Bioinformatic Analysis of Key Regulatory Genes in Adult Asthma and Prediction of Potential Drug Candidates
Abstract
1. Introduction
2. Results
2.1. Identification of DEGs
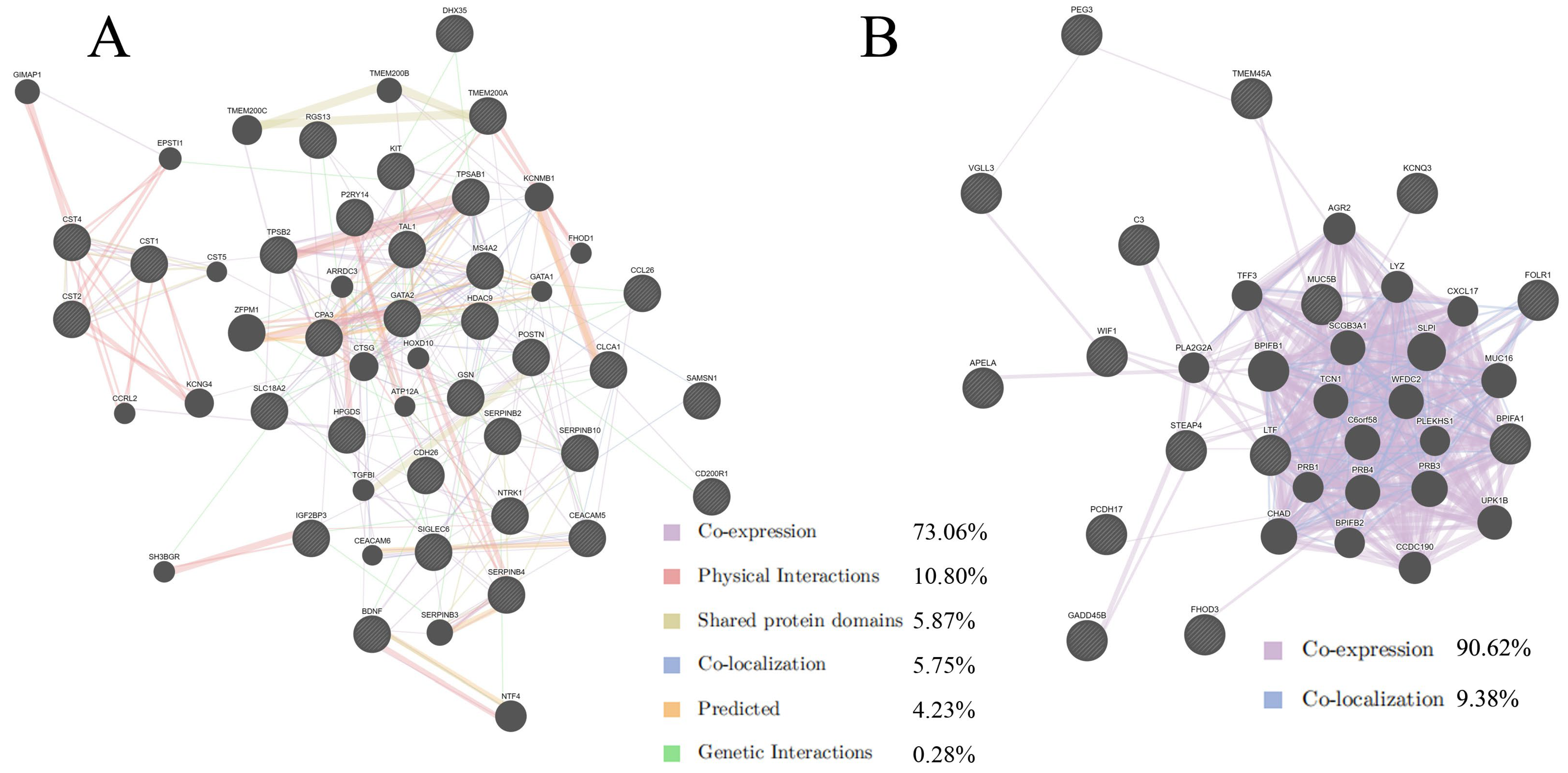
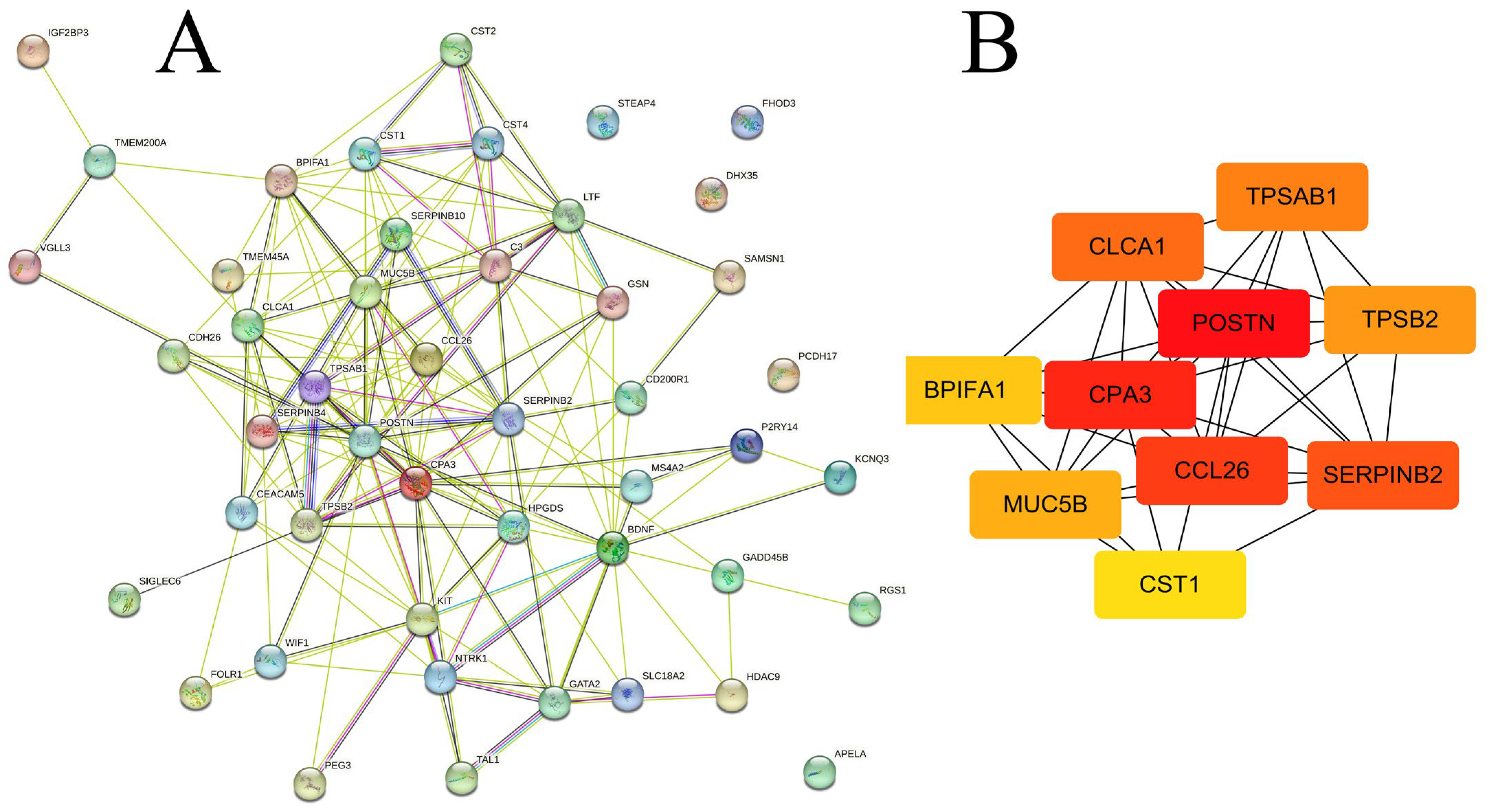
2.2. Protein–Protein Interaction Analysis by GeneMANIA
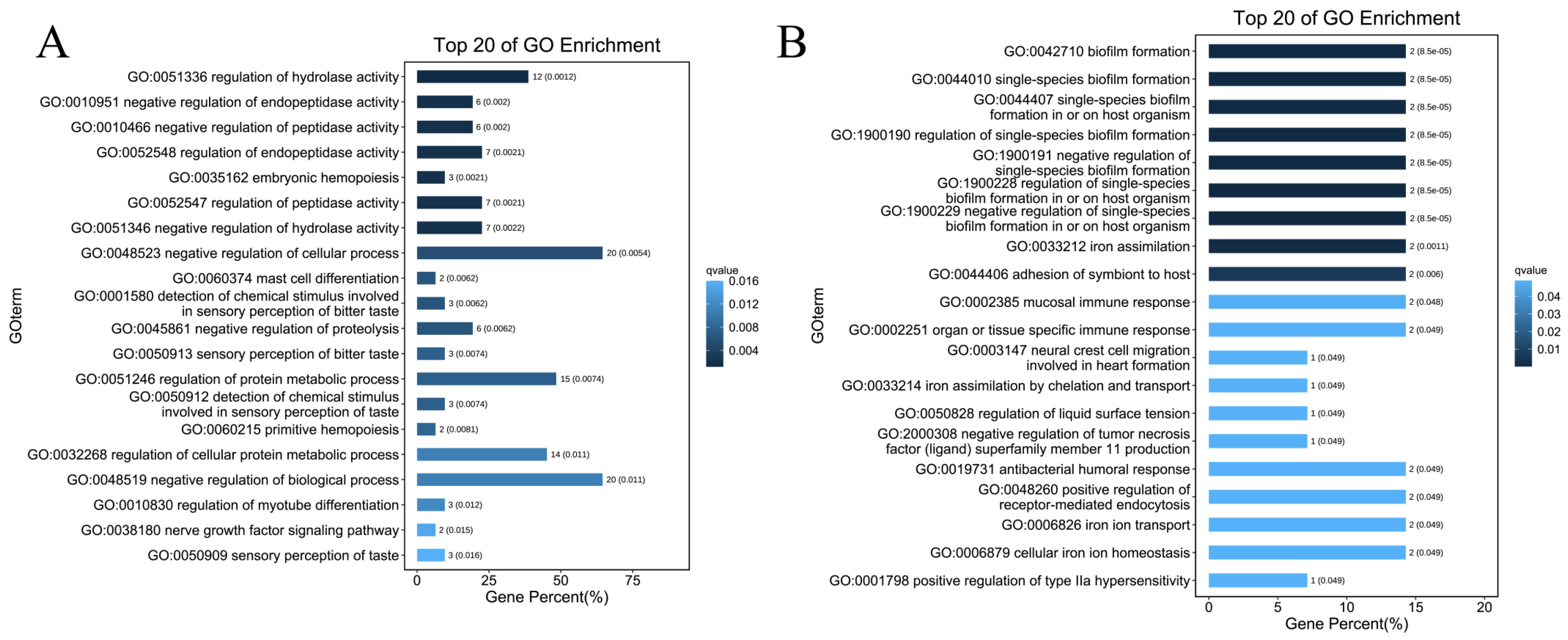
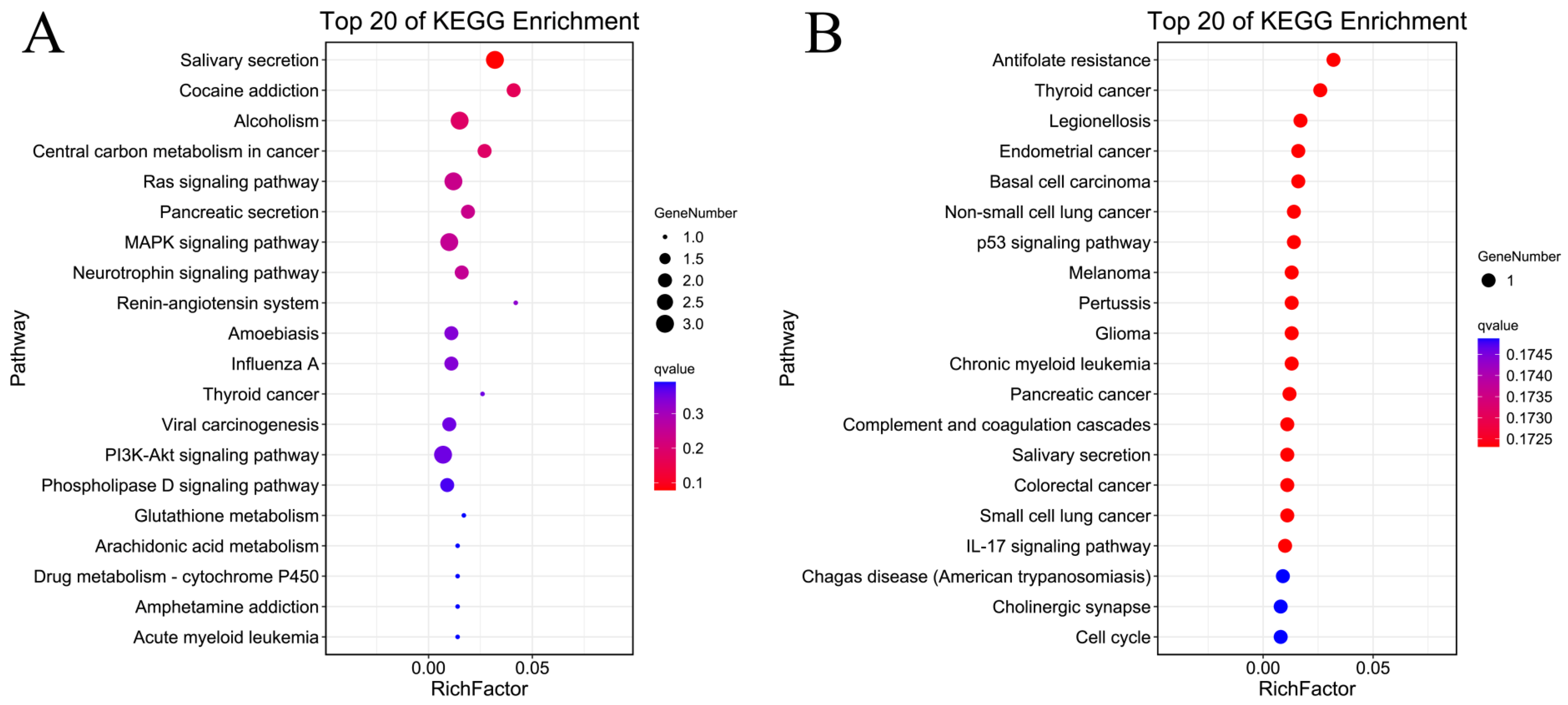
2.3. GO Enrichment and KEGG Enrichment Analyses
2.4. Identification of Hub Gene and Modules
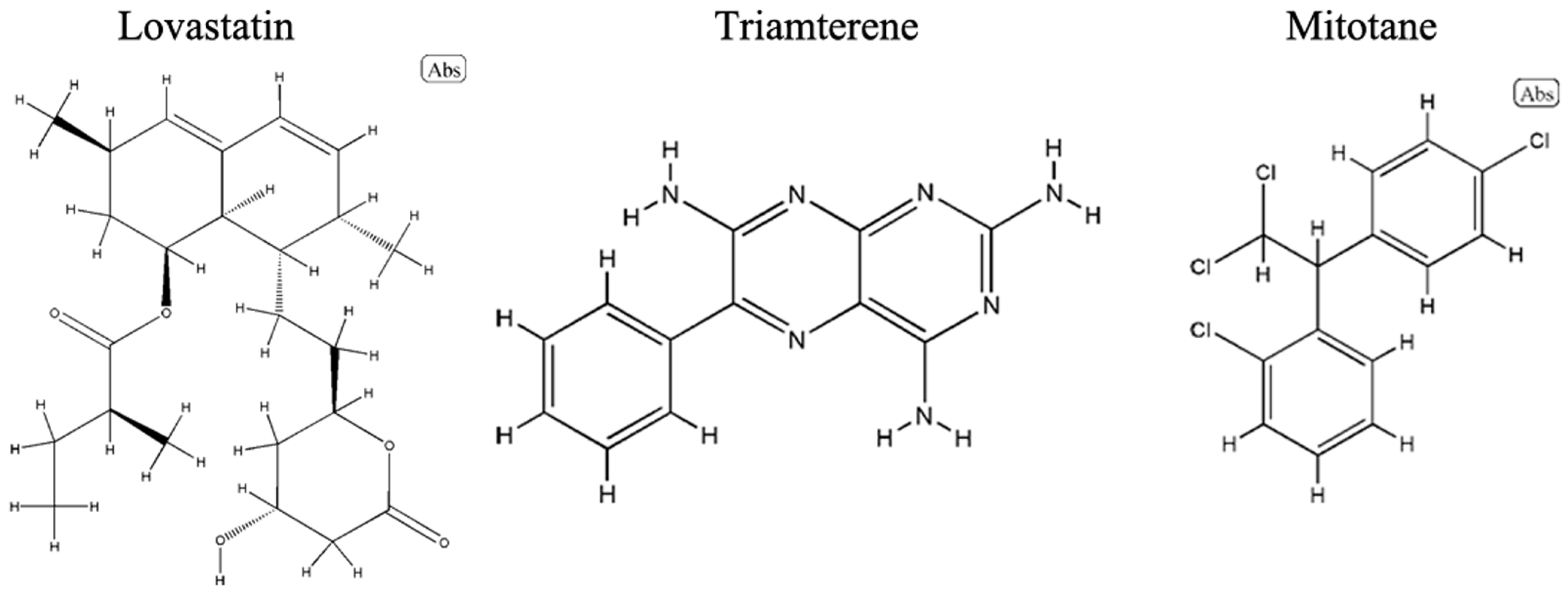
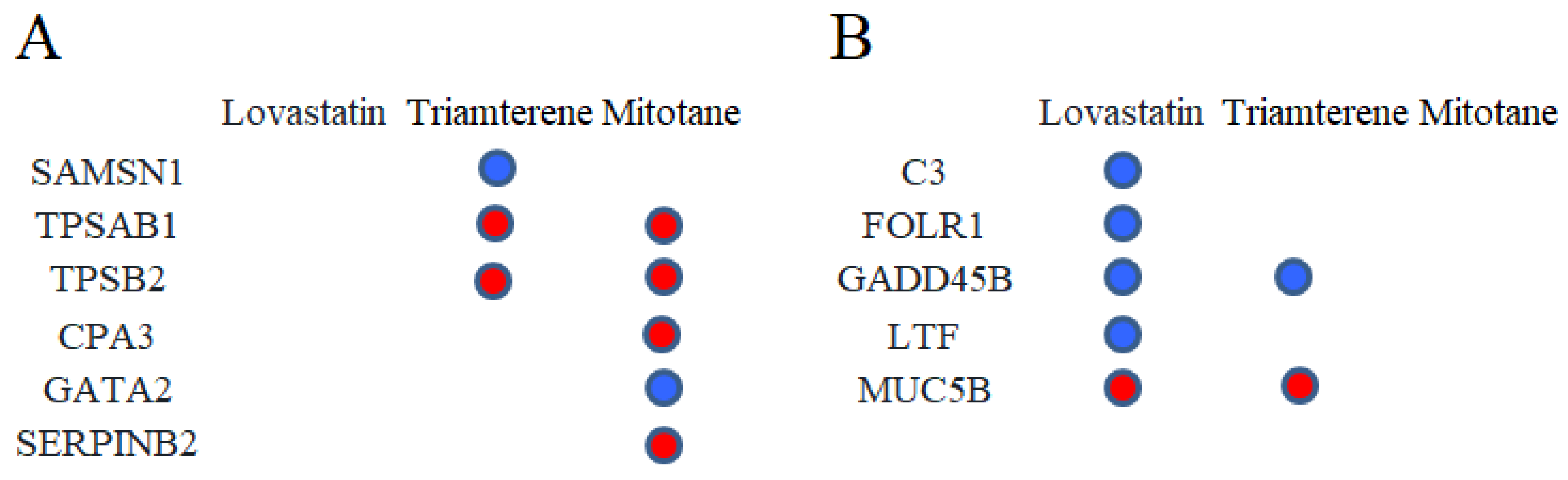
2.5. Drug Prediction
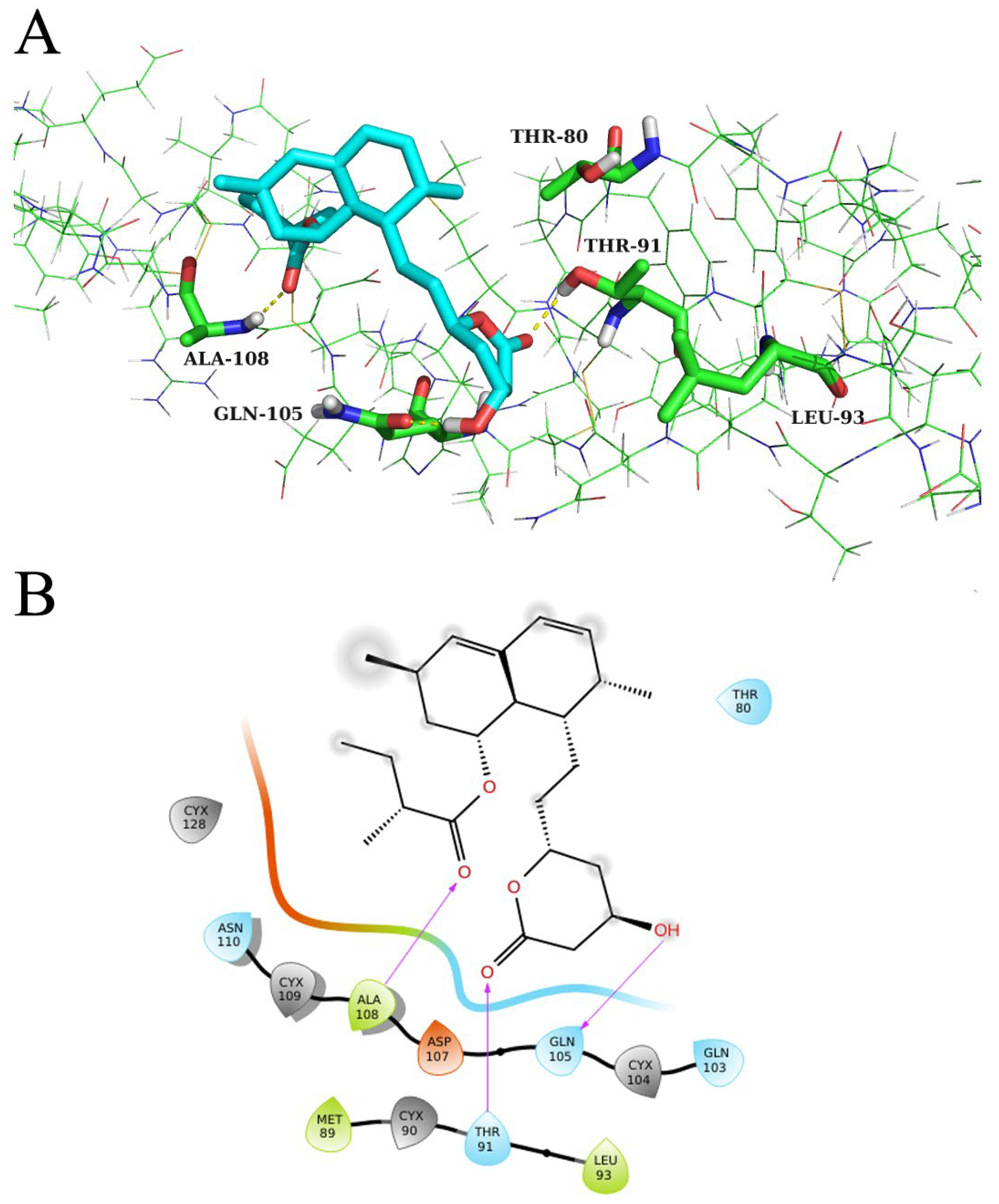
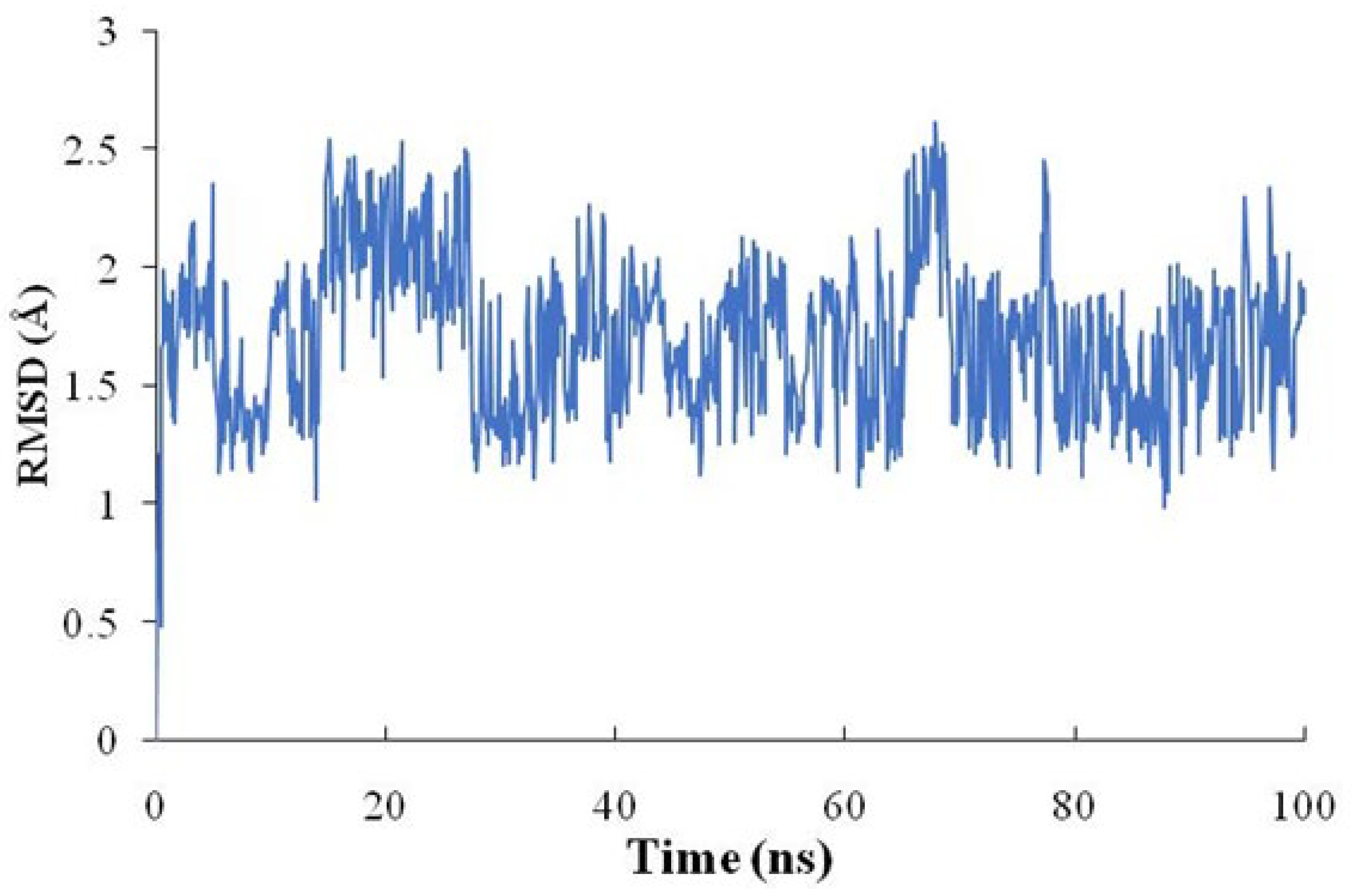
2.6. Molecular Docking Verification and Molecular Dynamics (MD) Simulation
2.7. Alanine Mutation of Key Residues
3. Discussion
4. Methods and Materials
4.1. Data Collection
4.2. Identification of DEGs
4.3. Protein–Protein Interaction, GO Enrichment, and KEGG Enrichment Analyses
4.4. Hub Gene Identification
4.5. Prediction of Potential Drugs
4.6. Molecular Docking and Molecular Dynamic Simulation
4.7. Alanine Mutation of Key Residues
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention, 2022; Global Initiative for Asthma: Fontana, WI, USA, 2022. [Google Scholar]
- Porsbjerg, C.; Melen, E.; Lehtimaki, L.; Shaw, D. Asthma. Lancet 2023, 401, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Hammad, H.; Lambrecht, B.N. The basic immunology of asthma. Cell 2021, 184, 2521–2522. [Google Scholar] [CrossRef] [PubMed]
- Stern, J.; Pier, J.; Litonjua, A.A. Asthma epidemiology and risk factors. Semin. Immunopathol. 2020, 42, 5–15. [Google Scholar] [CrossRef]
- Soriano, J.B.; Abajobir, A.A.; Abate, K.H.; Abera, S.F.; Agrawal, A.; Ahmed, M.B.; Aichour, A.N.; Aichour, I.; Aichour, M.T.E.; Alam, K.; et al. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir. Med. 2017, 5, 691–706. [Google Scholar] [CrossRef]
