Identification of Small Molecule Inhibitors of Human Cytomegalovirus pUL89 Endonuclease Using Integrated Computational Approaches
Abstract
1. Introduction
2. Results and Discussion
2.1. Structure and Ligand-Based Virtual Screening
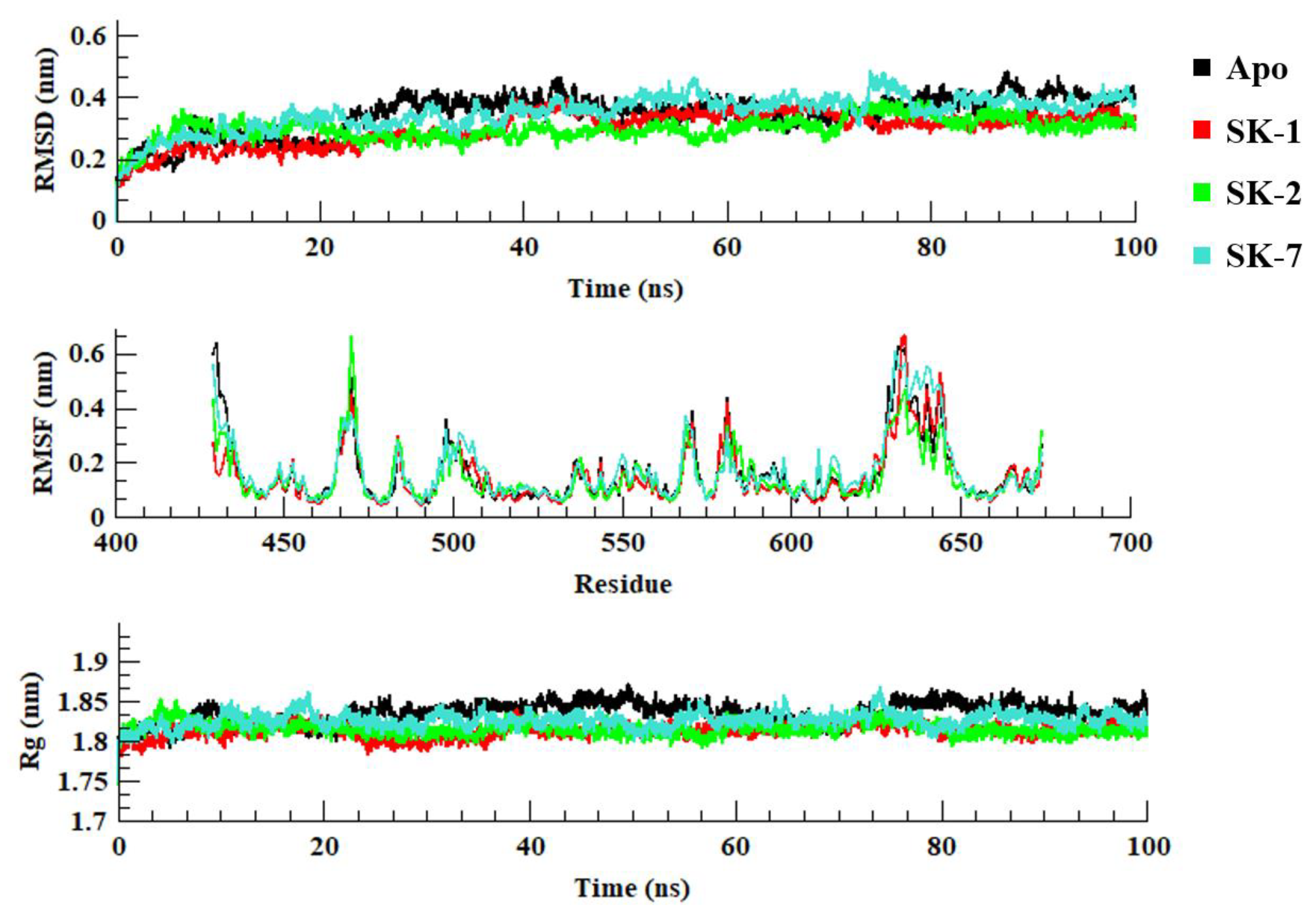
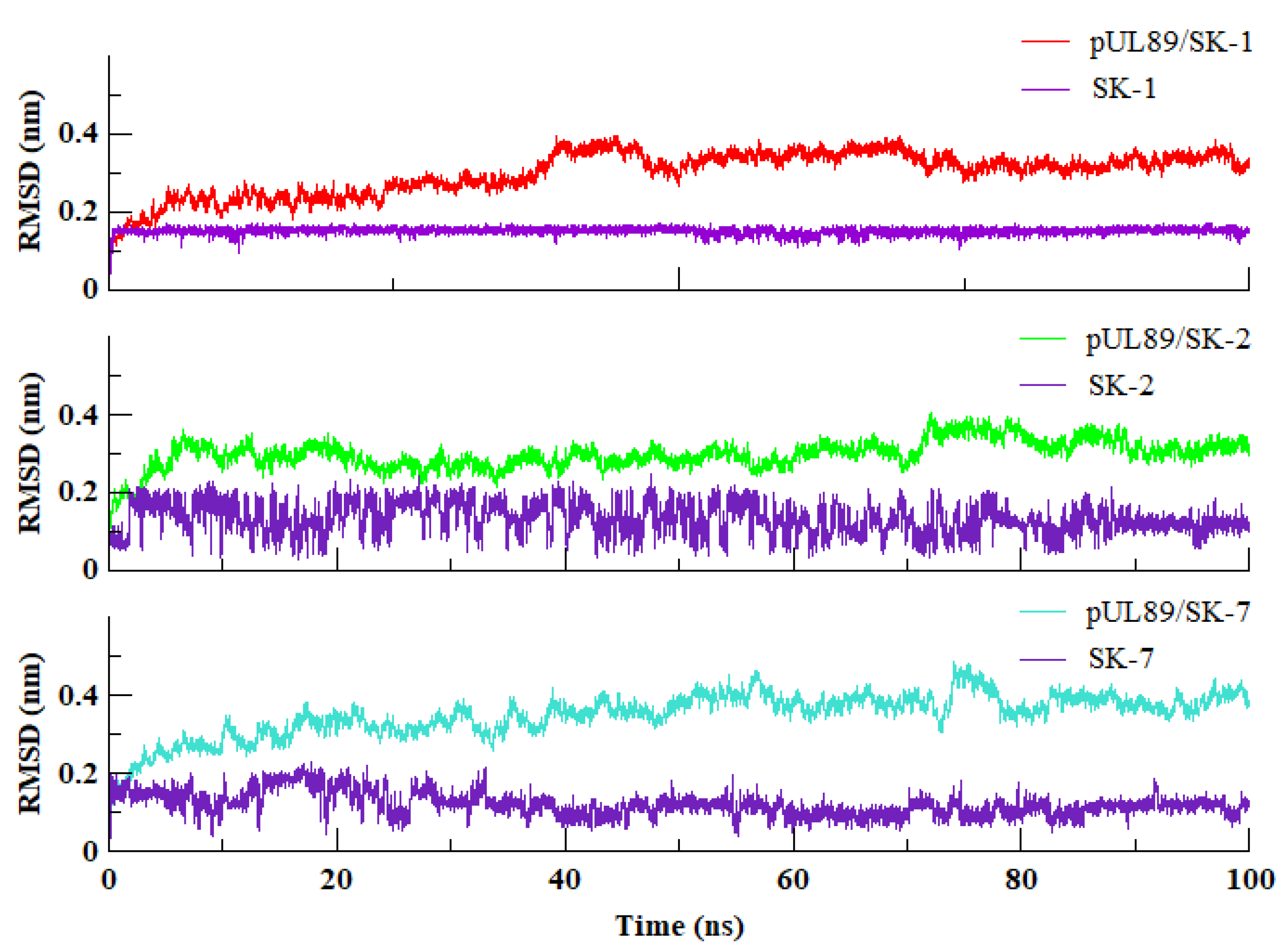
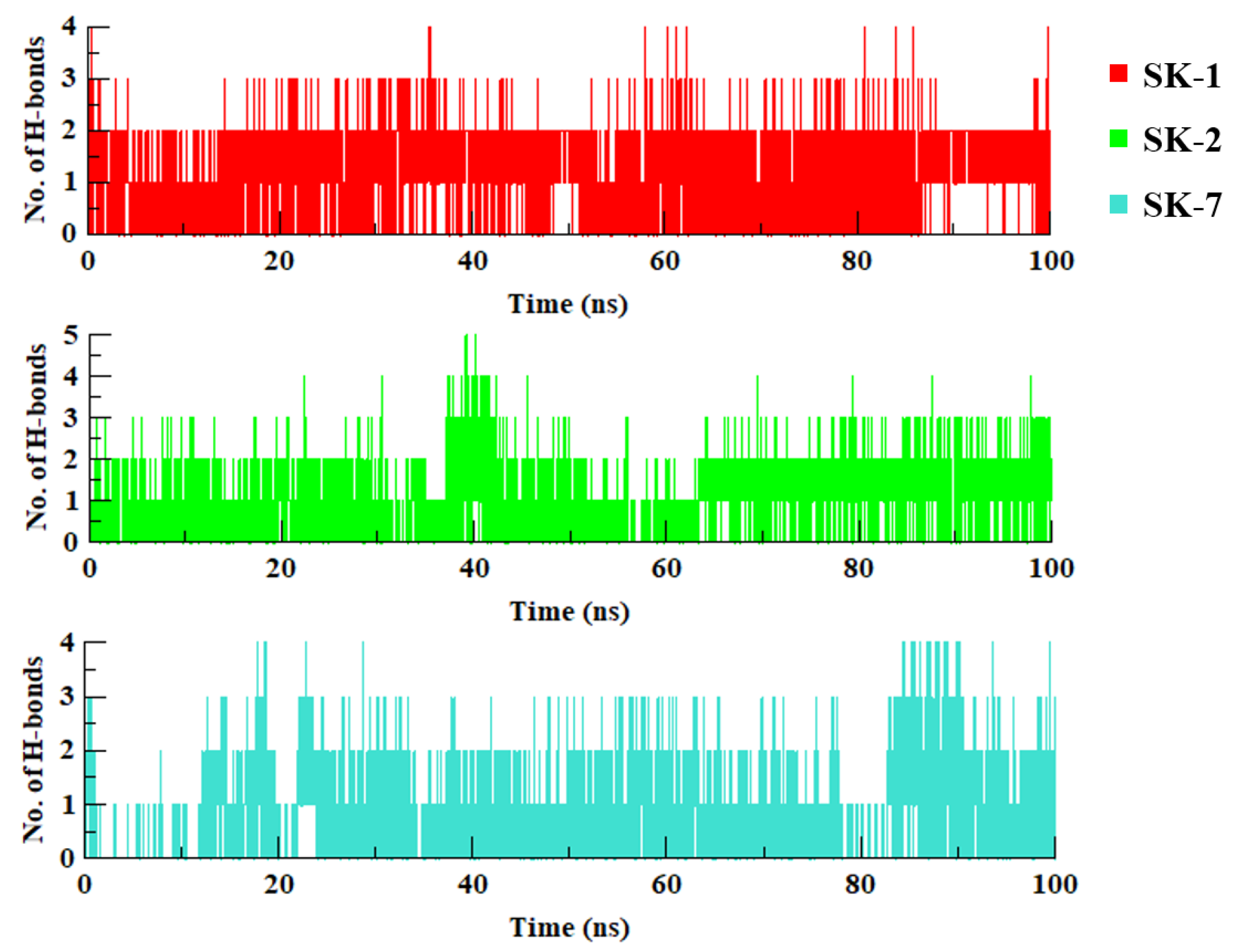
2.2. Molecular Dynamics Simulation
2.3. Binding Free Energy
3. Materials and Methods
3.1. Database Collection and Preparation
3.2. Target Protein Preparation
3.3. Structure and Ligand-Based Virtual Screening
3.4. Molecular Dynamics Simulation
3.5. Binding Free Energy Calculation
= ΔEMM + ΔGGB + ΔGSA − TΔS
= ΔEvdw + ΔEele + ΔGGB + ΔGSA − TΔS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zuhair, M.; Smit, G.S.A.; Wallis, G.; Jabbar, F.; Smith, C.; Devleesschauwer, B.; Griffiths, P. Estimation of the Worldwide Seroprevalence of Cytomegalovirus: A Systematic Review and Meta-Analysis. Rev. Med. Virol. 2019, 29, e2034. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.; Siddiqui, A.A.; Siddiqui, A.R.; Ahmed, W.; Moss, P.A.H.; Lalani, E.-N.M.A. Sociodemographic Factors Associated with IgG and IgM Seroprevalence for Human Cytomegalovirus Infection in Adult Populations of Pakistan: A Seroprevalence Survey. BMC Public Health 2016, 16, 1112. [Google Scholar] [CrossRef] [PubMed]
- Mozafarybazargany, M.; Khoshsirat, N.A. Severe Cytomegalovirus Encephalitis in an Immunocompetent Healthy Young Woman: A Case Report. IDCases 2022, 27, e01403. [Google Scholar] [CrossRef] [PubMed]
