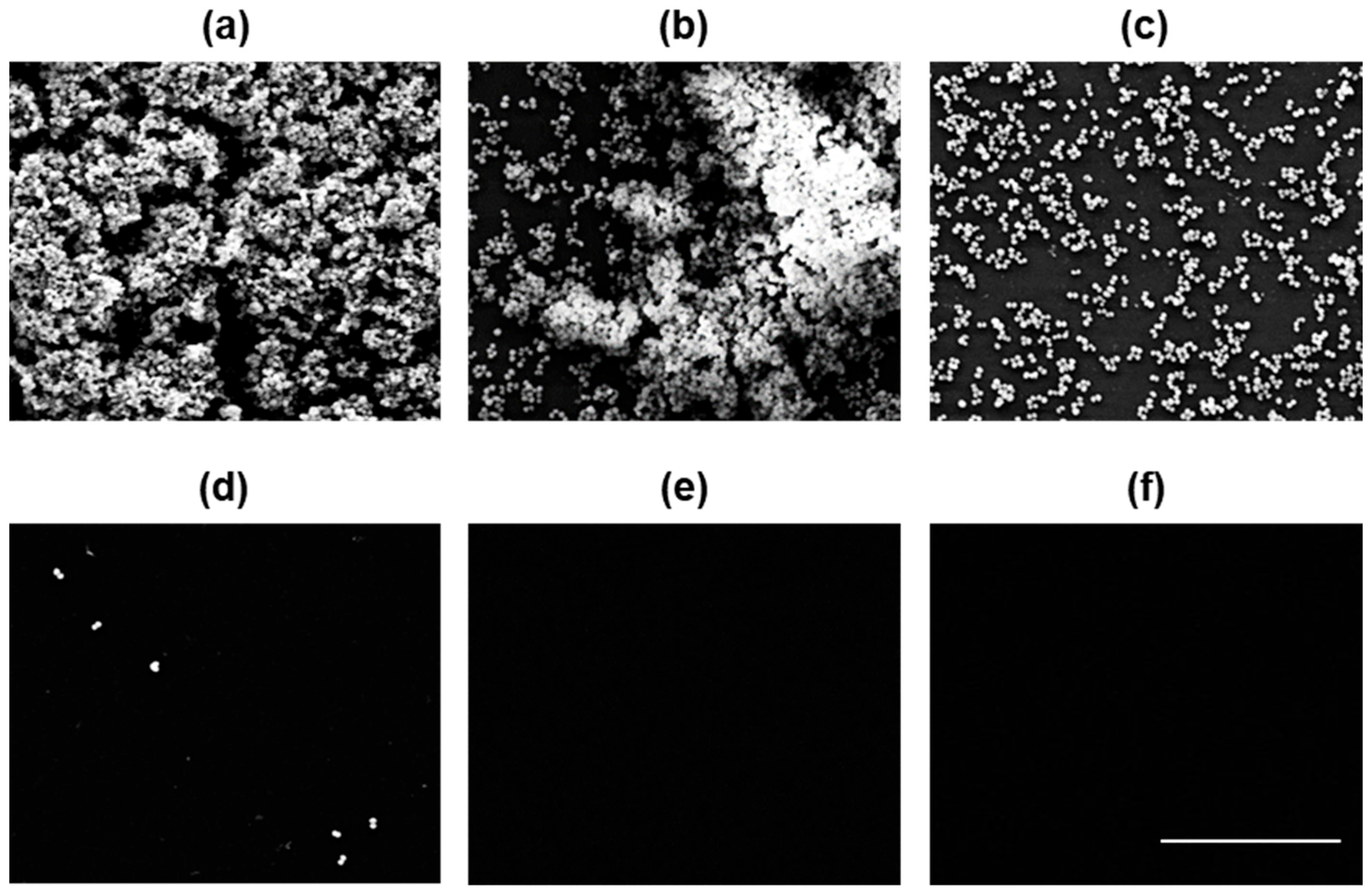
2.4. PKEO Has Germicidal Effects against MRSA
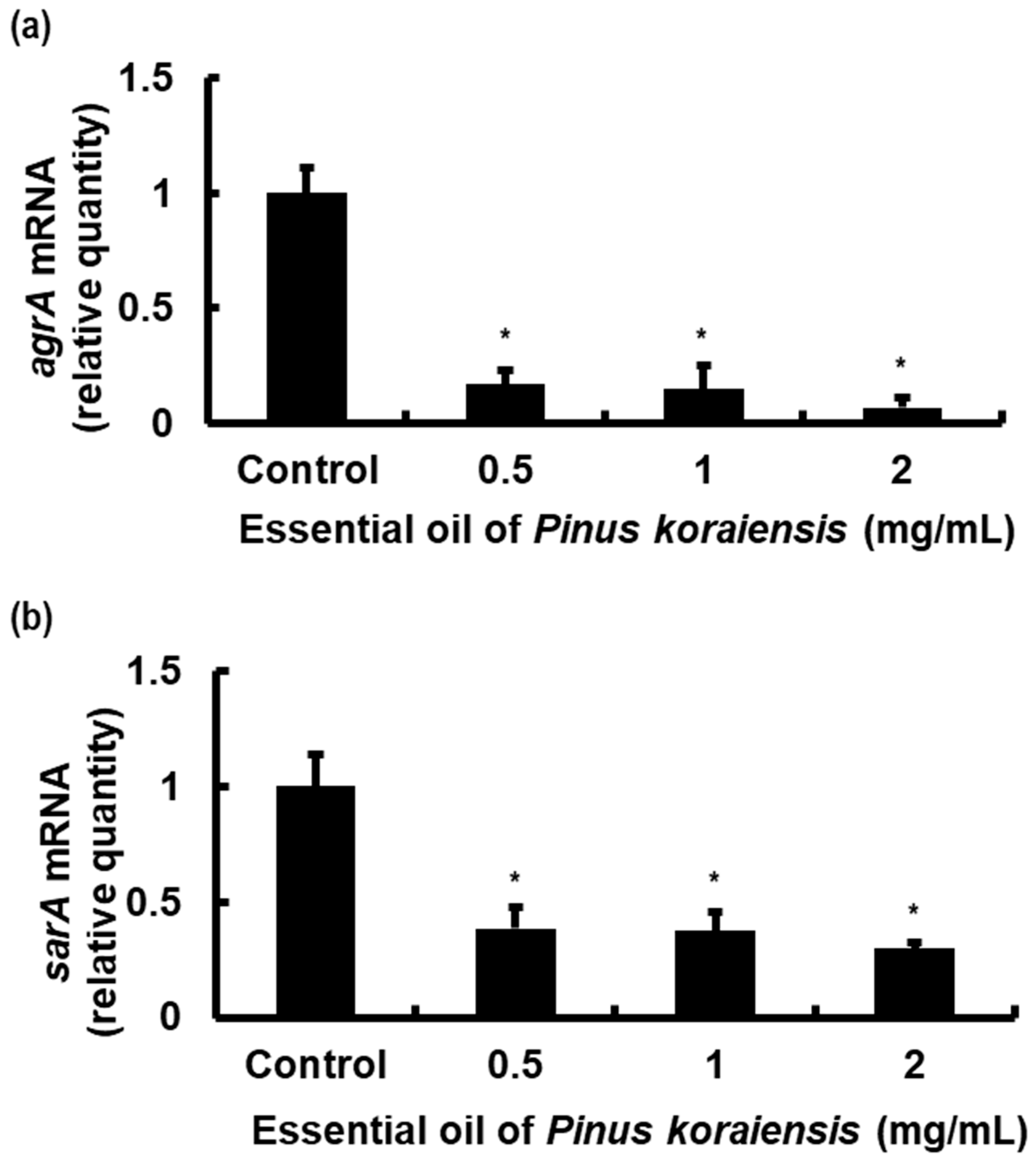
2.5. PKEO Represses Virulence Gene Expression in MRSA
3. Discussion
4. Materials and Methods
4.1. Isolation of PKEO
4.2. Analysis of PKEO
4.2.1. Gas Chromatography–Flame Ionization Detector Analysis
4.2.2. Gas Chromatography–Mass Spectrometry
4.3. Bacterial Culture
4.4. Measurement of MRSA Growth and Acidogenicity
4.5. Determination of MRSA Biofilm Formation
4.6. MRSA Viability Assay Using Confocal Laser Scanning Microscopy
4.7. Real-Time PCR Assay of MRSA Virulence Factors
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, Y.-M.; Chong, Y.P.; Kim, M.; Eom, Y.; Kim, E.S.; Kim, M.; Park, K.-H.; Kim, S.-H.; Lee, S.-O.; Choi, S.-H.; et al. Long-Term Methicillin-Resistant Staphylococcus aureus Bacteremia Persisting for More than 2 Weeks: Risk Factors and Outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Ortwine, J.K.; Bhavan, K. Morbidity, Mortality, and Management of Methicillin-Resistant S. Aureus Bacteremia in the USA: Update on Antibacterial Choices and Understanding. Hosp. Pract. 2018, 46, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, P.C. MRSA—The Tip of the Iceberg. Clin. Microbiol. Infect. 2006, 12, 3–10. [Google Scholar] [CrossRef] [PubMed]
- 2019 AR Threats Report. Antibiotic Resistance Threats in the United States: 2019. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 11 June 2023).