- Stikker, B.S.; Hendriks, R.W.; Stadhouders, R. Decoding the genetic and epigenetic basis of asthma. Allergy 2023, 78, 940–956. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Zhang, Y.; Qiu, C. Gene polymorphisms in asthma: A narrative review. Ann. Transl. Med. 2022, 10, 711. [Google Scholar] [CrossRef]
- Altman, M.C.; Gill, M.A.; Whalen, E.; Babineau, D.C.; Shao, B.; Liu, A.H.; Jepson, B.; Gruchalla, R.S.; O’Connor, G.T.; Pongracic, J.A.; et al. Transcriptome networks identify mechanisms of viral and nonviral asthma exacerbations in children. Nat. Immunol. 2019, 20, 637–651. [Google Scholar] [CrossRef]
- Gomez, J.L.; Chen, A.; Diaz, M.P.; Zirn, N.; Gupta, A.; Britto, C.; Sauler, M.; Yan, X.; Stewart, E.; Santerian, K.; et al. A Network of Sputum MicroRNAs Is Associated with Neutrophilic Airway Inflammation in Asthma. Am. J. Respir. Crit. Care Med. 2020, 202, 51–64. [Google Scholar] [CrossRef]
- Seumois, G.; Ramirez-Suastegui, C.; Schmiedel, B.J.; Liang, S.; Peters, B.; Sette, A.; Vijayanand, P. Single-cell transcriptomic analysis of allergen-specific T cells in allergy and asthma. Sci. Immunol. 2020, 5, aba6087. [Google Scholar] [CrossRef]
- O’Beirne, S.L.; Salit, J.; Kaner, R.J.; Crystal, R.G.; Strulovici-Barel, Y. Up-regulation of ACE2, the SARS-CoV-2 receptor, in asthmatics on maintenance inhaled corticosteroids. Respir. Res. 2021, 22, 200. [Google Scholar] [CrossRef]
- Vella, D.; Zoppis, I.; Mauri, G.; Mauri, P.; Di Silvestre, D. From protein-protein interactions to protein co-expression networks: A new perspective to evaluate large-scale proteomic data. EURASIP J. Bioinform. Syst. Biol. 2017, 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Kinoshita, K.; Nakamura, H. Hub promiscuity in protein-protein interaction networks. Int. J. Mol. Sci. 2010, 11, 1930–1943. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y.; Li, X.; Wu, J.; Zhao, L.; Li, W.; Liu, J. Dynamics-Based Discovery of Novel, Potent Benzoic Acid Derivatives as Orally Bioavailable Selective Estrogen Receptor Degraders for ERalpha+ Breast Cancer. J. Med. Chem. 2021, 64, 7575–7595. [Google Scholar] [CrossRef] [PubMed]
- Gohy, S.; Hupin, C.; Ladjemi, M.Z.; Hox, V.; Pilette, C. Key role of the epithelium in chronic upper airways diseases. Clin. Exp. Allergy 2020, 50, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Calven, J.; Ax, E.; Radinger, M. The Airway Epithelium-A Central Player in Asthma Pathogenesis. Int. J. Mol. Sci. 2020, 21, 8907. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, J.; Chen, J.; Zhang, X.; Guo, M.; Yu, G. A Literature Review of Gene Function Prediction by Modeling Gene Ontology. Front. Genet. 2020, 11, 400. [Google Scholar] [CrossRef]
- Pal, K.; Feng, X.; Steinke, J.W.; Burdick, M.D.; Shim, Y.M.; Sung, S.S.; Teague, W.G.; Borish, L. Leukotriene A4 Hydrolase Activation and Leukotriene B4 Production by Eosinophils in Severe Asthma. Am. J. Respir. Cell Mol. Biol. 2019, 60, 413–419. [Google Scholar] [CrossRef]
- Jiang, J.X.; Guan, Y.; Shen, H.J.; Jia, Y.L.; Shen, J.; Zhang, L.H.; Liu, Q.; Zhu, Y.L.; Xie, Q.M. Inhibition of soluble epoxide hydrolase attenuates airway remodeling in a chronic asthma model. Eur. J. Pharmacol. 2020, 868, 172874. [Google Scholar] [CrossRef]
- Flemming, H.C.; Baveye, P.; Neu, T.R.; Stoodley, P.; Szewzyk, U.; Wingender, J.; Wuertz, S. Who put the film in biofilm? The migration of a term from wastewater engineering to medicine and beyond. NPJ Biofilms Microbiomes 2021, 7, 10. [Google Scholar] [CrossRef]