- Fulkerson, H.L.; Nogalski, M.T.; Collins-McMillen, D.; Yurochko, A.D. Overview of Human Cytomegalovirus Pathogenesis. In Human Cytomegaloviruses: Methods and Protocols; Yurochko, A.D., Ed.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2021; pp. 1–18. ISBN 978-1-07-161111-1. [Google Scholar]
- Zahid, M.; Kumar, K.; Patel, H. Encephalitis Due to Co-Infection with Cytomegalovirus and Herpes Simplex Virus Type 2 in a Patient with Acquired Immunodeficiency Syndrome. Am. J. Case Rep. 2021, 22, e931821. [Google Scholar] [CrossRef] [PubMed]
- Piret, J.; Boivin, G. Antiviral Drugs Against Herpesviruses. In Antiviral Drug Discovery and Development; Liu, X., Zhan, P., Menéndez-Arias, L., Poongavanam, V., Eds.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2021; pp. 1–30. ISBN 9789811602672. [Google Scholar]
- Oiknine-Djian, E.; Bar-On, S.; Laskov, I.; Lantsberg, D.; Haynes, R.K.; Panet, A.; Wolf, D.G. Artemisone Demonstrates Synergistic Antiviral Activity in Combination with Approved and Experimental Drugs Active against Human Cytomegalovirus. Antivir. Res. 2019, 172, 104639. [Google Scholar] [CrossRef] [PubMed]
- El Helou, G.; Razonable, R.R. Safety Considerations with Current and Emerging Antiviral Therapies for Cytomegalovirus Infection in Transplantation. Expert Opin. Drug Saf. 2019, 18, 1017–1030. [Google Scholar] [CrossRef]
- Mercorelli, B.; Celegato, M.; Luganini, A.; Gribaudo, G.; Lepesheva, G.I.; Loregian, A. The Antifungal Drug Isavuconazole Inhibits the Replication of Human Cytomegalovirus (HCMV) and Acts Synergistically with Anti-HCMV Drugs. Antivir. Res. 2021, 189, 105062. [Google Scholar] [CrossRef]
