Unlocking the Conformational Changes of P2Y12: Exploring an Acridinone Compound’s Effect on Receptor Activity and Conformation
Abstract
1. Introduction
2. Results
3. Discussion
4. Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Najjar, B.O.; Saqallah, F.G.; Abbas, M.A.; Al-Hijazeen, S.Z.; Sibai, O.A. P2Y12 antagonists: Approved drugs, potential naturally isolated and synthesised compounds, and related in-silico studies. Eur. J. Med. Chem. 2022, 227, 113924. [Google Scholar] [CrossRef] [PubMed]
- Al-Najjar, B.O.; Abbas, M.A.; Sibai, O.A.; Saqallah, F.G.; Al-Kabariti, A.Y. QSAR, structure-based pharmacophore modelling and biological evaluation of novel platelet ADP receptor (P2Y12) antagonist. RSC Med. Chem. 2023, 14, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Al-Najjar, B.O. Investigation of 15-hydroxyprostaglandin dehydrogenase catalytic reaction mechanism by molecular dynamics simulations. J. Mol. Graph. Model. 2018, 80, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Al-Anazi, M.; Al-Najjar, B.O.; Khairuddean, M. Structure-Based Drug Design Studies Toward the Discovery of Novel Chalcone Derivatives as Potential Epidermal Growth Factor Receptor (EGFR) Inhibitors. Molecules 2018, 23, 3203. [Google Scholar] [CrossRef]
- Al-Anazi, M.; Khairuddean, M.; Al-Najjar, B.O.; Murwih Alidmat, M.; Nur Syazni Nik Mohamed Kamal, N.; Muhamad, M. Synthesis, anticancer activity and docking studies of pyrazoline and pyrimidine derivatives as potential epidermal growth factor receptor (EGFR) inhibitors. Arab. J. Chem. 2022, 15, 103864. [Google Scholar] [CrossRef]
- Xie, C.-L.; Zhang, D.; Guo, K.-Q.; Yan, Q.-X.; Zou, Z.-B.; He, Z.-H.; Wu, Z.; Zhang, X.-K.; Chen, H.-F.; Yang, X.-W. Meroterpenthiazole A, a unique meroterpenoid from the deep-sea-derived Penicillium allii-sativi, significantly inhibited retinoid X receptor (RXR)-α transcriptional effect. Chin. Chem. Lett. 2022, 33, 2057–2059. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, Y.; Zhang, X.; Chen, L.; Zheng, M.; Zhang, J.; Brust, P.; Deuther-Conrad, W.; Huang, Y.; Jia, H. Synthesis and characterization of the two enantiomers of a chiral sigma-1 receptor radioligand: (S)-(+)- and (R)-(-)-[18F]FBFP. Chin. Chem. Lett. 2022, 33, 3543–3548. [Google Scholar] [CrossRef]
- Li, M.; Xia, Q.; Lv, S.; Tong, J.; Wang, Z.; Nie, Q.; Yang, J. Enhanced CO2 capture for photosynthetic lycopene production in engineered Rhodopseudomonas palustris, a purple nonsulfur bacterium. Green Chem. 2022, 24, 7500–7518. [Google Scholar] [CrossRef]
- Harismah, K.; Da’i, M.; Azimzadeh-Sadeghi, S.; Poursafa, P.; Mirzaei, M.; Salarrezaei, E. Interactions of coumarin derivatives with monoamine oxidase biomarkers: In silico approach. Main Group Chem. 2022, 21, 641–650. [Google Scholar] [CrossRef]
- Green ChemistryProtoplasmaMirzaei, M.; Harismah, K.; Soleimani, M.; Mousavi, S. Inhibitory effects of curcumin on aldose reductase and cyclooxygenase-2 enzymes. J. Biomol. Struct. Dyn. 2021, 39, 6424–6430. [Google Scholar]
- Zhang, K.; Yang, Y.; Ge, H.; Wang, J.; Lei, X.; Chen, X.; Wan, F.; Feng, H.; Tan, L. Neurogenesis and Proliferation of Neural Stem/Progenitor Cells Conferred by Artesunate via FOXO3a/p27Kip1 Axis in Mouse Stroke Model. Mol. Neurobiol. 2022, 59, 4718–4729. [Google Scholar] [CrossRef] [PubMed]
- ChemSpider N,N′-Bis(9-oxo-9,10-dihydro-2-acridinyl)nonanediamide. Available online: http://www.chemspider.com/Chemical-Structure.387488.html (accessed on 29 April 2023).