- Antimicrobial Resistance Surveillance in Europe 2023–2021 Data. Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2023-2021-data (accessed on 19 June 2023).
- Annual Report of the China Antimicrobial Resistance Surveillance. 2021. Available online: http://www.carss.cn/Report/Details?aId=862 (accessed on 20 June 2023).
- Liu, C.; Bayer, A.; Cosgrove, S.E.; Daum, R.S.; Fridkin, S.K.; Gorwitz, R.J.; Kaplan, S.L.; Karchmer, A.W.; Levine, D.P.; Murray, B.E.; et al. Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections in Adults and Children. Clin. Infect. Dis. 2011, 52, e18–e55. [Google Scholar] [CrossRef] [PubMed]
- Vora, S. Acute Renal Failure Due to Vancomycin Toxicity in the Setting of Unmonitored Vancomycin Infusion. In Baylor University Medical Center Proceedings; Taylor & Francis: Abingdon, UK, 2016; Volume 29, pp. 412–413. [Google Scholar] [CrossRef][Green Version]
- Elyasi, S.; Khalili, H.; Dashti-Khavidaki, S.; Mohammadpour, A. Vancomycin-Induced Nephrotoxicity: Mechanism, Incidence, Risk Factors and Special Populations. A Literature Review. Eur. J. Clin. Pharmacol. 2012, 68, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, Y.; VanBeek, M.J.; Greenlee, J.D.W. Red Man Syndrome Caused by Vancomycin Powder. J. Clin. Neurosci. 2018, 50, 149–150. [Google Scholar] [CrossRef]
- Sievert, D.M.; Rudrik, J.T.; Patel, J.B.; McDonald, L.C.; Wilkins, M.J.; Hageman, J.C. Vancomycin-Resistant Staphylococcus aureus in the United States, 2002–2006. Clin. Infect. Dis. 2008, 46, 668–674. [Google Scholar] [CrossRef]
- Shariati, A.; Dadashi, M.; Moghadam, M.T.; Van Belkum, A.; Yaslianifard, S.; Darban-Sarokhalil, D. Global Prevalence and Distribution of Vancomycin Resistant, Vancomycin Intermediate and Heterogeneously Vancomycin Intermediate Staphylococcus aureus Clinical Isolates: A Systematic Review and Meta-Analysis. Sci. Rep. 2020, 10, 12689. [Google Scholar] [CrossRef]
- Wu, Q.; Sabokroo, N.; Wang, Y.; Hashemian, M.; Karamollahi, S.; Kouhsari, E. Systematic Review and Meta-Analysis of the Epidemiology of Vancomycin-Resistance Staphylococcus aureus Isolates. Antimicrob. Resist. Infect. Control 2021, 10, 101. [Google Scholar] [CrossRef]
- Sasaki, T.; Takane, H.; Ogawa, K.; Isagawa, S.; Hirota, T.; Higuchi, S.; Horii, T.; Otsubo, K.; Ieiri, I. Population pharmacokinetic and pharmacodynamic analysis of linezolid and a hematologic side effect, thrombocytopenia, in Japanese patients. Antimicrob. Agents Chemother. 2011, 55, 1867–1873. [Google Scholar] [CrossRef]
- Osorio, C.; Garzón, L.; Jaimes, D.; Silva, E.; Bustos, R.H. Impact on antibiotic resistance, therapeutic success, and control of side effects in therapeutic drug monitoring (TDM) of daptomycin: A scoping review. Antibiotics 2021, 10, 263. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Li, C.; Dai, J.; Cui, H.; Lin, L. Antibacterial Activity and Mechanism of Litsea Cubeba Essential Oil against Methicillin-Resistant Staphylococcus aureus (MRSA). Ind. Crops Prod. 2019, 130, 34–41. [Google Scholar] [CrossRef]