- Bowman, L.J.; Melaragno, J.I.; Brennan, D.C. Letermovir for the Management of Cytomegalovirus Infection. Expert Opin. Investig. Drugs 2017, 26, 235–241. [Google Scholar] [CrossRef]
- Upadhyayula, S.; Michaels, M.G. Ganciclovir, Foscarnet, and Cidofovir: Antiviral Drugs Not Just for Cytomegalovirus. J. Pediatr. Infect. Dis. Soc. 2013, 2, 286–290. [Google Scholar] [CrossRef]
- Popping, S.; Dalm, V.A.S.H.; Lübke, N.; di Cristanziano, V.; Kaiser, R.; Boucher, C.A.B.; Van Kampen, J.J.A. Emergence and Persistence of Letermovir-Resistant Cytomegalovirus in a Patient With Primary Immunodeficiency. Open Forum Infect. Dis. 2019, 6, ofz375. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.; Song, K.; Wu, J.; Bo, T.; Crumpacker, C. Drug Resistance Mutations and Associated Phenotypes Detected in Clinical Trials of Maribavir for Treatment of Cytomegalovirus Infection. J. Infect. Dis. 2022, 226, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Dobbins, G.C.; Kimberlin, D.W.; Ross, S.A. Cytomegalovirus Variation among Newborns Treated with Valganciclovir. Antivir. Res. 2022, 203, 105326. [Google Scholar] [CrossRef]
- Loregian, A.; Coen, D.M. Selective Anti-Cytomegalovirus Compounds Discovered by Screening for Inhibitors of Subunit Interactions of the Viral Polymerase. Chem. Biol. 2006, 13, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Loregian, A.; Rigatti, R.; Murphy, M.; Schievano, E.; Palu, G.; Marsden, H.S. Inhibition of Human Cytomegalovirus DNA Polymerase by C-Terminal Peptides from the UL54 Subunit. J. Virol. 