- Martin-Rodriguez, A.J. Respiration-induced biofilm formation as a driver for bacterial niche colonization. Trends Microbiol. 2023, 31, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Liu, L.; Jiao, L.; Wen, X.; Liu, J.; Wang, N. Bioinformatics Analysis and Identification of Underlying Biomarkers Potentially Linking Allergic Rhinitis and Asthma. Med. Sci. Monit. 2020, 26, e924934. [Google Scholar] [CrossRef] [PubMed]
- Santri, I.N.; Irham, L.M.; Djalilah, G.N.; Perwitasari, D.A.; Wardani, Y.; Phiri, Y.V.A.; Adikusuma, W. Identification of Hub Genes and Potential Biomarkers for Childhood Asthma by Utilizing an Established Bioinformatic Analysis Approach. Biomedicines 2022, 10, 2311. [Google Scholar] [CrossRef]
- Wang, Z.; An, J.; Zhu, D.; Chen, H.; Lin, A.; Kang, J.; Liu, W.; Kang, X. Periostin: An emerging activator of multiple signaling pathways. J. Cell Commun. Signal. 2022, 16, 515–530. [Google Scholar] [CrossRef] [PubMed]
- Alzobaidi, N.; Rehman, S.; Naqvi, M.; Gulati, K.; Ray, A. Periostin: A Potential Biomarker and Therapeutic Target in Pulmonary Diseases. J. Pharm. Pharm. Sci. 2022, 25, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H. Roles of Periostin in Asthma. Adv. Exp. Med. Biol. 2019, 1132, 145–159. [Google Scholar] [CrossRef]
- Siddhuraj, P.; Clausson, C.M.; Sanden, C.; Alyamani, M.; Kadivar, M.; Marsal, J.; Wallengren, J.; Bjermer, L.; Erjefalt, J.S. Lung Mast Cells Have a High Constitutive Expression of Carboxypeptidase A3 mRNA That Is Independent from Granule-Stored CPA3. Cells 2021, 10, 309. [Google Scholar] [CrossRef]
- Waern, I.; Taha, S.; Lorenzo, J.; Montpeyo, D.; Covaleda-Cortes, G.; Aviles, F.X.; Wernersson, S. Carboxypeptidase inhibition by NvCI suppresses airway hyperreactivity in a mouse asthma model. Allergy 2021, 76, 2234–2237. [Google Scholar] [CrossRef]