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Von Bertalanffy, L. Acridine orange fluorescence in cell physiology, cytochemistry and medicine. Protoplasma 1963, 57, 51–83. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, K.; Gao, Z.-G.; Paoletta, S.; Zhang, D.; Han, G.W.; Li, T.; Ma, L.; Zhang, W.; Müller, C.E.; et al. Agonist-bound structure of the human P2Y12 receptor. Nature 2014, 509, 119–122. [Google Scholar] [CrossRef]
- Dandan, Z.; Qiang, Z.; Beili, W. Structural Studies of G Protein-Coupled Receptors. Mol. Cells 2015, 38, 836–842. [Google Scholar]
- Hao, M.; Li, Y.; Wang, Y.; Yan, Y.; Zhang, S. Combined 3D-QSAR, Molecular Docking, and Molecular Dynamics Study on Piperazinyl-Glutamate-Pyridines/Pyrimidines as Potent P2Y12 Antagonists for Inhibition of Platelet Aggregation. J. Chem. Inf. Model. 2011, 51, 2560–2572. [Google Scholar] [CrossRef]
- Paoletta, S.; Sabbadin, D.; Von Kügelgen, I.; Hinz, S.; Katritch, V.; Hoffmann, K.; Abdelrahman, A.; Straßburger, J.; Baqi, Y.; Zhao, Q.; et al. Modeling