2003, 77, 8336–8344. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Miller, H.; Knox, K.; Kundu, M.; Henrickson, K.J.; Arav-Boger, R. Inhibition of Human Coronaviruses by Antimalarial Peroxides. ACS Infect. Dis. 2021, 7, 1985–1995. [Google Scholar] [CrossRef]
- Jung, E.; Majima, R.; Edwards, T.C.; Soto-Acosta, R.; Geraghty, R.J.; Wang, Z. 8-Hydroxy-1,6-Naphthyridine-7-Carboxamides as Inhibitors of Human Cytomegalovirus PUL89 Endonuclease. ChemMedChem 2022, 17, e202200334. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Edwards, T.C.; Majima, R.; Jung, E.; Kankanala, J.; Xie, J.; Geraghty, R.J.; Wang, Z. Repurposing N-Hydroxy Thienopyrimidine-2,4-Diones (HtPD) as Inhibitors of Human Cytomegalovirus PUL89 Endonuclease: Synthesis and Biological Characterization. Bioorg. Chem. 2022, 129, 106198. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Edwards, T.C.; Xie, J.; Aihara, H.; Geraghty, R.J.; Wang, Z. 4,5-Dihydroxypyrimidine Methyl Carboxylates, Carboxylic Acids, and Carboxamides as Inhibitors of Human Cytomegalovirus PUL89 Endonuclease. J. Med. Chem. 2022, 65, 5830–5849. [Google Scholar] [CrossRef]
- Senaweera, S.; Edwards, T.C.; Kankanala, J.; Wang, Y.; Sahani, R.L.; Xie, J.; Geraghty, R.J.; Wang, Z. Discovery of N-Benzyl Hydroxypyridone Carboxamides as a Novel and Potent Antiviral Chemotype against Human Cytomegalovirus (HCMV). Acta Pharm. Sin. B 2022, 12, 1671–1684. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, J.; Wang, Z.; Geraghty, R.J. Metal-Chelating 3-Hydroxypyrimidine-2,4-Diones Inhibit Human Cytomegalovirus PUL89 Endonuclease Activity and Virus Replication. Antivir. Res. 2018, 152, 10–17. [Google Scholar] [CrossRef]