- Dougherty, R.H.; Sidhu, S.S.; Raman, K.; Solon, M.; Solberg, O.D.; Caughey, G.H.; Woodruff, P.G.; Fahy, J.V. Accumulation of intraepithelial mast cells with a unique protease phenotype in T(H)2-high asthma. J. Allergy Clin. Immunol. 2010, 125, 1046–1053. [Google Scholar] [CrossRef]
- Akula, S.; Riihimaki, M.; Waern, I.; Abrink, M.; Raine, A.; Hellman, L.; Wernersson, S. Quantitative Transcriptome Analysis of Purified Equine Mast Cells Identifies a Dominant Mucosal Mast Cell Population with Possible Inflammatory Functions in Airways of Asthmatic Horses. Int. J. Mol. Sci. 2022, 23, 13976. [Google Scholar] [CrossRef]
- Duan, Q.; Reid, S.P.; Clark, N.R.; Wang, Z.; Fernandez, N.F.; Rouillard, A.D.; Readhead, B.; Tritsch, S.R.; Hodos, R.; Hafner, M.; et al. L1000CDS(2): LINCS L1000 characteristic direction signatures search engine. NPJ Syst. Biol. Appl. 2016, 2, 16015. [Google Scholar] [CrossRef] [PubMed]
- Zeki, A.A.; Elbadawi-Sidhu, M. Innovations in asthma therapy: Is there a role for inhaled statins? Expert Rev. Respir. Med. 2018, 12, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Tobert, J.A. Lovastatin and beyond: The history of the HMG-CoA reductase inhibitors. Nat. Rev. Drug Discov. 2003, 2, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Arima, J.; Sakai, H.; Misawa, M. Lovastatin inhibits bronchial hyperresponsiveness by reducing RhoA signaling in rat allergic asthma. Am. J. Physiol. Lung Cell Mol. Physiol. 2008, 294, L705–L713. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Sato, S.; Misawa, M. Upregulation of geranylgeranyltransferase I in bronchial smooth muscle of mouse experimental asthma: Its inhibition by lovastatin. J. Smooth Muscle Res. 2010, 46, 57–64. [Google Scholar] [CrossRef]