- Okwu, M.U.; Olley, M.; Akpoka, A.O.; Izevbuwa, O.E. Methicillin-Resistant Staphylococcus aureus (MRSA) and Anti-MRSA Activities of Extracts of Some Medicinal Plants: A Brief Review. AIMS Microbiol. 2019, 5, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Lim, T.K. Pinus koraiensis. In Edible Medicinal and Non-Medicinal Plants; Springer: Dordrecht, The Netherlands, 2012; pp. 297–303. ISBN 978-94-007-4052-5. [Google Scholar]
- Li, X.; Liu, X.-T.; Wei, J.-T.; Li, Y.; Tigabu, M.; Zhao, X.-Y. Genetic Improvement of Pinus koraiensis in China: Current Situation and Future Prospects. Forests 2020, 11, 148. [Google Scholar] [CrossRef]
- Li, K.; Li, Q.; Li, J.; Zhang, T.; Han, Z.; Gao, D.; Zheng, F. Antitumor Activity of the Procyanidins from Pinus koraiensis Bark on Mice Bearing U14 Cervical Cancer. Yakugaku Zasshi 2007, 127, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xin, C.; Cheng, C.; Wang, Z. Antitumor Activity of Nanoemulsion Based on Essential Oil of Pinus koraiensis Pinecones in MGC-803 Tumor-Bearing Nude Mice. Arab. J. Chem. 2020, 13, 8226–8238. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, H.; Zhang, Y.; Zhao, H.; Dong, A.; Xu, D.; Yang, L.; Ma, Y.; Wang, J. Analysis of the Essential Oils of Pine Cones of Pinus koraiensis Steb. Et Zucc. and P. sylvestris L. from China. J. Essent. Oil Res. 2010, 22, 446–448. [Google Scholar] [CrossRef]
- Hong, E.-J.; Na, K.-J.; Choi, I.-G.; Choi, K.-C.; Jeung, E.-B. Antibacterial and Antifungal Effects of Essential Oils from Coniferous Trees. Biol. Pharm. Bull. 2004, 27, 863–866. [Google Scholar] [CrossRef]
- Junxing, L.; Zhiwei, Y.; Fei, W.; Lin, W.; Hongwei, N. Antimicrobial Effects of Essential Oil from Pinus koraiensis Sieb. et Zucc. Needles in the Bioflims. Afr. J. Microbiol. Res. 2013, 7, 3078–3084. [Google Scholar] [CrossRef][Green Version]
- Aryee, A.; Edgeworth, J.D. Carriage, Clinical Microbiology and Transmission of Staphylococcus aureus. In Staphylococcus aureus; Bagnoli, F., Rappuoli, R., Grandi, G., Eds.; Current Topics in Microbiology and Immunology; Springer International Publishing: Cham, Switzerland, 2016; Volume 409, pp. 1–19. ISBN 978-3-319-72061-6. [Google Scholar]
- Romero, D.V.; Treston, J.; O’Sullivan, A.L. Hand-to-Hand Combat: Preventing MRSA Infection. Adv. Ski. Wound Care 2006, 19, 328–333. [Google Scholar] [CrossRef]
- Leonard, F.C.; Markey, B.K. Meticillin-Resistant Staphylococcus aureus in Animals: A Review. Vet. J. 2008, 175, 27–36. [Google Scholar] [CrossRef]
- Ferreira, M.T.; Manso, A.S.; Gaspar, P.; Pinho, M.G.; Neves, A.R. Effect of Oxygen on Glucose Metabolism: Utilization of Lactate in Staphylococcus aureus as Revealed by In Vivo NMR Studies. PLoS ONE 2013, 8, e58277. [Google Scholar] [CrossRef] [PubMed]
- Wallin-Carlquist, N.; Cao, R.; Márta, D.; Da Silva, A.S.; Schelin, J.; Rådström, P. Acetic Acid Increases the Phage-Encoded Enterotoxin a Expression in Staphylococcus aureus. BMC Microbiol. 2010, 10, 147. [Google Scholar] [CrossRef] [PubMed]
- Graf, A.C.; Leonard, A.; Schäuble, M.; Rieckmann, L.M.; Hoyer, J.; Maass, S.; Lalk, M.; Becher, D.; Pané-Farré, J.; Riedel, K. Virulence Factors Produced by Staphylococcus aureus Biofilms Have a Moonlighting Function Contributing to Biofilm Integrity. Mol. Cell. Proteom. 2019, 18, 1036–1053. [Google Scholar] [CrossRef] [PubMed]