- Wang, L.; Edwards, T.C.; Sahani, R.L.; Xie, J.; Aihara, H.; Geraghty, R.J.; Wang, Z. Metal Binding 6-Arylthio-3-Hydroxypyrimidine-2,4-Diones Inhibited Human Cytomegalovirus by Targeting the PUL89 Endonuclease of the Terminase Complex. Eur. J. Med. Chem. 2021, 222, 113640. [Google Scholar] [CrossRef] [PubMed]
- Ligat, G.; Cazal, R.; Hantz, S.; Alain, S. The Human Cytomegalovirus Terminase Complex as an Antiviral Target: A Close-up View. FEMS Microbiol. Rev. 2018, 42, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Neuber, S.; Wagner, K.; Goldner, T.; Lischka, P.; Steinbrueck, L.; Messerle, M.; Borst, E.M. Mutual Interplay between the Human Cytomegalovirus Terminase Subunits PUL51, PUL56, and PUL89 Promotes Terminase Complex Formation. J. Virol. 2017, 91, e02384-16. [Google Scholar] [CrossRef] [PubMed]
- Nadal, M.; Mas, P.J.; Blanco, A.G.; Arnan, C.; Solà, M.; Hart, D.J.; Coll, M. Structure and Inhibition of Herpesvirus DNA Packaging Terminase Nuclease Domain. Proc. Natl. Acad. Sci. USA 2010, 107, 16078–16083. [Google Scholar] [CrossRef]
- Wang, Y.; Mao, L.; Kankanala, J.; Wang, Z.; Geraghty, R.J. Inhibition of Human Cytomegalovirus PUL89 Terminase Subunit Blocks Virus Replication and Genome Cleavage. J. Virol. 2017, 91, e02152-16. [Google Scholar] [CrossRef]