ligand recognition at the P2Y12 receptor in light of X-ray structural information. J. Comput. Aided Mol. Des. 2015, 29, 737–756. [Google Scholar] [CrossRef]
- Haghighi, F. Prediction of Ticagrelor’s Effect on the Lipid Composition and the P2Y12 Receptor of Platelet’s Membrane by Molecular Dynamic and Docking. Ph.D. Thesis, Université de Franche-Comté, Besançon, France, 2019. [Google Scholar]
- Parravicini, C.; Ranghino, G.; Abbracchio, M.P.; Fantucci, P. GPR17: Molecular modeling and dynamics studies of the 3-D structure and purinergic ligand binding features in comparison with P2Y receptors. BMC Bioinform. 2008, 9, 263. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Sato, M.; Kimura, M.; Ichinohe, T.; Tazaki, M.; Shibukawa, Y. Expression and function of purinergic P2Y12 receptors in rat trigeminal ganglion neurons. Neurosci. Res. 2015, 98, 17–27. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham III, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER 2018; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Dassault-Systèmes. BIOVIA, Discovery Studio Modeling Environment; 16.1; Dassault Systèmes Biovia: San Diego, CA, USA, 2016. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Jakalian, A.; Bush, B.L.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J. Comput. Chem. 2000, 21, 132–146. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Leach, A.R. Molecular Modelling: Principles and Applications; Prentice Hall: Hoboken, NJ, USA, 2001. [Google Scholar]
- Li, P.; Song, L.F.; Merz, K.M., Jr. Systematic Parameterization of Monovalent Ions Employing the Nonbonded Model. J. Chem. Theory Comput. 2015, 11, 1645–1657. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Durrant, J.D.; Votapka, L.; Sørensen, J.; Amaro, R.E. POVME 2.0: An Enhanced Tool for Determining Pocket Shape and Volume Characteristics. J. Chem. Theory Comput. 2014, 10, 5047–5056. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
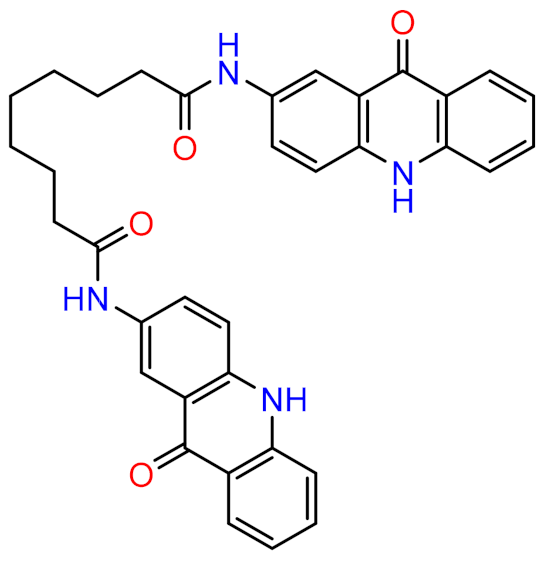
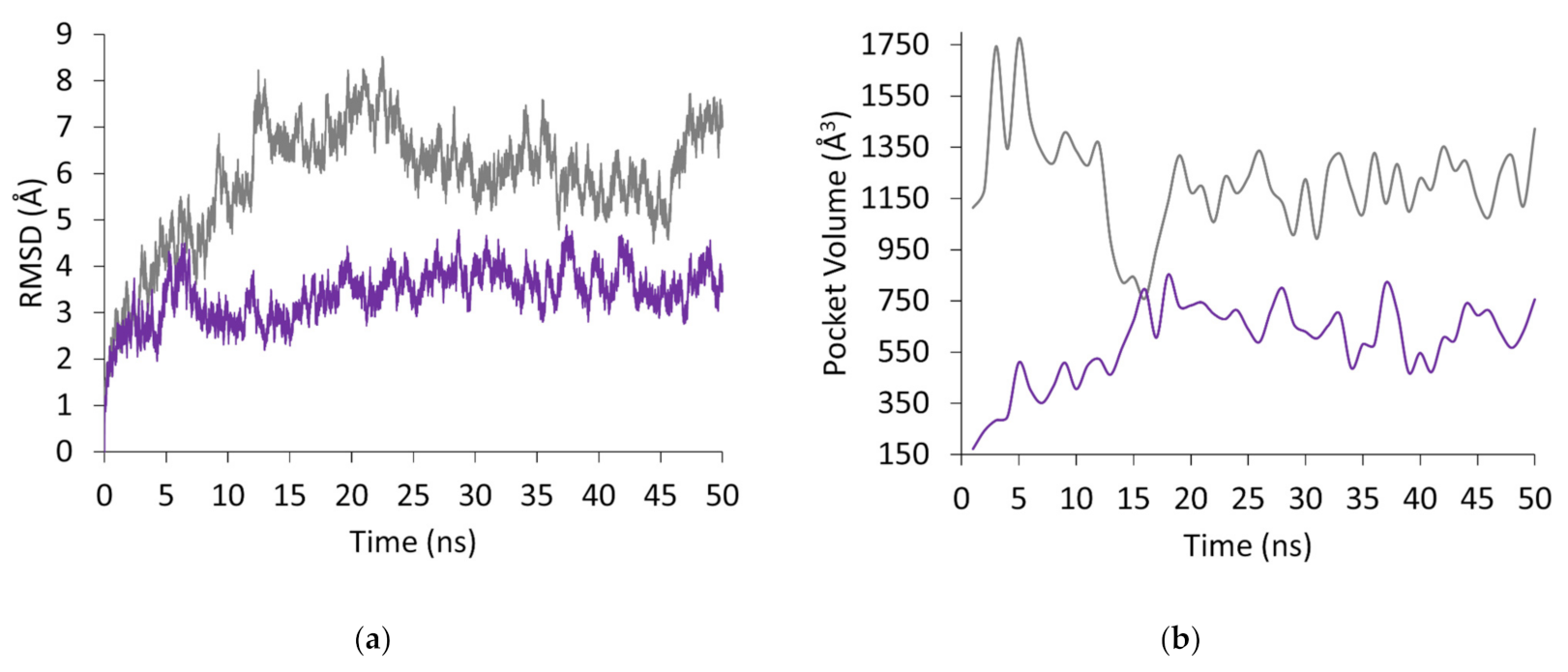
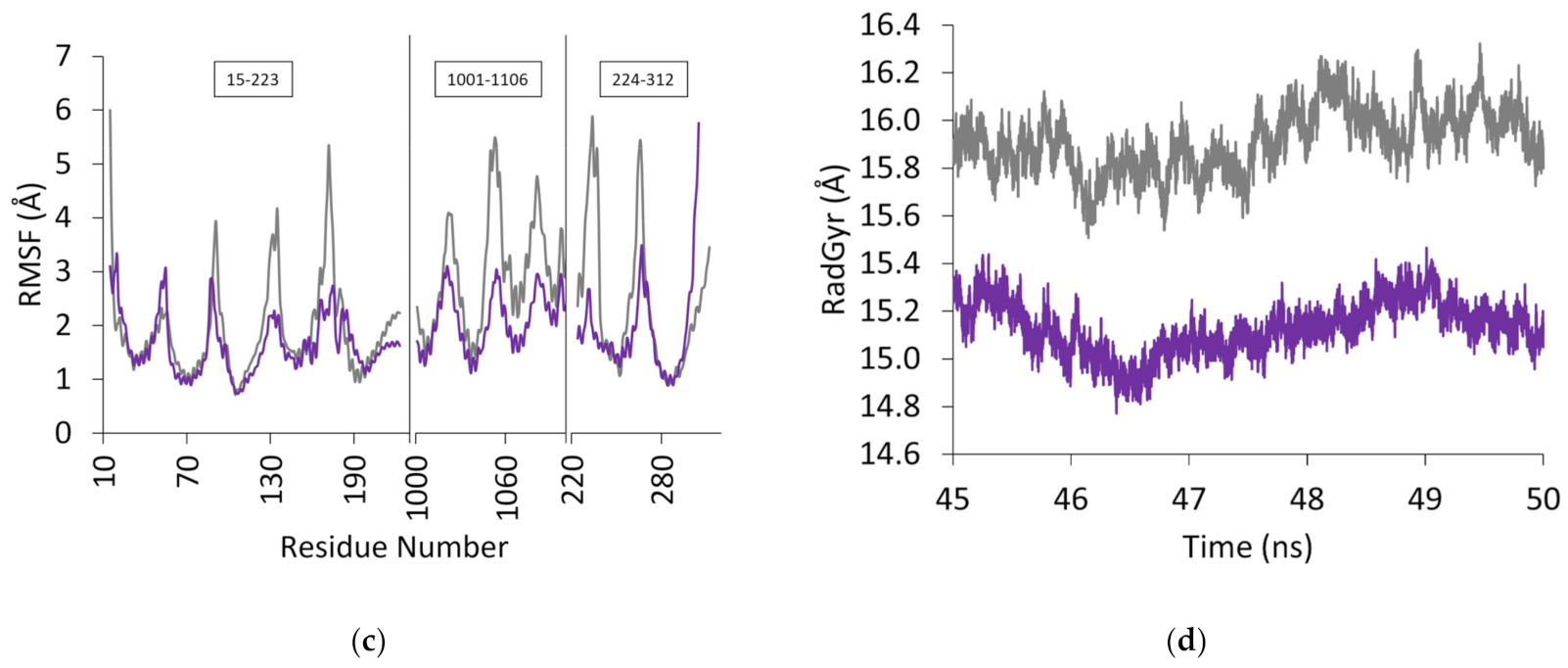