- Liou, C.J.; Cheng, P.Y.; Huang, W.C.; Chan, C.C.; Chen, M.C.; Kuo, M.L.; Shen, J.J. Oral lovastatin attenuates airway inflammation and mucus secretion in ovalbumin-induced murine model of asthma. Allergy Asthma Immunol. Res. 2014, 6, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Michalik, M.; Soczek, E.; Kosinska, M.; Rak, M.; Wojcik, K.A.; Lasota, S.; Pierzchalska, M.; Czyz, J.; Madeja, Z. Lovastatin-induced decrease of intracellular cholesterol level attenuates fibroblast-to-myofibroblast transition in bronchial fibroblasts derived from asthmatic patients. Eur. J. Pharmacol. 2013, 704, 23–32. [Google Scholar] [CrossRef]
- Thornton, D.J.; Rousseau, K.; McGuckin, M.A. Structure and function of the polymeric mucins in airways mucus. Annu. Rev. Physiol. 2008, 70, 459–486. [Google Scholar] [CrossRef]
- Bonser, L.R.; Erle, D.J. Airway Mucus and Asthma: The Role of MUC5AC and MUC5B. J Clin. Med. 2017, 6, 112. [Google Scholar] [CrossRef]
- Li, X.; Li, H.; Christenson, S.A.; Castro, M.; Denlinger, L.C.; Erzurum, S.C.; Fahy, J.V.; Gaston, B.M.; Israel, E.; Jarjour, N.N.; et al. Genetic analyses of chr11p15.5 region identify MUC5AC-MUC5B associated with asthma-related phenotypes. J. Asthma 2023. online ahead of print. [Google Scholar] [CrossRef]
- Lee, E.J.; Song, K.J.; Kwon, J.H.; Park, A.Y.; Jo, K.H.; Kim, K.S. Chronic cholesterol depletion by lovastatin suppresses MUC5AC gene expression in human airway epithelial cells. Am. J. Rhinol. Allergy 2014, 28, e125–e129. [Google Scholar] [CrossRef] [PubMed]
- Soleymani, F.; Paquet, E.; Viktor, H.; Michalowski, W.; Spinello, D. Protein-protein interaction prediction with deep learning: A comprehensive review. Comput. Struct. Biotechnol. J. 2022, 20, 5316–5341. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
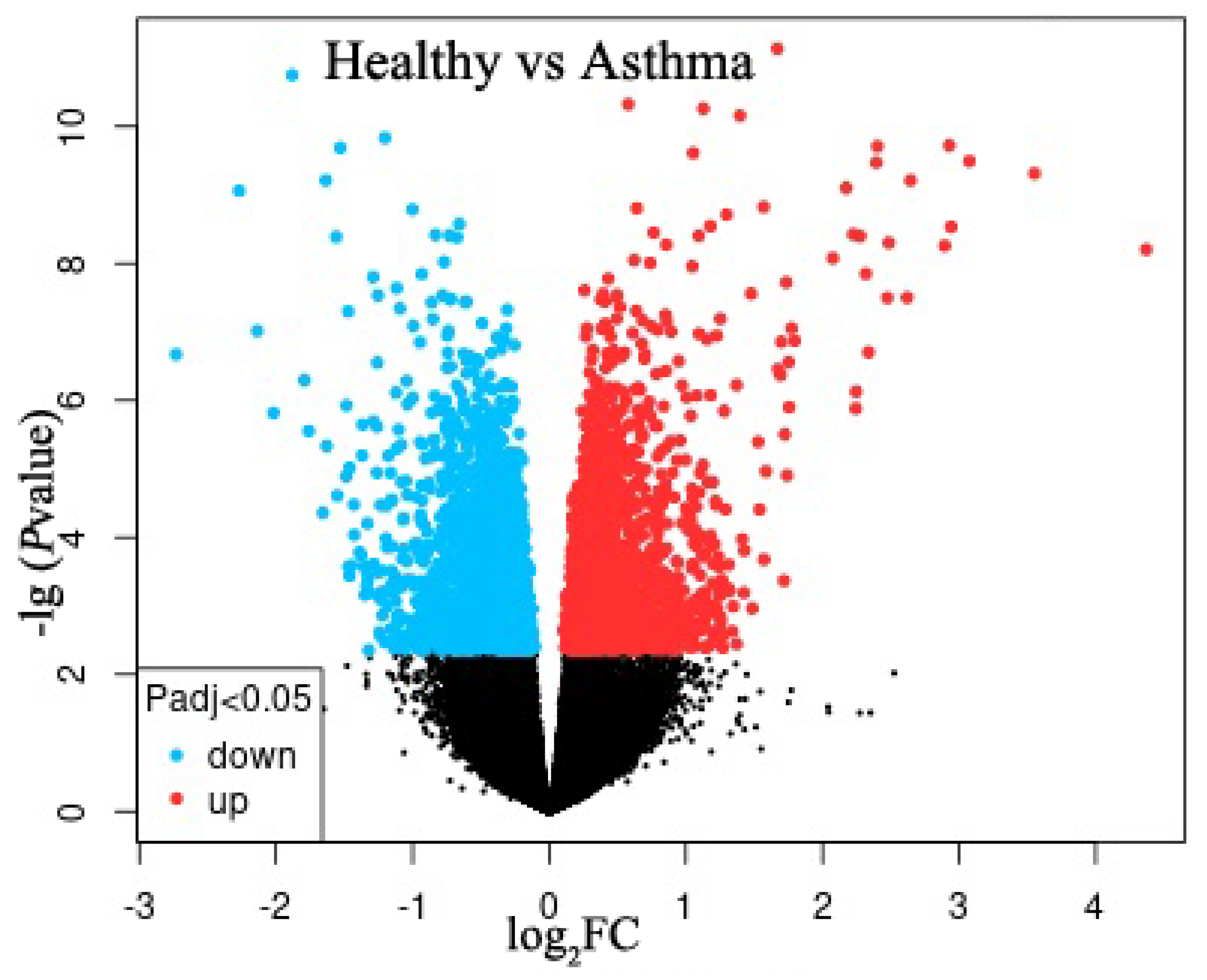

| DEGs | Gene | logFC | Adj.p.Val |
|---|---|---|---|
| Upregulated | CLCA1 | 4.37 | 9.50 × 10−6 |
| CST1 | 3.551 | 2.05 × 10−6 | |