- Kankanala, J.; Wang, Y.; Geraghty, R.J.; Wang, Z. Hydroxypyridonecarboxylic Acids as Inhibitors of Human Cytomegalovirus PUL89 Endonuclease. ChemMedChem 2018, 13, 1658–1663. [Google Scholar] [CrossRef]
- Bongarzone, S.; Nadal, M.; Kaczmarska, Z.; Machón, C.; Álvarez, M.; Albericio, F.; Coll, M. Structure-Driven Discovery of α,γ-Diketoacid Inhibitors Against UL89 Herpesvirus Terminase. ACS Omega 2018, 3, 8497–8505. [Google Scholar] [CrossRef]
- Tan, L.; Geppert, H.; Sisay, M.T.; Gütschow, M.; Bajorath, J. Integrating Structure- and Ligand-Based Virtual Screening: Comparison of Individual, Parallel, and Fused Molecular Docking and Similarity Search Calculations on Multiple Targets. ChemMedChem 2008, 3, 1566–1571. [Google Scholar] [CrossRef]
- Krüger, D.M.; Evers, A. Comparison of Structure- and Ligand-Based Virtual Screening Protocols Considering Hit List Complementarity and Enrichment Factors. ChemMedChem 2010, 5, 148–158. [Google Scholar] [CrossRef]
- Svensson, F.; Karlén, A.; Sköld, C. Virtual Screening Data Fusion Using Both Structure- and Ligand-Based Methods. J. Chem. Inf. Model. 2012, 52, 225–232. [Google Scholar] [CrossRef]
- Khan, S.A.; Khan, A.; Zia, K.; Shawish, I.; Barakat, A.; Ul-Haq, Z. Cheminformatics-Based Discovery of Potential Chemical Probe Inhibitors of Omicron Spike Protein. Int. J. Mol. Sci. 2022, 23, 10315. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Zia, K.; Ashraf, S.; Uddin, R.; Ul-Haq, Z. Identification of Chymotrypsin-like Protease Inhibitors of SARS-CoV-2 via Integrated Computational Approach. J. Biomol. Struct. Dyn. 2021, 39, 2607–2616. [Google Scholar] [CrossRef] [PubMed]