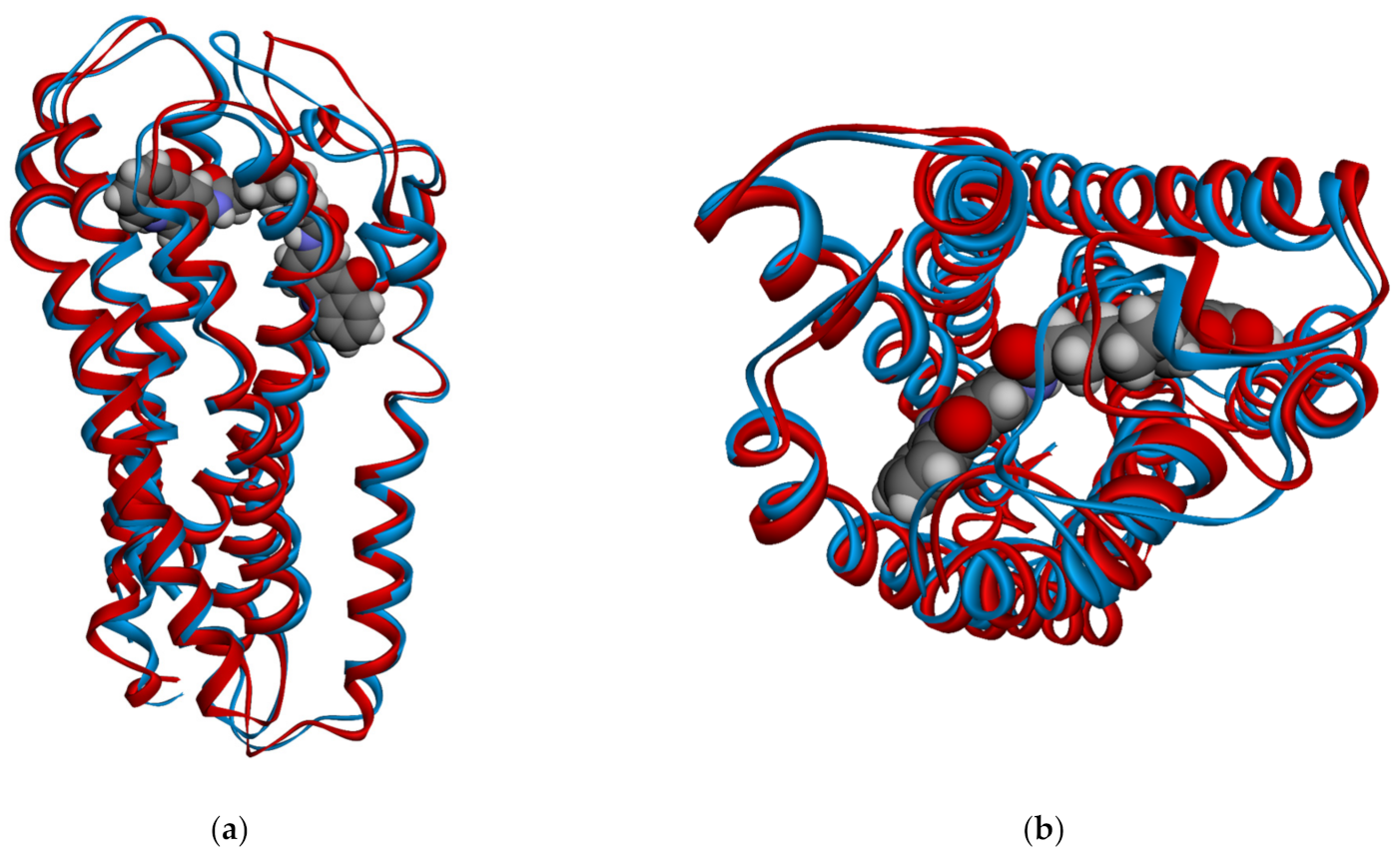


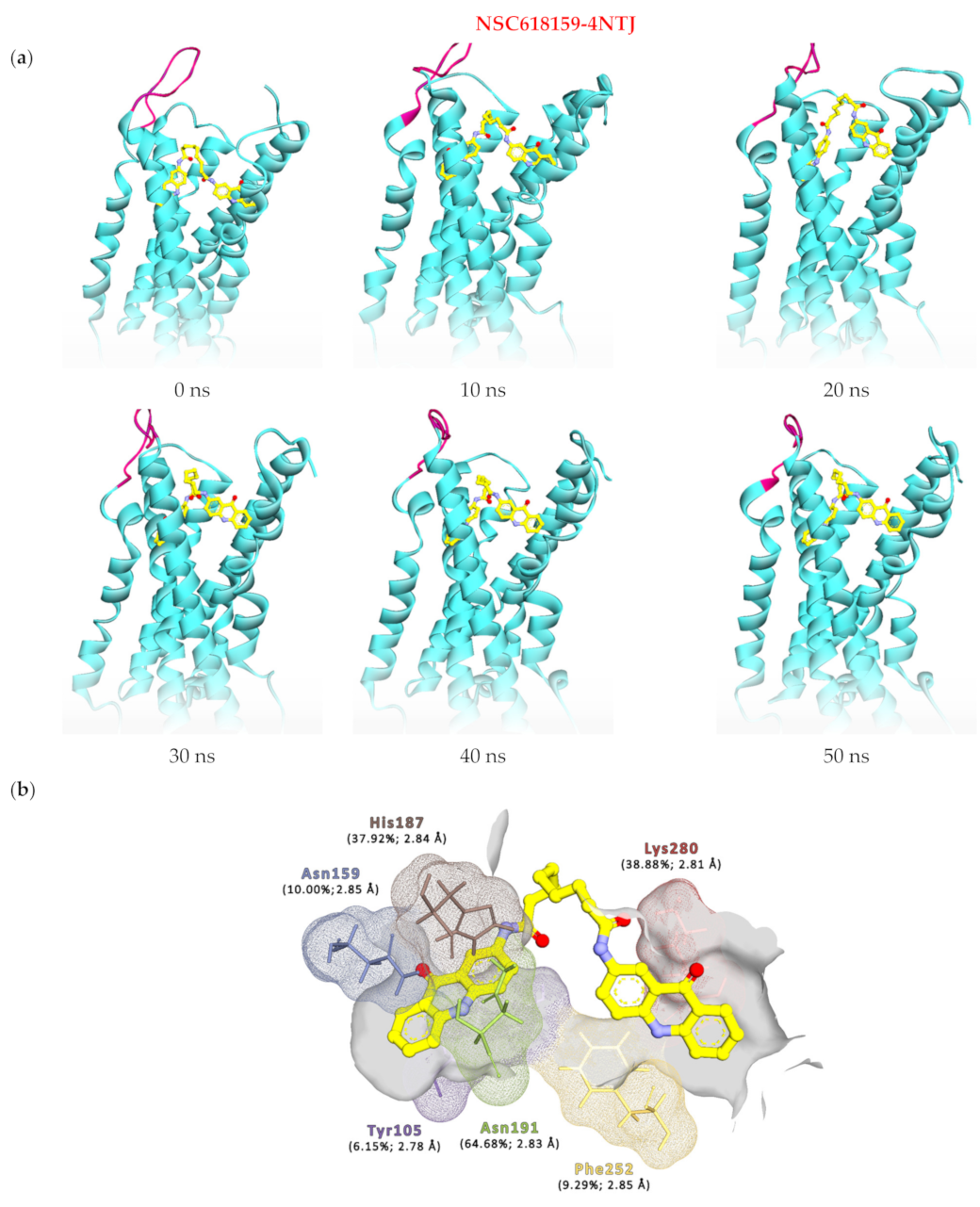

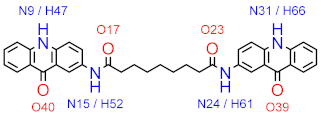
| Complex | Acceptor | Donor | Occupancy (%) | Average Distance (Å) | Average Angle (Å) |
|---|---|---|---|---|---|
| 4PXZ | GLU281@OE2 | NSC159@H47/N9 * | 90.93 | 2.74 | 154.18 |
| NSC159@O39 * | ASN159@HD22/ND2 | 68.41 | 2.84 | 159.27 | |
| NSC159@O17 * | TYR105@HH/OH | 31.94 | 2.75 | 160.15 | |
| NSC159@O17 * | TYR259@HH/OH | 17.84 | 2.77 | 162.74 | |
| NSC159@O23 * | LYS179@HZ3/NZ | 16.03 | 2.81 | 154.93 | |
| NSC159@O23 * | LYS179@HZ2/NZ | 15.73 | 2.81 | 155.21 | |
| NSC159@O23 * | LYS179@HZ1/NZ | 15.24 | 2.81 | 155.19 | |
| CYS79@O | NSC159@H61/N24 * | 11.17 | 2.88 | 161.19 | |
| ASN191@OD1 | NSC159@H66/N31 * | 7.98 | 2.83 | 151.56 | |
| NSC159@O17 * | GLN263@HE21/NE2 | 6.59 | 2.86 | 159.53 | |
| SER101@O | NSC159@H61/N24 * | 5.93 | 2.88 | 156.16 | |
| 4NTJ | ASN191@OD1 | NSC159@H66/N31 * | 64.68 | 2.83 | 162.28 |
| NSC159@O17 * | HIS187@HE2/NE2 | 37.92 | 2.84 | 155.02 | |