| CPA3 | 3.075 | 1.54 × 10−6 | |
| TPSAB1 | 2.941 | 6.70 × 10−6 | |
| TPSB2 | 2.928 | 1.24 × 10−6 | |
| POSTN | 2.645 | 2.24 × 10−6 | |
| SIGLEC6 | 2.475 | 3.11 × 10−5 | |
| CEACAM5 | 2.402 | 1.24 × 10−6 | |
| MS4A2 | 2.394 | 1.54 × 10−6 | |
| SERPINB2 | 2.317 | 1.82 × 10−5 | |
| CST2 | 2.246 | 2.52 × 10−4 | |
| CD200R1 | 2.244 | 3.70 × 10−4 | |
| RGS13 | 2.173 | 2.71 × 10−6 | |
| CCL26 | 1.795 | 7.43 × 10−5 | |
| SIGLEC17P | 1.773 | 5.95 × 10−5 | |
| NTRK1 | 1.754 | 1.21 × 10−4 | |
| SERPINB10 | 1.733 | 2.26 × 10−5 | |
| CST4 | 1.692 | 1.69 × 10−4 | |
| SLC18A2 | 1.676 | 1.45 × 10−4 | |
| P2RY14 | 1.57 | 4.48 × 10−6 | |
| GATA2 | 1.396 | 7.60 × 10−7 | |
| BDNF | 1.371 | 2.24 × 10−4 | |
| HPGDS | 1.299 | 5.05 × 10−6 | |
| SAMSN1 | 1.28 | 3.80 × 10−4 | |
| DHX35 | 1.252 | 4.94 × 10−5 | |
| GSN | 1.227 | 6.83 × 10−5 | |
| KIT | 1.178 | 6.70 × 10−6 | |
| CDH26 | 1.128 | 7.53 × 10−7 | |
| TAL1 | 1.095 | 7.14 × 10−6 | |
| TMEM200A | 1.095 | 6.53 × 10−5 | |
| IGF2BP3 | 1.081 | 2.76 × 10−4 | |
| HDAC9 | 1.047 | 1.47 × 10−5 | |
| SERPINB4 | 1.036 | 4.03 × 10−4 | |
| PP14571 | 1.006 | 2.87 × 10−4 | |
| Downregulated | PEG3 | −1.036 | 3.34 × 10−4 |
| STEAP4 | −1.043 | 2.02 × 10−4 | |
| KCNQ3 | −1.093 | 3.83 × 10−5 | |
| FOLR1 | −1.116 | 2.67 × 10−5 | |
| GADD45B | −1.121 | 2.56 × 10−4 | |
| PCDH17 | −1.254 | 3.03 × 10−5 | |
| FHOD3 | −1.259 | 1.22 × 10−4 | |
| VGLL3 | −1.286 | 4.63 × 10−4 | |
| C3 | −1.289 | 1.97 × 10−5 | |
| LTF | −1.635 | 2.24 × 10−6 | |
| APELA | −1.791 | 1.99 × 10−4 | |
| TMEM45A | −1.881 | 4.87 × 10−7 | |
| WIF1 | −2.018 | 3.87 × 10−4 | |
| MUC5B | −2.137 | 6.17 × 10−5 | |
| BPIFA1 | −2.731 | 1.03 × 10−4 |
| Perturbation | Drug Name | Drugbank ID | Summary |
|---|---|---|---|