- Zia, K.; Khan, S.A.; Ashraf, S.; Nur-e-Alam, M.; Ahmed, S.; Ul-Haq, Z. Probing CAS Database as Prospective Antiviral Agents against SARS-CoV-2 Main Protease. J. Mol. Struct. 2021, 1231, 129953. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A Free Tool to Discover Chemistry for Biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. G_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
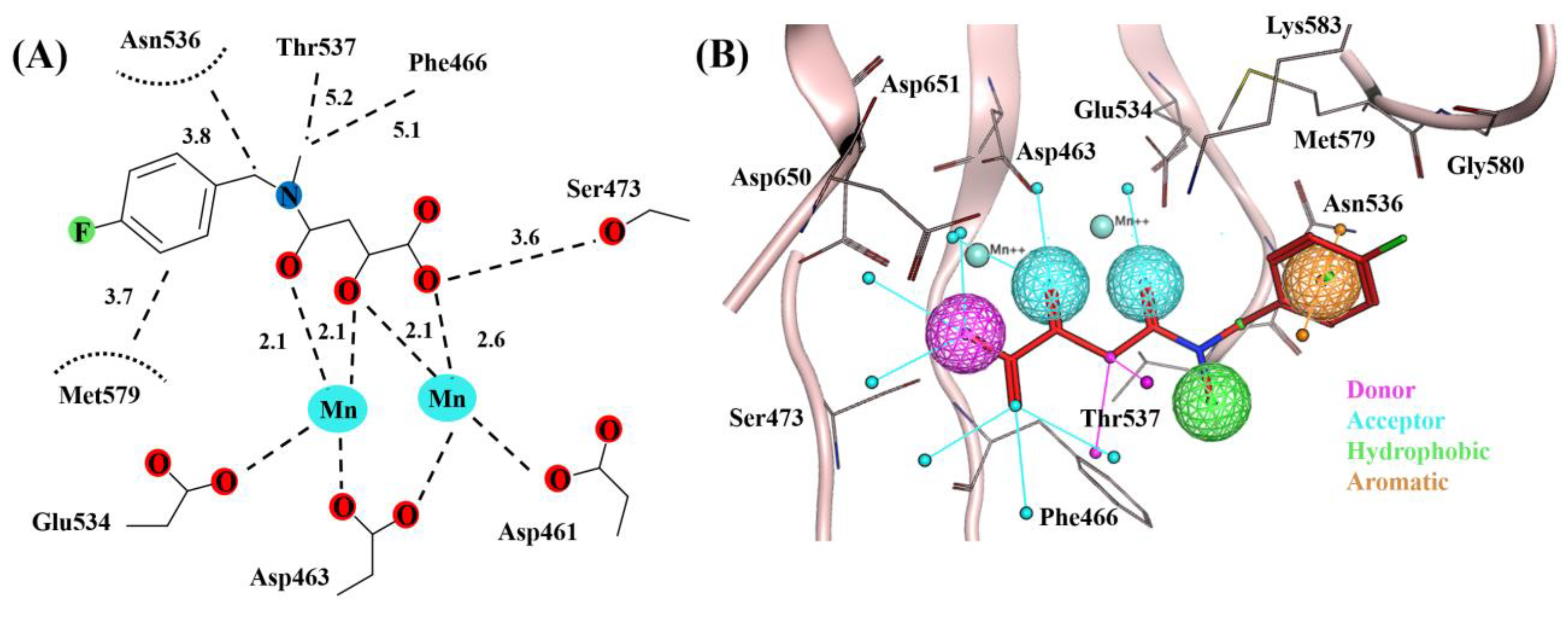
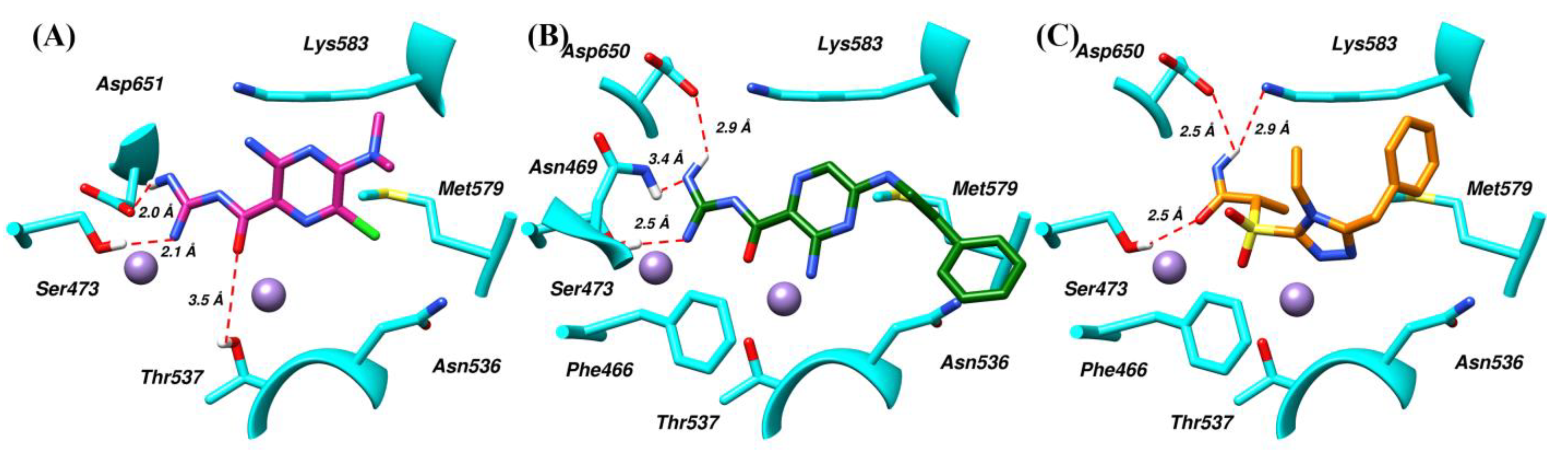

| System | Type of Interaction | Residue | Distance with Highest Occupancy (Å) | % Occupancy |
|---|---|---|---|---|
| SK-1 | Hydrogen bonding | Ser473 | 2.5 | 63 |
| Thr537 | 3.5 | 69 | ||
| Asp651 | 2.5 | 78 | ||
| Hydrophobic | Asn536 | 5.0 | 35 | |
| Met579 | -- | -- | ||
| Lys583 | 4.6 | 88 | ||
| SK-2 | Hydrogen bonding | Ser473 | 2.2 | 57 |
| Asn469 | 3.6 | 54 | ||
| Asp650 | -- | -- | ||
| Hydrophobic | Phe466 | 4.4 | 81 | |
| Asn536 | 4.1 | 98 | ||
| Thr537 | 3.5 | 66 | ||
| Met579 | 5.2 | 84 | ||
| SK-7 | Hydrogen bonding | Ser473 | 3.5 | 51 |
| Lys583 | 3.1 | 76 | ||
| Asp650 | 2.3 | 80 | ||
| Hydrophobic | Phe466 | 4.4 | 74 | |
| Asn536 | 4.1 | 62 | ||
| Thr537 | -- | -- | ||
| Met579 | 4.8 | 91 | ||
| Lys583 | 4.1 | 72 |
| Code | Structure | Database ID | Binding Score (kcal/mol) | Binding Energy (kJ/mol) |
|---|---|---|---|---|
| SK-1 | 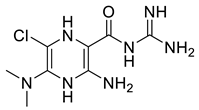 | ChEMBL465179 | −6.10 | −51.99 |
| SK-2 |  | ChEMBL1962850 | −6.15 | −46.63 |