| NSC159@O23 * | LYS280@HZ2/NZ | 14.10 | 2.81 | 154.99 | |
| NSC159@O23 * | LYS280@HZ1/NZ | 12.73 | 2.81 | 154.62 | |
| NSC159@O23 * | LYS280@HZ3/NZ | 12.05 | 2.81 | 154.49 | |
| NSC159@O39 * | ASN159@HD22/ND2 | 10.00 | 2.85 | 159.42 | |
| PHE252@O | NSC159@H47/N9 * | 9.29 | 2.85 | 159.95 | |
| NSC159@O23 * | TYR105@HH/OH | 6.15 | 2.78 | 159.14 |
 .
.| Energy Component (kcal/mol) | Energy Value (kcal/mol) per Complex | |
|---|---|---|
| NSC618159-4PXZ | NSC618159-4NTJ | |
| van der Waals Energy (ΔEvdW) | −71.783 | −64.022 |
| ±4.0481 | ±3.4897 | |
| Electrostatic Energy (ΔEEL) | −49.4273 | −54.907 |
| ±6.2724 | ±7.715 | |
| Polar Solvation Energy (ΔEPB) | 84.6431 | 83.1979 |
| ±8.5487 | ±9.1691 | |
| Non-Polar Solvation Energy (ΔENPOLAR) | −6.9489 | −5.9511 |
| ±0.1474 | ±0.2037 | |
| Total Gas Phase Free Energy (ΔGgas) | −121.2103 | −118.929 |
| ±6.9472 | ±7.9284 | |
| Total Solvation Free Energy (ΔGsolv) | 77.6942 | 77.2468 |
| ±8.5099 | ±9.1647 | |
| Total Energy (ΔGbind) | −43.5161 | −41.6822 |
| ±6.0448 | ±7.3535 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Najjar, B.O.; Saqallah, F.G. Unlocking the Conformational Changes of P2Y12: Exploring an Acridinone Compound’s Effect on Receptor Activity and Conformation. Molecules 2023, 28, 3878. https://doi.org/10.3390/molecules28093878
Al-Najjar BO, Saqallah FG. Unlocking the Conformational Changes of P2Y12: Exploring an Acridinone Compound’s Effect on Receptor Activity and Conformation. Molecules. 2023; 28(9):3878. https://doi.org/10.3390/molecules28093878
Chicago/Turabian StyleAl-Najjar, Belal O., and Fadi G. Saqallah. 2023. "Unlocking the Conformational Changes of P2Y12: Exploring an Acridinone Compound’s Effect on Receptor Activity and Conformation" Molecules 28, no. 9: 3878. https://doi.org/10.3390/molecules28093878
APA StyleAl-Najjar, B. O., & Saqallah, F. G. (2023). Unlocking the Conformational Changes of P2Y12: Exploring an Acridinone Compound’s Effect on Receptor Activity and Conformation. Molecules, 28(9), 3878. https://doi.org/10.3390/molecules28093878