- Craft, K.M.; Nguyen, J.M.; Berg, L.J.; Townsend, S.D. Methicillin-Resistant Staphylococcus aureus (MRSA): Antibiotic-Resistance and the Biofilm Phenotype. Med. Chem. Commun. 2019, 10, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, H.; Rudkin, J.K.; Black, N.S.; Gallagher, L.; O’Neill, E.; O’Gara, J.P. Methicillin Resistance and the Biofilm Phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2015, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Tang, X.; Dong, W.; Sun, N.; Yuan, W. A review of biofilm formation of Staphylococcus aureus and its regulation mechanism. Antibiotics 2022, 12, 12. [Google Scholar] [CrossRef]
- McGoverin, C.; Robertson, J.; Jonmohamadi, Y.; Swift, S.; Vanholsbeeck, F. Species Dependence of SYTO 9 Staining of Bacteria. Front. Microbiol. 2020, 11, 545419. [Google Scholar] [CrossRef]
- Ferrer-Espada, R.; Liu, X.; Goh, X.S.; Dai, T. Antimicrobial Blue Light Inactivation of Polymicrobial Biofilms. Front. Microbiol. 2019, 10, 721. [Google Scholar] [CrossRef]
- Vivero-Escoto, J.L.; Slowing, I.I.; Lin, V.S.-Y. Tuning the Cellular Uptake and Cytotoxicity Properties of Oligonucleotide Intercalator-Functionalized Mesoporous Silica Nanoparticles with Human Cervical Cancer Cells HeLa. Biomaterials 2010, 31, 1325–1333. [Google Scholar] [CrossRef]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2019, 7, 7.2.29. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Manna, A.C.; Pan, C.-H.; Kriksunov, I.A.; Thiel, D.J.; Cheung, A.L.; Zhang, G. Structural and Function Analyses of the Global Regulatory Protein SarA from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2006, 103, 2392–2397. [Google Scholar] [CrossRef] [PubMed]
- Reyes, D.; Andrey, D.O.; Monod, A.; Kelley, W.L.; Zhang, G.; Cheung, A.L. Coordinated Regulation by AgrA, SarA, and SarR To Control Agr Expression in Staphylococcus aureus. J. Bacteriol. 2011, 193, 6020–6031. [Google Scholar] [CrossRef] [PubMed]
- Le, K.Y.; Otto, M. Quorum-Sensing Regulation in Staphylococci—An Overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef] [PubMed]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription Profiling-Based Identification of Staphylococcus aureus Genes Regulated by the Agr and/or sarA Loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef] [PubMed]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.-H.L.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-Independent Target Gene Control by the Agr Quorum-Sensing System: Insight into the Evolution of Virulence Regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef]
- Kim, B.; Lee, H.-J.; Jo, S.-H.; Kim, M.-G.; Lee, Y.; Lee, W.; Kim, W.; Joo, H.-S.; Kim, Y.-G.; Kim, J.-S.; et al. Study of SarA by DNA Affinity Capture Assay (DACA) Employing Three Promoters of Key Virulence and Resistance Genes in Methicillin-Resistant Staphylococcus aureus. Antibiotics 2022, 11, 1714. [Google Scholar] [CrossRef]
- Cheung, A.L.; Nishina, K.A.; Trotonda, M.P.; Tamber, S. The SarA Protein Family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 2008, 40, 355–361. [Google Scholar] [CrossRef]
- Chien, Y.; Manna, A.C.; Cheung, A.L. SarA Level Is a Determinant of Agr Activation in Staphylococcus aureus. Mol. Microbiol. 1998, 30, 991–1001. [Google Scholar] [CrossRef]