| BRD-K09416995 | Lovastatin | DB00227 | Lovastatin is an HMG-CoA reductase inhibitor used to lower LDL cholesterol and reduce the risk of cardiovascular disease and associated conditions, including myocardial infarction and stroke. |
| BRD-K92049597 | Triamterene | DB00384 | Triamterene is a potassium-sparing diuretic used in the treatment of edema and the management of hypertension. |
| BRD-A31204924 | Mitotane | DB00648 | Mitotane is an adrenal cortex inhibitor used to treat adrenocortical tumors and Cushing’s syndrome. |
| Contribution | Wild-Type | T80A | T91A | L93A | G105A |
|---|---|---|---|---|---|
| ΔGVDW a | −27.06 | −23.26 | −29.62 | −27.37 | −25.36 |
| ΔGele b | −5.87 | −3.35 | −2.41 | −6.85 | −4.30 |
| ΔGGB c | 10.70 | 8.49 | 8.03 | 11.43 | 8.30 |
| ΔGGA d | −39.70 | −32.54 | −36.15 | −36.79 | −31.86 |
| ΔGbind e | −59.84 | −49.01 | −58.37 | −57.90 | −51.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Lv, J.; Luo, Y.; Chen, H.; Ma, S.; Zhang, L. Bioinformatic Analysis of Key Regulatory Genes in Adult Asthma and Prediction of Potential Drug Candidates. Molecules 2023, 28, 4100. https://doi.org/10.3390/molecules28104100
Chen S, Lv J, Luo Y, Chen H, Ma S, Zhang L. Bioinformatic Analysis of Key Regulatory Genes in Adult Asthma and Prediction of Potential Drug Candidates. Molecules. 2023; 28(10):4100. https://doi.org/10.3390/molecules28104100
Chicago/Turabian StyleChen, Shaojun, Jiahao Lv, Yiyuan Luo, Hongjiang Chen, Shuwei Ma, and Lihua Zhang. 2023. "Bioinformatic Analysis of Key Regulatory Genes in Adult Asthma and Prediction of Potential Drug Candidates" Molecules 28, no. 10: 4100. https://doi.org/10.3390/molecules28104100
APA StyleChen, S., Lv, J., Luo, Y., Chen, H., Ma, S., & Zhang, L. (2023). Bioinformatic Analysis of Key Regulatory Genes in Adult Asthma and Prediction of Potential Drug Candidates. Molecules, 28(10), 4100. https://doi.org/10.3390/molecules28104100