| SK-7 |  | ZINC79951807 | −6.33 | −48.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almehmadi, M.; Haq, I.U.; Alsaiari, A.A.; Alshabrmi, F.M.; Abdulaziz, O.; Allahyani, M.; Aladhadh, M.; Shafie, A.; Aljuaid, A.; Alotaibi, R.T.; et al. Identification of Small Molecule Inhibitors of Human Cytomegalovirus pUL89 Endonuclease Using Integrated Computational Approaches. Molecules 2023, 28, 3938. https://doi.org/10.3390/molecules28093938
Almehmadi M, Haq IU, Alsaiari AA, Alshabrmi FM, Abdulaziz O, Allahyani M, Aladhadh M, Shafie A, Aljuaid A, Alotaibi RT, et al. Identification of Small Molecule Inhibitors of Human Cytomegalovirus pUL89 Endonuclease Using Integrated Computational Approaches. Molecules. 2023; 28(9):3938. https://doi.org/10.3390/molecules28093938
Chicago/Turabian StyleAlmehmadi, Mazen, Ihtisham Ul Haq, Ahad Amer Alsaiari, Fahad M. Alshabrmi, Osama Abdulaziz, Mamdouh Allahyani, Mohammed Aladhadh, Alaa Shafie, Abdulelah Aljuaid, Rema Turki Alotaibi, and et al. 2023. "Identification of Small Molecule Inhibitors of Human Cytomegalovirus pUL89 Endonuclease Using Integrated Computational Approaches" Molecules 28, no. 9: 3938. https://doi.org/10.3390/molecules28093938
APA StyleAlmehmadi, M., Haq, I. U., Alsaiari, A. A., Alshabrmi, F. M., Abdulaziz, O., Allahyani, M., Aladhadh, M., Shafie, A., Aljuaid, A., Alotaibi, R. T., Ullah, J., & Alharthi, N. S. (2023). Identification of Small Molecule Inhibitors of Human Cytomegalovirus pUL89 Endonuclease Using Integrated Computational Approaches. Molecules, 28(9), 3938. https://doi.org/10.3390/molecules28093938