- Chan, P.F.; Foster, S.J. Role of SarA in Virulence Determinant Production and Environmental Signal Transduction in Staphylococcus aureus. J. Bacteriol. 1998, 180, 6232–6241. [Google Scholar] [CrossRef]
- Dumitrescu, O.; Choudhury, P.; Boisset, S.; Badiou, C.; Bes, M.; Benito, Y.; Wolz, C.; Vandenesch, F.; Etienne, J.; Cheung, A.L.; et al. β-Lactams Interfering with PBP1 Induce Panton-Valentine Leukocidin Expression by Triggering sarA and Rot Global Regulators of Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 3261–3271. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.; Borges, A.; Simões, M. Staphylococcus aureus Toxins and Their Molecular Activity in Infectious Diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef] [PubMed]
- Mrak, L.N.; Zielinska, A.K.; Beenken, K.E.; Mrak, I.N.; Atwood, D.N.; Griffin, L.M.; Lee, C.Y.; Smeltzer, M.S. saeRS and sarA Act Synergistically to Repress Protease Production and Promote Biofilm Formation in Staphylococcus aureus. PLoS ONE 2012, 7, e38453. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cheung, A.; Bayer, A.S.; Chen, L.; Abdelhady, W.; Kreiswirth, B.N.; Yeaman, M.R.; Xiong, Y.Q. The Global Regulon sarA Regulates β-Lactam Antibiotic Resistance in Methicillin-Resistant Staphylococcus aureus In Vitro and in Endovascular Infections. J. Infect. Dis. 2016, 214, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
- Riordan, J.T.; O’Leary, J.O.; Gustafson, J.E. Contributions of sigB and sarA to Distinct Multiple Antimicrobial Resistance Mechanisms of Staphylococcus aureus. Int. J. Antimicrob. Agents 2006, 28, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.C.; Barroso, J.G.; Pedro, L.G.; Scheffer, J.J.C. Factors Affecting Secondary Metabolite Production in Plants: Volatile Components and Essential Oils. Flavour Fragr. J. 2008, 23, 213–226. [Google Scholar] [CrossRef]
- Hwang, H.J.; Yu, J.S.; Lee, H.Y.; Kwon, D.J.; Han, W.; Heo, S.I.; Kim, S.Y. Evaluations on Deodorization Effect and Anti-Oral Microbial Activity of Essential Oil from Pinus koraiensis. Korean J. Plant Resour. 2014, 27, 1–10. [Google Scholar] [CrossRef]
- De Araújo, A.C.J.; Freitas, P.R.; Dos Santos Barbosa, C.R.; Muniz, D.F.; De Almeida, R.S.; Alencar De Menezes, I.R.; Ribeiro-Filho, J.; Tintino, S.R.; Coutinho, H.D.M. In Vitro and In Silico Inhibition of Staphylococcus aureus Efflux Pump NorA by α-Pinene and Limonene. Curr. Microbiol. 2021, 78, 3388–3393. [Google Scholar] [CrossRef]
- Han, Y.; Chen, W.; Sun, Z. Antimicrobial activity and mechanism of limonene against Staphylococcus aureus. J. Food Saf. 2021, 41, e12918. [Google Scholar] [CrossRef]
- Rocha, P.M.D.M.; Rodilla, J.M.; Díez, D.; Elder, H.; Guala, M.S.; Silva, L.A.; Pombo, E.B. Synergistic antibacterial activity of the essential oil of aguaribay (Schinus molle L.). Molecules 2012, 17, 12023–12036. [Google Scholar] [CrossRef]
- Yuan, W.; Yuk, H.G. Antimicrobial efficacy of Syzygium antisepticum plant extract against Staphylococcus aureus and methicillin-resistant S. aureus and its application potential with cooked chicken. Food Microbiol. 2018, 72, 176–184. [Google Scholar] [CrossRef] [PubMed]
- NIST Standard Reference Database. Number 69 Gas Chromatography—Retention Indices. Available online: https://webbook.nist.gov/cgi/cbook.cgi?ID=64-19-7 (accessed on 11 April 2023).
- Vitko, N.P.; Richardson, A.R. Laboratory Maintenance of Methicillin-Resistant Staphylococcus aureus (MRSA). Curr. Protoc. Microbiol. 2013, 28, 9C-2. [Google Scholar] [CrossRef] [PubMed]
- Mirani, Z.A.; Jamil, N. Effect of Sub-Lethal Doses of Vancomycin and Oxacillin on Biofilm Formation by Vancomycin Intermediate Resistant Staphylococcus aureus. J. Basic Microbiol. 2011, 51, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wu, D.; Wang, L.; Lin, C.; Ma, C.; Xi, X.; Zhou, M.; Duan, J.; Bininda-Emonds, O.R.P.; Chen, T.; et al. Targeted Modification of a Novel Amphibian Antimicrobial Peptide from Phyllomedusa Tarsius to Enhance Its Activity against MRSA and Microbial Biofilm. Front. Microbiol. 2017, 8, 628. [Google Scholar] [CrossRef] [PubMed]
- Seidl, K.; Goerke, C.; Wolz, C.; Mack, D.; Berger-Bächi, B.; Bischoff, M. Staphylococcus aureus CcpA Affects Biofilm Formation. Infect Immun. 2008, 76, 2044–2050. [Google Scholar] [CrossRef]
- Di Poto, A.; Sbarra, M.S.; Provenza, G.; Visai, L.; Speziale, P. The Effect of Photodynamic Treatment Combined with Antibiotic Action or Host Defence Mechanisms on Staphylococcus aureus Biofilms. Biomaterials 2009, 30, 3158–3166. [Google Scholar] [CrossRef]
- Lee, J.-W.; Bannerman, D.D.; Paape, M.J.; Huang, M.-K.; Zhao, X. Characterization of Cytokine Expression in Milk Somatic Cells during Intramammary Infections with Escherichia coli or Staphylococcus aureus by Real-Time PCR. Vet. Res. 2006, 37, 219–229. [Google Scholar] [CrossRef]


| Compound Name | RI a | Area [%] |
|---|---|---|
| α-thujene | 1007 | 0.83 |
| α-pinene | 1021 | 21.32 |
| Camphene | 1063 | 6.22 |
| n-hexanal | 1085 | 0.07 |
| β-pinene | 1103 | 2.53 |
| Sabinene | 1120 | 0.03 |
| δ-3-carene | 1140 | 10.32 |
| Myrcene | 1171 | 4.60 |
| α-phellandrene | 1183 | 0.28 |
| Limonene | 1196 | 5.17 |
| β-phellandrene | 1211 | 1.73 |
| trans-2-hexenal | 1222 | 0.79 |
| r-terpinene | 1247 | 0.24 |
| trans-β-ocimene | 1257 | 0.06 |
| ρ-cymene | 1273 | 0.05 |
| Terpinolene | 1283 | 7.20 |
| n-hexanol | 1380 | 0.11 |
| cis-3-hexen-1-ol | 1385 | 0.92 |
| β-thujone | 1438 | 0.02 |
| α-cubebene | 1463 | 0.11 |
| α-copaene | 1487 | 0.32 |
| Camphor | 1516 | 0.03 |
| iso-pinocamphone | 1566 | 0.12 |
| cis-α-Bergamotene | 1569 | 0.24 |
| Bornyl acetate | 1576 | 3.85 |
| β-caryophyllene | 1591 | 4.69 |
| Aromandendrene | 1605 | 0.15 |
| Terpinen-4-ol | 1614 | 0.16 |
| β-gurjunene | 1617 | 0.13 |
| γ-elemene | 1633 | 0.08 |
| α-humulene | 1669 | 0.86 |
| Neryl acetate | 1685 | 0.10 |
| α-terpinyl acetate | 1692 | 1.28 |
| α-terpineol | 1697 | 11.03 |
| Borneol | 1703 | 0.33 |
| α-muurolene | 1725 | 0.47 |
| γ-bisabolene | 1762 | 0.99 |
| δ-cadinene | 1674 | 4.38 |
| β-sesquiphellandrene | 1770 | 0.25 |
| α-cadinene | 1792 | 0.14 |
| Caryophyllene oxide | 1980 | 0.20 |
| Ledol | 2028 | 0.15 |
| α-cedrol | 2109 | 0.08 |
| τ-cadinol | 2175 | 0.47 |
| α-cadinol | 2183 | 0.55 |
| Spathulenol | 2188 | 0.12 |
| Stachene | 2208 | 1.32 |
| trans,trans-farnesol | 2350 | 0.17 |
| Dihydroabietanone | 2480 | 0.12 |
| Dodecanoic acid | 2516 | 0.99 |
| Phytol | 2600 | 0.10 |
| Total | 96.47 |
| Concentration [mg/mL] | pH (Before Cultivation) | pH (After Cultivation) |
|---|---|---|
| Control | 7.38 ± 0.00 | 5.94 ± 0.04 |
| 0.25 | 7.39 ± 0.00 | 6.16 ± 0.02 * |
| 0.5 | 7.37 ± 0.00 | 6.46 ± 0.03 * |
| 1 | 7.37 ± 0.00 | 7.09 ± 0.01 * |
| 2 | 7.38 ± 0.00 | 7.32 ± 0.00 * |
| 2 μg/mL vancomycin | 7.37 ± 0.00 | 7.39 ± 0.00 * |
| Genes | Sequence (5′-3′) | Length a | Tm [°C] |
|---|---|---|---|
| agrA | Forward: 5′-TGATAATCCTTATGAGGTGCTT-3′ Reverse: 5′-CACTGTGACTCGTAACGAAAA-3′ | 22 | 50 |
| sarA | Forward: 5′-TGTTATCAATGGTCACTTATGCTG-3′ Reverse: 5′-TCTTTGTTTTCGCTGATGTATGTC-3′ | 24 | 53 |
| 16s rRNA | Forward: 5′-ACTGGGATAACTTCGGGAAA-3′ Reverse: 5′-CGTTGCCTTGGTAAGCC-3′ | 20 | 52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-H.; Kim, Y.-H.; Park, B.-I.; Choi, N.-Y.; Kim, K.-J. Pinus koraiensis Essential Oil Attenuates the Pathogenicity of Superbacteria by Suppressing Virulence Gene Expression. Molecules 2024, 29, 37. https://doi.org/10.3390/molecules29010037
Kim J-H, Kim Y-H, Park B-I, Choi N-Y, Kim K-J. Pinus koraiensis Essential Oil Attenuates the Pathogenicity of Superbacteria by Suppressing Virulence Gene Expression. Molecules. 2024; 29(1):37. https://doi.org/10.3390/molecules29010037
Chicago/Turabian StyleKim, Ji-Hee, Young-Hoi Kim, Bog-Im Park, Na-Young Choi, and Kang-Ju Kim. 2024. "Pinus koraiensis Essential Oil Attenuates the Pathogenicity of Superbacteria by Suppressing Virulence Gene Expression" Molecules 29, no. 1: 37. https://doi.org/10.3390/molecules29010037
APA StyleKim, J.-H., Kim, Y.-H., Park, B.-I., Choi, N.-Y., & Kim, K.-J. (2024). Pinus koraiensis Essential Oil Attenuates the Pathogenicity of Superbacteria by Suppressing Virulence Gene Expression. Molecules, 29(1), 37. https://doi.org/10.3390/molecules29010037





